Construction and Characterization of a Botrytis Virus F Infectious Clone
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. RNA Next Generation Sequencing (NGS) and In Vivo Detection of Mycoviruses
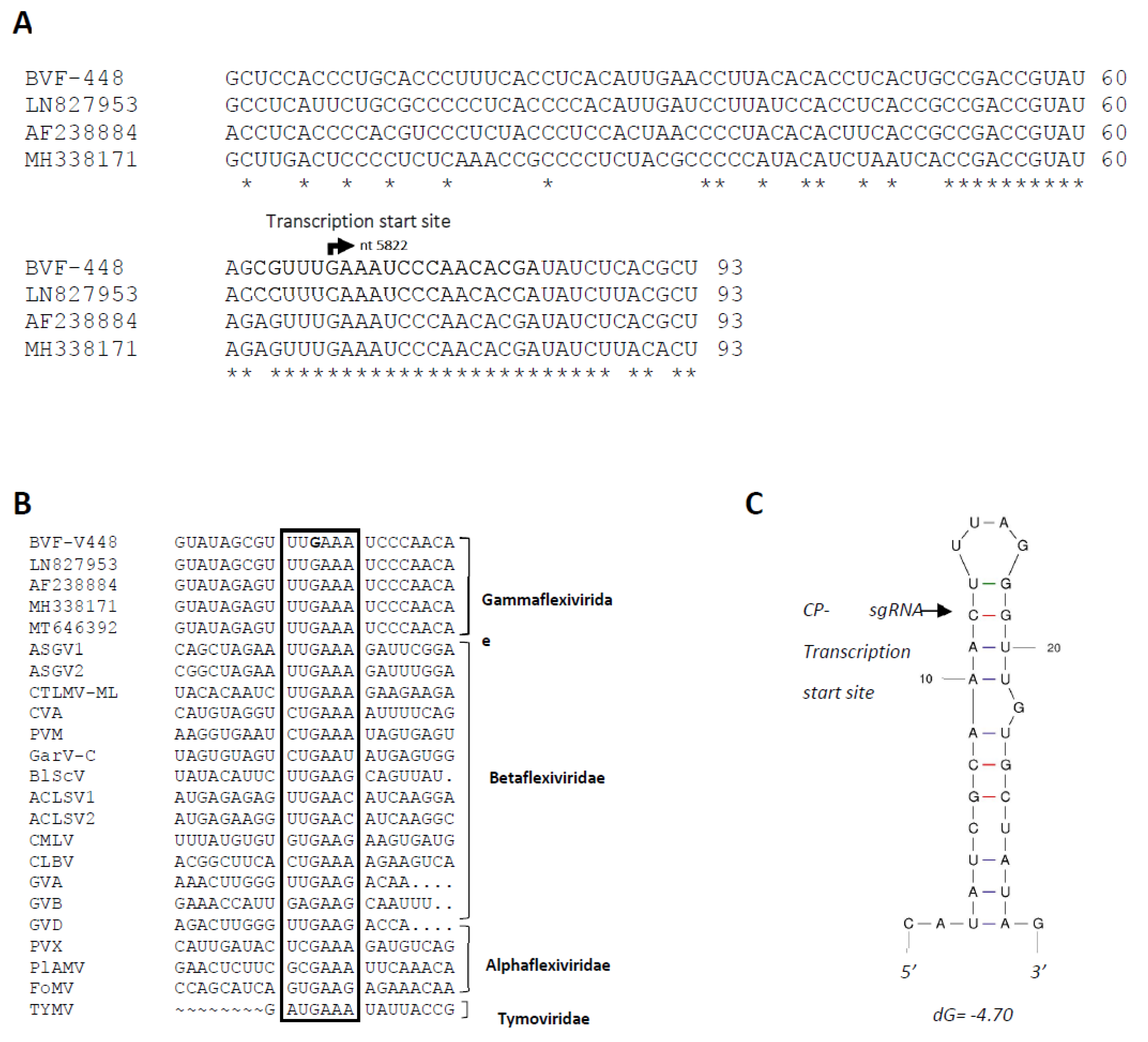
2.3. Determination of 5′ and 3′ Ends, Full Length Genome of Botrytis Virus F Genome, and the Transcription Initiation Site of Capsid Protein (CP) Subgenomic RNA (sgRNA)
2.4. In Silico Prediction of RNA Secondary Structure
2.5. Construction of a Full-Length cDNA Clone of BVF
2.6. Transfection of Botrytis cinerea Protoplasts with BVF RNA Transcripts
2.7. Total RNA Extraction and RT-PCR for BVF Detection
2.8. Northern Blot Hybridization of BVF RNAs
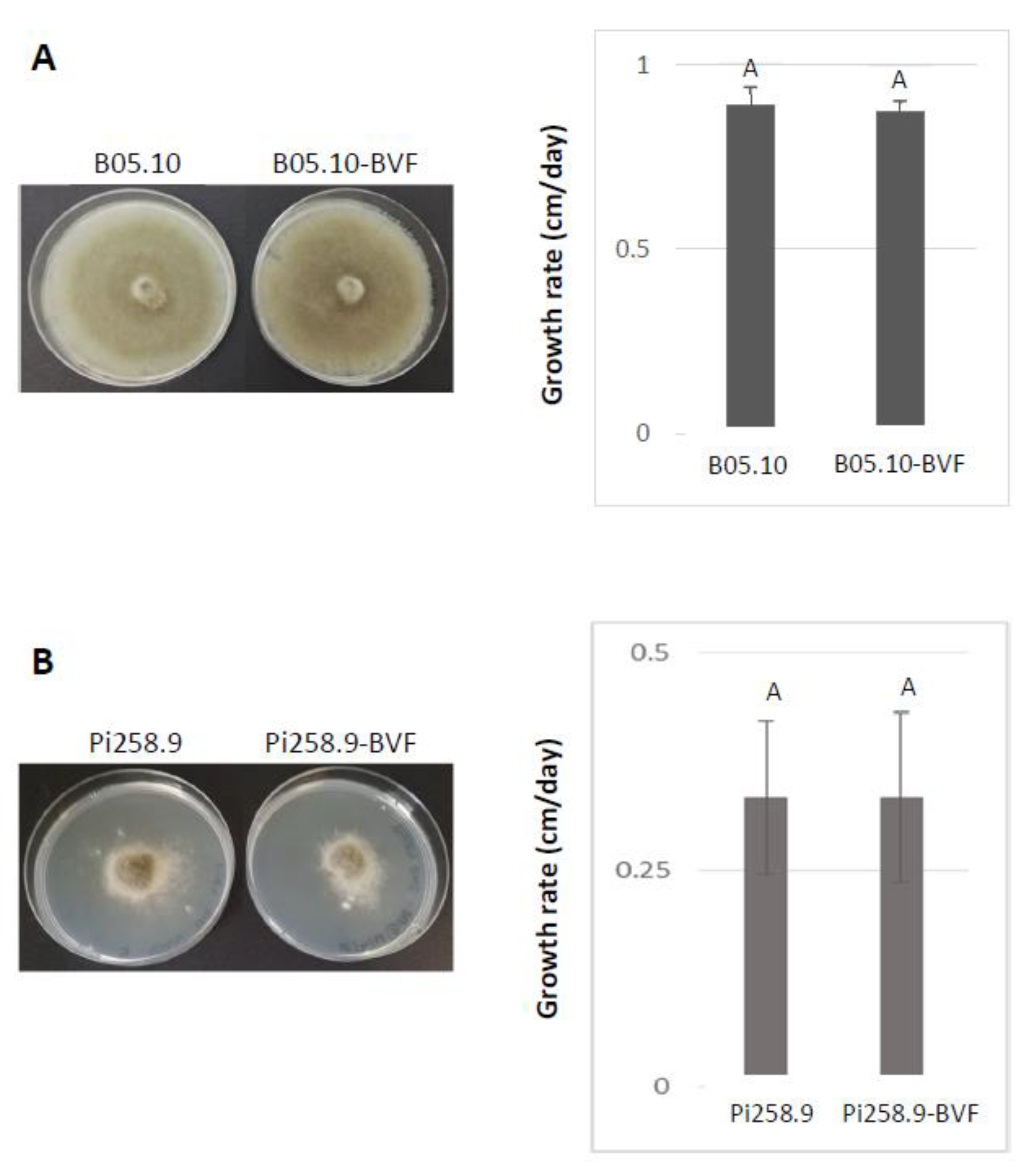
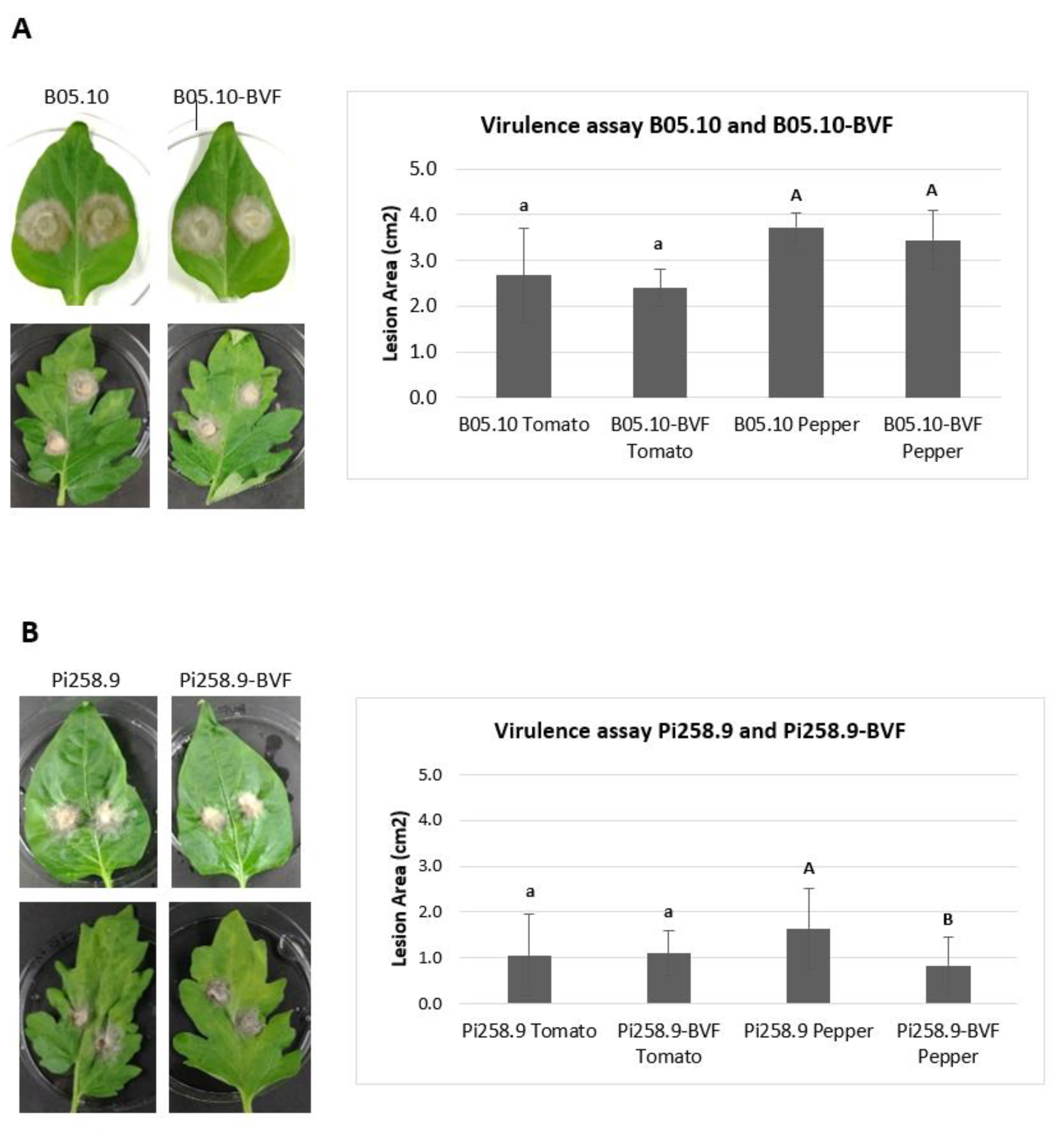
2.9. Determination of Mycelial Growth and Virulence Assays
3. Results
3.1. Determination of the BVF Genome Ends and Transcription Initiation Site of CP sgRNA
3.2. Infectivity of Full-Length BVF Clone in Botrytis cinerea
3.3. Effect of BVF on Fungal Growth and Virulence
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghabrial, S.A.; Suzuki, N. Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 2009, 47, 353–384. [Google Scholar] [CrossRef] [PubMed]
- Pearson, M.N.; Beever, R.E.; Boine, B.; Arthur, K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol. Plant Pathol. 2009, 10, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Jiang, D. New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu. Rev. Phytopathol. 2014, 52, 45–68. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Pedrajas, M.D.; Canizares, M.C.; Sarmiento-Villamil, J.L.; Jacquat, A.G.; Dambolena, J.S. Mycoviruses in Biological Control: From Basic Research to Field Implementation. Phytopathology 2019, 109, 1828–1839. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Caston, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef] [Green Version]
- Son, M.; Yu, J.; Kim, K.H. Five Questions about Mycoviruses. PLoS Pathog. 2015, 11, e1005172. [Google Scholar] [CrossRef]
- Yu, X.; Li, B.; Fu, Y.; Jiang, D.; Ghabrial, S.A.; Li, G.; Peng, Y.; Xie, J.; Cheng, J.; Huang, J.; et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc. Natl. Acad. Sci. USA 2010, 107, 8387–8392. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. USA 2014, 111, 12205–12210. [Google Scholar] [CrossRef] [Green Version]
- Khalifa, M.E.; MacDiarmid, R.M. A Mechanically Transmitted DNA Mycovirus Is Targeted by the Defence Machinery of Its Host, Botrytis cinerea. Viruses 2021, 13, 1315. [Google Scholar] [CrossRef]
- Ruiz-Padilla, A.; Rodriguez-Romero, J.; Gomez-Cid, I.; Pacifico, D.; Ayllon, M.A. Novel Mycoviruses Discovered in the Mycovirome of a Necrotrophic Fungus. mBio 2021, 12, e03705-20. [Google Scholar] [CrossRef]
