Clinical Aspergillus Signatures in COPD and Bronchiectasis
Abstract
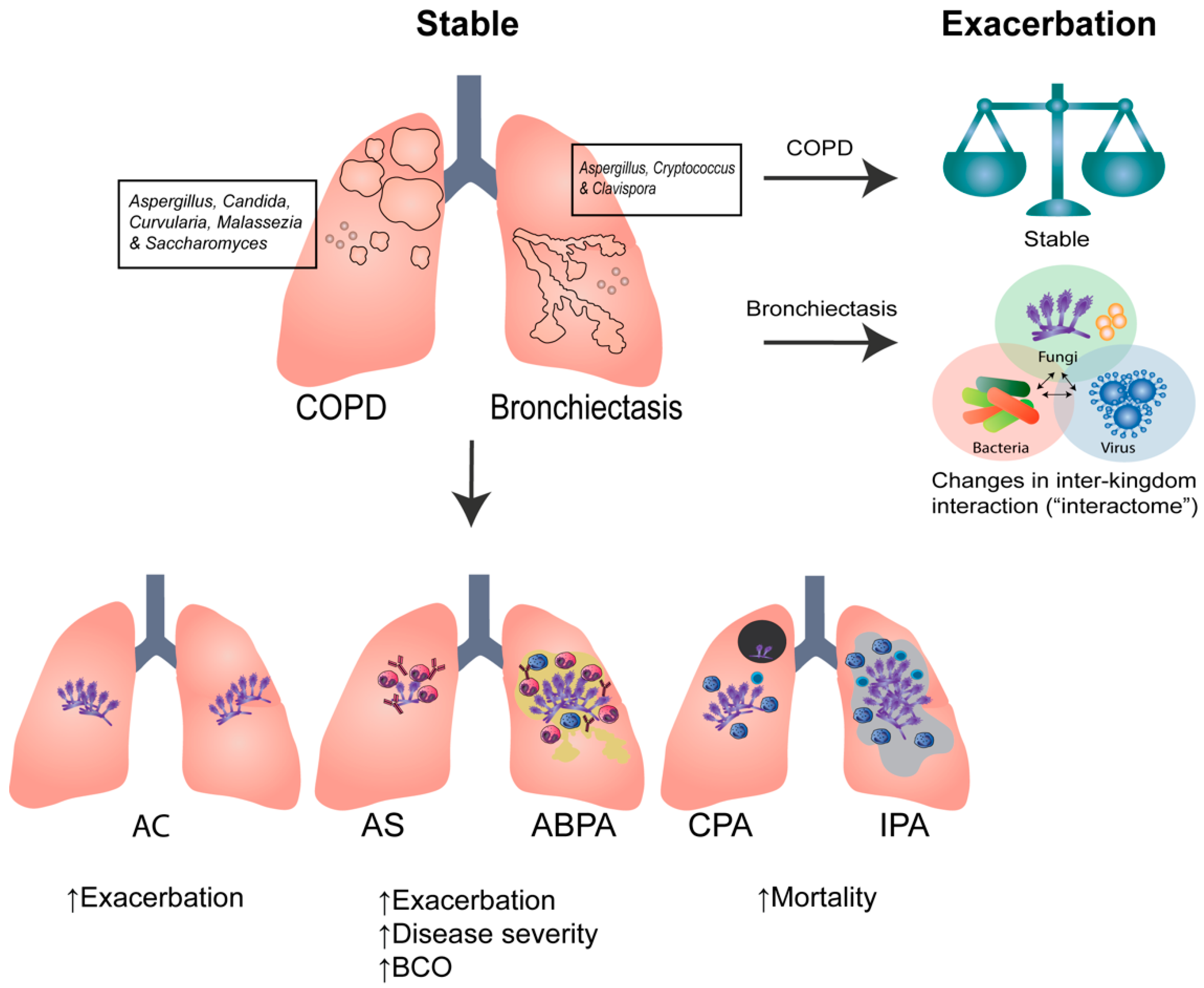
:1. Introduction
2. Clinical Aspergillus Signatures in COPD and Bronchiectasis
3. The Challenges of Sequencing the Mycobiome
4. The Pulmonary Mycobiome in Health, COPD, and Bronchiectasis
4.1. The Pulmonary Mycobiome in Healthy Individuals
4.2. COPD
4.3. Bronchiectasis
5. Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalmers, J.D.; Chang, A.B.; Chotirmall, S.H.; Dhar, R.; McShane, P.J. Bronchiectasis. Nat. Rev. Dis. Primers 2018, 4, 45. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.M.; Tiew, P.Y.; Mac Aogain, M.; Budden, K.F.; Yong, V.F.; Thomas, S.S.; Pethe, K.; Hansbro, P.M.; Chotirmall, S.H. The role of acute and chronic respiratory colonization and infections in the pathogenesis of COPD. Respirology 2017, 22, 634–650. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S. Infection as a comorbidity of COPD. Eur. Respir. J. 2010, 35, 1209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goh, K.J.; Yii, A.C.A.; Lapperre, T.S.; Chan, A.K.; Chew, F.T.; Chotirmall, S.H.; Koh, M.S. Sensitization to Aspergillus species is associated with frequent exacerbations in severe asthma. J. Asthma Allergy 2017, 10, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Tiew, P.Y.; Mac Aogáin, M.; Ter, S.K.; Aliberti, S.; Chalmers, J.D.; Chotirmall, S.H. Respiratory Mycoses in COPD and Bronchiectasis. Mycopathologia 2021, 186, 623–638. [Google Scholar] [CrossRef]
- Jaggi, T.K.; Ter, S.K.; Mac Aogain, M.; Chotirmall, S.H. Aspergillus-Associated Endophenotypes in Bronchiectasis. Semin. Respir. Crit. Care Med. 2021, 42, 556–566. [Google Scholar] [CrossRef]
- Nguyen, L.D.N.; Viscogliosi, E.; Delhaes, L. The lung mycobiome: An emerging field of the human respiratory microbiome. Front. Microbiol. 2015, 6, 89. [Google Scholar] [CrossRef]
- Mac Aogáin, M.; Chandrasekaran, R.; Lim, A.Y.; Low, T.B.; Tan, G.L.; Hassan, T.; Ong, T.H.; Ng, A.H.; Bertrand, D.; Koh, J.Y.; et al. Immunological corollary of the pulmonary mycobiome in bronchiectasis: The CAMEB study. Eur. Respir. J. 2018, 52, 1800766. [Google Scholar] [CrossRef]
- Mac Aogáin, M.; Narayana, J.K.; Tiew, P.Y.; Ali, N.; Yong, V.F.L.; Jaggi, T.K.; Lim, A.Y.H.; Keir, H.R.; Dicker, A.J.; Thng, K.X. Integrative microbiomics in bronchiectasis exacerbations. Nat. Med. 2021, 27, 688–699. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Dicker, A.J.; Keir, H.R.; Poh, M.E.; Pang, S.L.; Mac Aogain, M.; Chua, B.Q.Y.; Tan, J.L.; Xu, H.; Koh, M.S.; et al. A high-risk airway mycobiome is associated with frequent exacerbation and mortality in COPD. Eur. Respir. J. 2021, 57, 20020502002050. [Google Scholar] [CrossRef]