- Hao, F.; Wu, M.; Li, G. Characterization of a novel genomovirus in the phytopathogenic fungus Botrytis cinerea. Virology 2021, 553, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, S.; Zhang, L.; Qiu, D.; Zhou, X.; Guo, L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci. Adv. 2020, 6, eaay9634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiapello, M.; Rodriguez-Romero, J.; Ayllon, M.A.; Turina, M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020, 6, veaa058. [Google Scholar] [CrossRef] [PubMed]
- Bissegger, M.; Rigling, D.; Heiniger, U. Population Structure and Disease Development of Cryphonectria parasitica in European Chestnut Forests in the Presence of Natural Hypovirulence. Phytopathology 1997, 87, 50–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuss, D.L. Biological control of chestnut blight: An example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 1992, 56, 561–576. [Google Scholar] [CrossRef]
- Suzuki, N.; Cornejo, C.; Aulia, A.; Shahi, S.; Hillman, B.I.; Rigling, D. In-Tree Behavior of Diverse Viruses Harbored in the Chestnut Blight Fungus, Cryphonectria parasitica. J. Virol. 2021, 95, e01962-20. [Google Scholar] [CrossRef]
- Marzano, S.Y.; Hobbs, H.A.; Nelson, B.D.; Hartman, G.L.; Eastburn, D.M.; McCoppin, N.K.; Domier, L.L. Transfection of Sclerotinia sclerotiorum with in vitro transcripts of a naturally occurring interspecific recombinant of Sclerotinia sclerotiorum hypovirus 2 significantly reduces virulence of the fungus. J. Virol. 2015, 89, 5060–5071. [Google Scholar] [CrossRef] [Green Version]
- Choi, G.H.; Nuss, D.L. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science 1992, 257, 800–803. [Google Scholar] [CrossRef]
- Moleleki, N.; van Heerden, S.W.; Wingfield, M.J.; Wingfield, B.D.; Preisig, O. Transfection of Diaporthe perjuncta with Diaporthe RNA virus. Appl. Environ. Microbiol. 2003, 69, 3952–3956. [Google Scholar] [CrossRef] [Green Version]
- Craven, M.G.; Pawlyk, D.M.; Choi, G.H.; Nuss, D.L. Papain-like protease p29 as a symptom determinant encoded by a hypovirulence-associated virus of the chestnut blight fungus. J. Virol. 1993, 67, 6513–6521. [Google Scholar] [CrossRef] [Green Version]
- Esteban, R.; Fujimura, T. Launching the yeast 23S RNA Narnavirus shows 5′ and 3′ cis-acting signals for replication. Proc. Natl. Acad. Sci. USA 2003, 100, 2568–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, R.; Vega, L.; Fujimura, T. Launching of the yeast 20 s RNA narnavirus by expressing the genomic or antigenomic viral RNA in vivo. J. Biol. Chem. 2005, 280, 33725–33734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Hisano, S.; Tani, A.; Kondo, H.; Kanematsu, S.; Suzuki, N. A capsidless ssRNA virus hosted by an unrelated dsRNA virus. Nat. Microbiol. 2016, 1, 15001. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mu, F.; Xie, J.; Cheng, J.; Fu, Y.; Jiang, D. A Single ssRNA Segment Encoding RdRp Is Sufficient for Replication, Infection, and Transmission of Ourmia-Like Virus in Fungi. Front. Microbiol. 2020, 11, 379. [Google Scholar] [CrossRef] [Green Version]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konstantinou, S.; Veloukas, T.; Leroch, M.; Menexes, G.; Hahn, M.; Karaoglanidis, G. Population Structure, Fungicide Resistance Profile, and sdhB Mutation Frequency of Botrytis cinerea from Strawberry and Greenhouse-Grown Tomato in Greece. Plant Dis. 2015, 99, 240–248. [Google Scholar] [CrossRef] [Green Version]
- Kretschmer, M.; Leroch, M.; Mosbach, A.; Walker, A.S.; Fillinger, S.; Mernke, D.; Schoonbeek, H.J.; Pradier, J.M.; Leroux, P.; De Waard, M.A.; et al. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 2009, 5, e1000696. [Google Scholar] [CrossRef]
- Rodríguez-García, C.; Medina, V.; Alonso, A.; Ayllón, M.A. Mycoviruses of Botrytis cinerea isolates from different hosts. Ann. Appl. Biol. 2014, 164, 46–61. [Google Scholar] [CrossRef]
- Donaire, L.; Ayllon, M.A. Deep sequencing of mycovirus-derived small RNAs from Botrytis species. Mol. Plant Pathol. 2017, 18, 1127–1137. [Google Scholar] [CrossRef]
- Donaire, L.; Pagan, I.; Ayllon, M.A. Characterization of Botrytis cinerea negative-stranded RNA virus 1, a new mycovirus related to plant viruses, and a reconstruction of host pattern evolution in negative-sense ssRNA viruses. Virology 2016, 499, 212–218. [Google Scholar] [CrossRef]
- Donaire, L.; Rozas, J.; Ayllon, M.A. Molecular characterization of Botrytis ourmia-like virus, a mycovirus close to the plant pathogenic genus Ourmiavirus. Virology 2016, 489, 158–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Fu, Y.; Xie, J.; Cheng, J.; Ghabrial, S.A.; Li, G.; Peng, Y.; Yi, X.; Jiang, D. Evolutionary genomics of mycovirus-related dsRNA viruses reveals cross-family horizontal gene transfer and evolution of diverse viral lineages. BMC Evol. Biol. 2012, 12, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayllon, M.A.; Turina, M.; Xie, J.; Nerva, L.; Marzano, S.L.; Donaire, L.; Jiang, D.; ICTV Report Consortium. ICTV Virus Taxonomy Profile: Botourmiaviridae. J. Gen. Virol. 2020, 101, 454–455. [Google Scholar] [CrossRef] [PubMed]