- Aliberti, S.; Masefield, S.; Polverino, E.; De Soyza, A.; Loebinger, M.R.; Menendez, R.; Ringshausen, F.C.; Vendrell, M.; Powell, P.; Chalmers, J.D. Research priorities in bronchiectasis: A consensus statement from the EMBARC Clinical Research Collaboration. Eur. Respir. J. 2016, 48, 632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, A.; Armstrong-James, D.; Chotirmall, S.H. Respiratory Mycoses: A Call to Action to Recognize, Educate and Invest. Mycopathologia 2021, 186, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Pashley, C.; Hartl, D.; Wardlaw, A.; Godet, C.; Del Giacco, S.; Delhaes, L.; Sergejeva, S. Fungal allergy in asthma-state of the art and research needs. Clin. Transl. Allergy 2014, 4, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agusti, C.; Rano, A.; Aldabo, I.; Torres, A. Fungal pneumonia, chronic respiratory diseases and glucocorticoids. Med. Mycol. 2006, 44, S207–S211. [Google Scholar] [CrossRef] [Green Version]
- Palmieri, F.; Koutsokera, A.; Bernasconi, E.; Junier, P.; von Garnier, C.; Ubags, N. Recent Advances in Fungal Infections: From Lung Ecology to Therapeutic Strategies With a Focus on Aspergillus spp. Front. Med. 2022, 9, 832510. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Martin-Gomez, M.T. Aspergillus Species in Bronchiectasis: Challenges in the Cystic Fibrosis and Non-cystic Fibrosis Airways. Mycopathologia 2018, 183, 45–59. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Al-Alawi, M.; Mirkovic, B.; Lavelle, G.; Logan, P.M.; Greene, C.M.; McElvaney, N.G. Aspergillus-associated airway disease, inflammation, and the innate immune response. BioMed Res. Int. 2013, 2013, 723129. [Google Scholar] [CrossRef] [Green Version]
- Gusareva, E.S.; Acerbi, E.; Lau, K.J.; Luhung, I.; Premkrishnan, B.N.; Kolundžija, S.; Purbojati, R.W.; Wong, A.; Houghton, J.N.; Miller, D. Microbial communities in the tropical air ecosystem follow a precise diel cycle. Proc. Natl. Acad. Sci. USA 2019, 116, 23299–23308. [Google Scholar] [CrossRef] [Green Version]
- Tiew, P.Y.; Mac Aogáin, M.; Chotirmall, S.H. The current understanding and future directions for sputum microbiome profiling in chronic obstructive pulmonary disease. Curr. Opin. Pulm. Med. 2022, 28, 121–133. [Google Scholar] [CrossRef]
- Yii, A.C.; Koh, M.S.; Lapperre, T.S.; Tan, G.L.; Chotirmall, S.H. The emergence of Aspergillus species in chronic respiratory disease. Front. Biosci. 2017, 9, 127–138. [Google Scholar] [CrossRef]
- Chandrasekaran, R.; Mac Aogain, M.; Chalmers, J.D.; Elborn, S.J.; Chotirmall, S.H. Geographic variation in the aetiology, epidemiology and microbiology of bronchiectasis. BMC Pulm. Med. 2018, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Choi, J.H.; Jeon, H.G.; Cha, H.E.; Hwang, Y.J.; Chung, Y.S. Comparison between polymerase chain reaction and fungal culture for the detection of fungi in patients with chronic sinusitis and normal controls. Acta Otolaryngol. 2005, 125, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The microbiome of the lung. Transl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budden, K.F.; Shukla, S.D.; Rehman, S.F.; Bowerman, K.L.; Keely, S.; Hugenholtz, P.; Armstrong-James, D.P.H.; Adcock, I.M.; Chotirmall, S.H.; Chung, K.F.; et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir. Med. 2019, 7, 907–920. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Jaggi, T.K.; Chan, L.L.Y.; Chotirmall, S.H. The airway microbiome in COPD, bronchiectasis and bronchiectasis-COPD overlap. Clin. Respir. J. 2021, 15, 123–133. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Mac Aogain, M.; Ali, N.; Thng, K.X.; Goh, K.; Lau, K.J.X.; Chotirmall, S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia 2020, 185, 207–231. [Google Scholar] [CrossRef]
- Cui, L.; Lucht, L.; Tipton, L.; Rogers, M.B.; Fitch, A.; Kessinger, C.; Camp, D.; Kingsley, L.; Leo, N.; Greenblatt, R.M.; et al. Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am. J. Respir. Crit. Care Med. 2015, 191, 932–942. [Google Scholar] [CrossRef]
- Su, J.; Liu, H.-Y.; Tan, X.-L.; Ji, Y.; Jiang, Y.-X.; Prabhakar, M.; Rong, Z.-H.; Zhou, H.-W.; Zhang, G.-X. Sputum Bacterial and Fungal Dynamics during Exacerbations of Severe COPD. PLoS ONE 2015, 10, e0130736. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Zhang, S.-Y.; Yang, W.-Y.; Su, X.-F.; He, Y.; Zhou, H.-W.; Su, J. Oropharyngeal and Sputum Microbiomes Are Similar Following Exacerbation of Chronic Obstructive Pulmonary Disease. Front. Microbiol. 2017, 8, 1163. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Mirkovic, B.; Lavelle, G.M.; McElvaney, N.G. Immunoevasive Aspergillus virulence factors. Mycopathologia 2014, 178, 363–370. [Google Scholar] [CrossRef]
- Coughlan, C.A.; Chotirmall, S.H.; Renwick, J.; Hassan, T.; Low, T.B.; Bergsson, G.; Eshwika, A.; Bennett, K.; Dunne, K.; Greene, C.M.; et al. The effect of Aspergillus fumigatus infection on vitamin D receptor expression in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2012, 186, 999–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, M.A.; Chotirmall, S.H.; McCullagh, B.; Branagan, P.; McElvaney, N.G.; Logan, P.M. Radiological abnormalities associated with Aspergillus colonization in a cystic fibrosis population. Eur. J. Radiol. 2012, 81, e197–e202. [Google Scholar] [CrossRef] [PubMed]
- Chotirmall, S.H.; Gellatly, S.L.; Budden, K.F.; Mac Aogain, M.; Shukla, S.D.; Wood, D.L.; Hugenholtz, P.; Pethe, K.; Hansbro, P.M. Microbiomes in respiratory health and disease: An Asia-Pacific perspective. Respirology 2017, 22, 240–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalmers, J.D.; Chotirmall, S.H. Bronchiectasis: New therapies and new perspectives. Lancet Respir. Med. 2018, 6, 715–726. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.; Ivan, F.X.; Mac Aogain, M.; Narayana, J.K.; Lee, S.Y.; Lim, C.L.; Chotirmall, S.H. The Healthy Airway Mycobiome in Individuals of Asian Descent. Chest 2021, 159, 544–548. [Google Scholar] [CrossRef]
- Tiew, P.Y.; Ko, F.W.S.; Pang, S.L.; Matta, S.A.; Sio, Y.Y.; Poh, M.E.; Lau, K.J.X.; Mac Aogain, M.; Jaggi, T.K.; Ivan, F.X.; et al. Environmental fungal sensitisation associates with poorer clinical outcomes in COPD. Eur. Respir. J. 2020, 56, 2000418. [Google Scholar] [CrossRef]