- Howitt, R.L.J.; Beever, R.E.; Pearson, M.N.; Forster, R.L.S. Genome characterization of Botrytis virus F, a flexuous rod-shaped mycovirus resembling plant ‘potex-like’ viruses. J. Gen. Virol. 2001, 82, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Wu, M.; Li, G. Molecular Characterization and Geographic Distribution of a Mymonavirus in the Population of Botrytis cinerea. Viruses 2018, 10, 432. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Zhang, L.; Li, G.; Jiang, D.; Ghabrial, S.A. Genome characterization of a debilitation-associated mitovirus infecting the phytopathogenic fungus Botrytis cinerea. Virology 2010, 406, 117–126. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.D.; Zhang, L.; Li, G.Q.; Jiang, D.H.; Hou, M.S.; Huang, H.C. Hypovirulence and Double-Stranded RNA in Botrytis cinerea. Phytopathology 2007, 97, 1590–1599. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Jin, F.; Zhang, J.; Yang, L.; Jiang, D.; Li, G. Characterization of a novel bipartite double-stranded RNA mycovirus conferring hypovirulence in the phytopathogenic fungus Botrytis porri. J. Virol. 2012, 86, 6605–6619. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Sang, W.; Wu, M.D.; Zhang, J.; Yang, L.; Zhou, Y.J.; Chen, W.D.; Li, G.Q. Novel hypovirulence-associated RNA mycovirus in the plant-pathogenic fungus Botrytis cinerea: Molecular and biological characterization. Appl. Environ. Microbiol. 2015, 81, 2299–2310. [Google Scholar] [CrossRef] [Green Version]
- Potgieter, C.A.; Castillo, A.; Castro, M.; Cottet, L.; Morales, A. A wild-type Botrytis cinerea strain co-infected by double-stranded RNA mycoviruses presents hypovirulence-associated traits. Virol. J. 2013, 10, 220. [Google Scholar] [CrossRef] [Green Version]
- Hao, F.; Ding, T.; Wu, M.; Zhang, J.; Yang, L.; Chen, W.; Li, G. Two Novel Hypovirulence-Associated Mycoviruses in the Phytopathogenic Fungus Botrytis cinerea: Molecular Characterization and Suppression of Infection Cushion Formation. Viruses 2018, 10, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, S.L.; Nagata, T.; Junqueira, B.R.; Zanardo, L.G.; Paiva, A.C.; Carvalho, C.M. Construction of a full-length infectious cDNA clone of Cowpea mild mottle virus. Virus Genes 2017, 53, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Bin, Y.; Yan, J.; Mei, P.; Li, Z.; Zhou, C.; Song, Z. Development of Infectious cDNA Clones of Citrus Yellow Vein Clearing Virus Using a Novel and Rapid Strategy. Phytopathology 2018, 108, 1212–1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dizadji, A.; Koohi-Habibi, M.; Izadpanah, K.; Dietrich, C.; Mossahebi, G.H.; Winter, S. Characterisation of lettuce virus X, a new potexvirus infecting lettuce in Iran. Arch. Virol. 2008, 153, 1867–1875. [Google Scholar] [CrossRef]
- Hasiow-Jaroszewska, B.; Borodynko, N. Characterization of the necrosis determinant of the European genotype of pepino mosaic virus by site-specific mutagenesis of an infectious cDNA clone. Arch. Virol. 2012, 157, 337–341. [Google Scholar] [CrossRef]
- Li, X.; Hataya, T. Construction and characterization of an infectious cDNA clone of potato virus S developed from selected populations that survived genetic bottlenecks. Virol. J. 2019, 16, 18. [Google Scholar] [CrossRef] [Green Version]
- Mathews, D.H.; Sabina, J.; Zuker, M.; Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999, 288, 911–940. [Google Scholar] [CrossRef] [Green Version]
- Lu, Z.J.; Turner, D.H.; Mathews, D.H. A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation. Nucleic Acids Res. 2006, 34, 4912–4924. [Google Scholar] [CrossRef] [Green Version]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Zuker, M.; Turner, D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef] [Green Version]
- Xia, T.; SantaLucia, J., Jr.; Burkard, M.E.; Kierzek, R.; Schroeder, S.J.; Jiao, X.; Cox, C.; Turner, D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 1998, 37, 14719–14735. [Google Scholar] [CrossRef] [Green Version]
- Churchill, A.C.L.; Ciuffetti, L.M.; Hansen, D.R.; Van Etten, H.D.; Van Alfen, N.K. Transformation of the fungal pathogen Cryphonectria parasitica with a variety of heterologous plasmids. Curr. Genet. 1990, 17, 25–31. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; p. 3. [Google Scholar]
- Svanella-Dumas, L.; Marais, A.; Faure, C.; Theil, S.; Lefebvre, M.; Candresse, T. Genome characterization of a divergent isolate of the mycovirus Botrytis virus F from a grapevine metagenome. Arch. Virol. 2018, 163, 3181–3183. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gong, C.; Zhao, Y.; Shen, L. Transcriptome and Resistance-Related Genes Analysis of Botrytis cinerea B05.10 Strain to Different Selective Pressures of Cyprodinil and Fenhexamid. Front. Microbiol. 2018, 9, 2591. [Google Scholar] [CrossRef]
- Mu, F.; Xie, J.; Cheng, S.; You, M.P.; Barbetti, M.J.; Jia, J.; Wang, Q.; Cheng, J.; Fu, Y.; Chen, T.; et al. Virome Characterization of a Collection of S. sclerotiorum from Australia. Front. Microbiol. 2017, 8, 2540. [Google Scholar] [CrossRef] [Green Version]
- Hartwick, E.W.; Costantino, D.A.; MacFadden, A.; Nix, J.C.; Tian, S.; Das, R.; Kieft, J.S. Ribosome-induced RNA conformational changes in a viral 3′-UTR sense and regulate translation levels. Nat. Commun. 2018, 9, 5074. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H.; Brown, J.K.M.; Lomonossoff, G.P. Improving plant transient expression through the rational design of synthetic 5′ and 3′ untranslated regions. Plant Methods 2019, 15, 108. [Google Scholar] [CrossRef]
- Dreher, T.W. Functions of the 3′-Untranslated Regions of Positive Strand Rna Viral Genomes. Annu. Rev. Phytopathol. 1999, 37, 151–174. [Google Scholar] [CrossRef]