- Mac Aogain, M.; Tiew, P.Y.; Lim, A.Y.H.; Low, T.B.; Tan, G.L.; Hassan, T.; Ong, T.H.; Pang, S.L.; Lee, Z.Y.; Gwee, X.W.; et al. Distinct “Immunoallertypes” of Disease and High Frequencies of Sensitization in Non-Cystic Fibrosis Bronchiectasis. Am. J. Respir. Crit. Care Med. 2019, 199, 842–853. [Google Scholar] [CrossRef]
- Chotirmall, S.H.; Chalmers, J.D. RESPIRE: Breathing new life into bronchiectasis. Eur. Respir. J. 2018, 51, 1702444. [Google Scholar] [CrossRef]
- Gernez, Y.; Waters, J.; Mirkovic, B.; Lavelle, G.M.; Dunn, C.E.; Davies, Z.A.; Everson, C.; Tirouvanziam, R.; Silver, E.; Wallenstein, S.; et al. Blood basophil activation is a reliable biomarker of allergic bronchopulmonary aspergillosis in cystic fibrosis. Eur. Respir. J. 2016, 47, 177–185. [Google Scholar] [CrossRef]
- Mirkovic, B.; Lavelle, G.M.; Azim, A.A.; Helma, K.; Gargoum, F.S.; Molloy, K.; Gernez, Y.; Dunne, K.; Renwick, J.; Murphy, P.; et al. The basophil surface marker CD203c identifies Aspergillus species sensitization in patients with cystic fibrosis. J. Allergy Clin. Immunol. 2016, 137, 436–443. [Google Scholar] [CrossRef]
- Hector, A.; Chotirmall, S.H.; Lavelle, G.M.; Mirkovic, B.; Horan, D.; Eichler, L.; Mezger, M.; Singh, A.; Ralhan, A.; Berenbrinker, S.; et al. Chitinase activation in patients with fungus-associated cystic fibrosis lung disease. J. Allergy Clin. Immunol. 2016, 138, 1183–1189.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poh, T.Y.; Tiew, P.Y.; Hou Lim, A.Y.; Thng, K.X.; Binte Mohamed Ali, N.A.; Narayana, J.K.; Aogain, M.M.; Tien, Z.; Chew, W.M.; Wai Chan, A.K.; et al. Increased Chitotriosidase Is Associated with Aspergillus and Frequent Exacerbations in South-East Asians with Bronchiectasis. Chest 2020, 158, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Mac Aogain, M.; Chotirmall, S.H. Microbiology and the Microbiome in Bronchiectasis. Clin. Chest Med. 2022, 43, 23–34. [Google Scholar] [CrossRef]
- Poh, T.Y.; Mac Aogain, M.; Chan, A.K.; Yii, A.C.; Yong, V.F.; Tiew, P.Y.; Koh, M.S.; Chotirmall, S.H. Understanding COPD-overlap syndromes. Expert Rev. Respir. Med. 2017, 11, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Tiew, P.Y.; Lim, A.Y.H.; Keir, H.R.; Dicker, A.J.; Mac Aogain, M.; Pang, S.L.; Low, T.B.; Hassan, T.M.; Poh, M.E.; Xu, H.; et al. High Frequency of Allergic Bronchopulmonary Aspergillosis in Bronchiectasis-COPD Overlap. Chest 2022, 161, 40–53. [Google Scholar] [CrossRef]
- Aogain, M.M.; Jaggi, T.K.; Chotirmall, S.H. The Airway Microbiome: Present and Future Applications. Arch Bronconeumol. 2022, 58, 8–10. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogain, M.; Ali, N.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Similarity network fusion for the integration of multi-omics and microbiomes in respiratory disease. Eur. Respir. J. 2021, 58, 2101016. [Google Scholar] [CrossRef]