- Annamalai, P.; Hsu, Y.H.; Liu, Y.P.; Tsai, C.H.; Lin, N.S. Structural and mutational analyses of cis-acting sequences in the 5′-untranslated region of satellite RNA of bamboo mosaic potexvirus. Virology 2003, 311, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.J.; Chan, D.; Xiang, Y.; Williams, H.; Li, X.R.; Sniezko, R.A.; Sturrock, R.N. Characterization of Five Novel Mitoviruses in the White Pine Blister Rust Fungus Cronartium ribicola. PLoS ONE 2016, 11, e0154267. [Google Scholar] [CrossRef]
- Koev, G.; Miller, W.A. A positive-strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 2000, 74, 5988–5996. [Google Scholar] [CrossRef] [Green Version]
- Balmori, E.; Gilmer, D.; Richards, K.; Guilley, H.; Jonard, G. Mapping the promoter for subgenomic RNA synthesis on beet necrotic yellow vein virus RNA 3. Biochimie 1993, 75, 517–521. [Google Scholar] [CrossRef]
- Boccard, F.; Baulcombe, D. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology 1993, 193, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Haasnoot, P.C.; Brederode, F.T.; Olsthoorn, R.C.; Bol, J.F. A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 2000, 6, 708–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, J.C.; Rochon, D.M. Deletion analysis of the promoter for the cucumber necrosis virus 0.9-kb subgenomic RNA. Virology 1995, 214, 100–109. [Google Scholar] [CrossRef] [Green Version]
- Koev, G.; Mohan, B.R.; Miller, W.A. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 1999, 73, 2876–2885. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, K.; Hirata, H.; Fukagawa, T.; Yamaji, Y.; Okano, Y.; Ishikawa, K.; Adachi, T.; Maejima, K.; Hashimoto, M.; Namba, S. Infection of capilloviruses requires subgenomic RNAs whose transcription is controlled by promoter-like sequences conserved among flexiviruses. Virus Res. 2012, 167, 8–15. [Google Scholar] [CrossRef]
- Renovell, A.; Gago, S.; Ruiz-Ruiz, S.; Velazquez, K.; Navarro, L.; Moreno, P.; Vives, M.C.; Guerri, J. Mapping the subgenomic RNA promoter of the Citrus leaf blotch virus coat protein gene by Agrobacterium-mediated inoculation. Virology 2010, 406, 360–369. [Google Scholar] [CrossRef] [Green Version]
- Tatineni, S.; Afunian, M.R.; Gowda, S.; Hilf, M.E.; Bar-Joseph, M.; Dawson, W.O. Characterization of the 5′- and 3′-terminal subgenomic RNAs produced by a capillovirus: Evidence for a CP subgenomic RNA. Virology 2009, 385, 521–528. [Google Scholar] [CrossRef] [Green Version]
- Van der Vossen, E.A.; Notenboom, T.; Bol, J.F. Characterization of sequences controlling the synthesis of alfalfa mosaic virus subgenomic RNA in vivo. Virology 1995, 212, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Simon, A.E. Analysis of the two subgenomic RNA promoters for turnip crinkle virus in vivo and in vitro. Virology 1997, 232, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Zavriev, S.K.; Hickey, C.M.; Lommel, S.A. Mapping of the red clover necrotic mosaic virus subgenomic RNA. Virology 1996, 216, 407–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, S.J.; Lim, W.S.; Park, S.H.; Park, M.R.; Kim, K.H. Molecular characterization of a dsRNA mycovirus, Fusarium graminearum virus-DK21, which is phylogenetically related to hypoviruses but has a genome organization and gene expression strategy resembling those of plant potex-like viruses. Mol. Cells 2007, 23, 304–315. [Google Scholar] [CrossRef] [PubMed]
- Revill, P.A.; Davidson, A.D.; Wright, P.J. Identification of a subgenomic mRNA encoding the capsid protein of mushroom bacilliform virus, a single-stranded RNA mycovirus. Virology 1999, 260, 273–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- French, R.; Ahlquist, P. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 1988, 62, 2411–2420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, L.E.; Dreher, T.W.; Hall, T.C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988, 16, 981–995. [Google Scholar] [CrossRef]
- Zhang, G.; Slowinski, V.; White, K.A. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 1999, 5, 550–561. [Google Scholar] [CrossRef] [Green Version]
- Sit, T.L.; Vaewhongs, A.A.; Lommel, S.A. RNA-mediated trans-activation of transcription from a viral RNA. Science 1998, 281, 829–832. [Google Scholar] [CrossRef]
- Vives, M.C.; Galipienso, L.; Navarro, L.; Moreno, P.; Guerri, J. Characterization of two kinds of subgenomic RNAs produced by citrus leaf blotch virus. Virology 2002, 295, 328–336. [Google Scholar] [CrossRef] [Green Version]
- Scheets, K. Maize chlorotic mottle machlomovirus expresses its coat protein from a 1.47-kb subgenomic RNA and makes a 0.34-kb subgenomic RNA. Virology 2000, 267, 90–101. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.H.; Hemenway, C.L. Long-distance RNA-RNA interactions and conserved sequence elements affect potato virus X plus-strand RNA accumulation. RNA 1999, 5, 636–645. [Google Scholar] [CrossRef] [Green Version]
- Quidde, T.; Osbourn, A.E.; Tudzynski, P. Detoxification of a-tomatine by Botrytis cinerea. Physiol. Mol. Plant Pathol. 1998, 52, 151–165. [Google Scholar] [CrossRef]



| Primer Name | Sequence (5′ to 3′) a | Purpose |
|---|---|---|
| BVF_FwQ1 | GGCATGGTTGGAACAACCAG | RT-PCR detection |
| BVF_RvQ2 | TCATTCAAGTCGATGCAC | |
| BVF_XbaIT7 Fw | CTAGCTTCTCTAGATAATACGACTCACTATAGGGGATTAAATTCACATCCAACA | |
| New BVF738_Rv | CAGGCTCTTCATGGTCATCCAT | |
| BVF 5out | CTTTGCGGACAGCATATGGTGC | BVF 5’-RACE |
| BVF 5in | CAGGTATTCGTTTGTGGTAGGGC | |
| BVF 3out | CCAAAGTGGTCATACGATCACC | BVF 3′-RACE |
| BVF 3in | CATGGGATACTTTCAGGCATGC | |
| BVFsg_5out | CCTTACTGGTTGTTCCAACCATGC | Determination transcription initation site CP sgRNA |
| BVFsg_5in | CGAAAGCTCTTTTCAACGTGGGC | |
| BVF738_Fw | ATGGATGACTATGAAGAGCCTG | Determination of BVF complete genomic sequence |
| New BVF738_Rv | CAGGCTCTTCATGGTCATCCAT | |
| BVF 1550_Fw | CCCGATATGCACTTAAAGCTG | |
| NewBVF 1550_Rv | CAGCTTTAAGTGCGTATCGGG | |
| NewBVF 2348_Fw | TCATCCATGACACTGGCAAG | |
| NewBVF 2348_Rv | CTTGCCAGTGTCATGGATGA | |
| BVF 3149_Fw | CCAGCGCAGAAGCTGAACAAC | |
| BVF3628_fw | TATGCCCATGAGTACCGCCGCGTTAGTGAC | |
| BVF 3628_Rv | GCTGGCGATGGCTTCAGTGG | |
| New_BVF 3932_Fw | GAACCCTCCGCCTTGTCTAT | |
| New_BVF 3932_Rv | ATAGACAAGGCGGAGGGTTC | |
| New_BVF 4755_Fw | GCATCTCACAAGTAAACCGC | |
| New_BVF 4755_Rv | GCGGTTTACTTGTGAGATGC | |
| BVF 5279_Rv | GGTCCAAACCTCGCCAGAGA | |
| New_BVF 6374_Fw | TTGAATCAGCGCACAAAGTCC | |
| New_BVF 6374_Rv | GGACTTTGTGCGCTGATTCAA | |
| New_qPCR-intergenic-BVF_For | TTCACCTCACATTGAACCTT | |
| M13_Rev | gcggataacaatttcacacagg | |
| M13_For | gtaaaacgacggccagt | |
| BVF_XbaIT7 Fw | CTAGCTTCTCTAGATAATACGACTCACTATAGGGGATTAAATTCACATCCAACA | Construction of infectious clone |
| BVF_NotI/EcoRI Rev | TTGAACGGGAATTCGCGGCCGCTTTTTTTTTTTTTTTTTTGCCTCGTGTGCAACGAAG | |
| BVF_5847 Rv | AGCGTAAGATATCGTGTTGGGAT | |
| BVF_6756 Fw | CCTCCGCTTTCACCACCG | |
| BcOLV2-Fw | TGTGCTCAGGAACCGAAGAT | Detection of Pi258.9 mycoviruses |
| BcOLV2-Rv | GCTTATGAGTTTGGAGGCCG | |
| SclerotiniaOuMV4-Fw | CTGGCTGACTTTGGTATCGC | |
| SclerotiniaOuMV4-Rv | CCTCCATCTCTTCTGCCACA | |
| Umbra SsULV3b Fw | ACGAGCAGTTGGTTCTCAGA | |
| Umbra SsULV3b Rv | CAACCACAGCCAACATGACA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Córdoba, L.; Ruiz-Padilla, A.; Rodríguez-Romero, J.; Ayllón, M.A. Construction and Characterization of a Botrytis Virus F Infectious Clone. J. Fungi 2022, 8, 459. https://doi.org/10.3390/jof8050459
Córdoba L, Ruiz-Padilla A, Rodríguez-Romero J, Ayllón MA. Construction and Characterization of a Botrytis Virus F Infectious Clone. Journal of Fungi. 2022; 8(5):459. https://doi.org/10.3390/jof8050459
Chicago/Turabian StyleCórdoba, Laura, Ana Ruiz-Padilla, Julio Rodríguez-Romero, and María A. Ayllón. 2022. "Construction and Characterization of a Botrytis Virus F Infectious Clone" Journal of Fungi 8, no. 5: 459. https://doi.org/10.3390/jof8050459
APA StyleCórdoba, L., Ruiz-Padilla, A., Rodríguez-Romero, J., & Ayllón, M. A. (2022). Construction and Characterization of a Botrytis Virus F Infectious Clone. Journal of Fungi, 8(5), 459. https://doi.org/10.3390/jof8050459