- Narayana, J.K.; Mac Aogain, M.; Goh, W.W.B.; Xia, K.; Tsaneva-Atanasova, K.; Chotirmall, S.H. Mathematical-based microbiome analytics for clinical translation. Comput. Struct. Biotechnol. J. 2021, 19, 6272–6281. [Google Scholar] [CrossRef]
- Kozel, T.R.; Wickes, B. Fungal diagnostics. Cold Spring Harb. Perspect. Med. 2014, 4, a019299. [Google Scholar] [CrossRef]
- Ullmann, A.J.; Aguado, J.M.; Arikan-Akdagli, S.; Denning, D.W.; Groll, A.H.; Lagrou, K.; Lass-Flörl, C.; Lewis, R.E.; Munoz, P.; Verweij, P.E. Diagnosis and management of Aspergillus diseases: Executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clin. Microbiol. Infect. 2018, 24, e1–e38. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Li, H.; Zhang, Y.; Huang, M.; He, Q.; Li, P.; Zhang, F.; Shi, Y.; Su, X. Diagnostic Value of Galactomannan Antigen Test in Serum and Bronchoalveolar Lavage Fluid Samples from Patients with Nonneutropenic Invasive Pulmonary Aspergillosis. J. Clin. Microbiol. 2017, 55, 2153–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.B.; Chen, G.P.; Lin, Q.C.; Lin, X.; Zhang, H.Y.; Wang, J.H. Bronchoalveolar lavage fluid galactomannan detection for diagnosis of invasive pulmonary aspergillosis in chronic obstructive pulmonary disease. Med. Mycol. 2013, 51, 688–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meersseman, W.; Lagrou, K.; Maertens, J.; Wilmer, A.; Hermans, G.; Vanderschueren, S.; Spriet, I.; Verbeken, E.; Van Wijngaerden, E. Galactomannan in bronchoalveolar lavage fluid: A tool for diagnosing aspergillosis in intensive care unit patients. Am. J. Respir. Crit. Care Med. 2008, 177, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.; Page, I. Role of Serological Tests in the Diagnosis of Mold Infections. Curr. Fungal Infect. Rep. 2018, 12, 127–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otasevic, S.; Momcilovic, S.; Stojanovic, N.M.; Skvarc, M.; Rajkovic, K.; Arsic-Arsenijevic, V. Non-culture based assays for the detection of fungal pathogens. J. Mycol. Med. 2018, 28, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Tarrand, J.J.; Lichterfeld, M.; Warraich, I.; Luna, M.; Han, X.Y.; May, G.S.; Kontoyiannis, D.P. Diagnosis of invasive septate mold infections. A correlation of microbiological culture and histologic or cytologic examination. Am. J. Clin. Pathol. 2003, 119, 854–858. [Google Scholar] [CrossRef]
- Gabaldón, T. Recent trends in molecular diagnostics of yeast infections: From PCR to NGS. FEMS Microbiol. Rev. 2019, 43, 517–547. [Google Scholar] [CrossRef] [Green Version]
- Croxatto, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Palanivel, M.; Mac Aogáin, M.; Purbojati, R.W.; Uchida, A.; Aung, N.W.; Lim, S.B.Y.; Putra, A.; Drautz-Moses, D.I.; Seaton, S.; Rogers, T.R.; et al. Whole-Genome Sequencing of Aspergillus terreus Species Complex. Mycopathologia 2020, 185, 405–408. [Google Scholar] [CrossRef]
- Tsang, C.-C.; Teng, J.L.L.; Lau, S.K.P.; Woo, P.C.Y. Rapid Genomic Diagnosis of Fungal Infections in the Age of Next-Generation Sequencing. J. Fungi 2021, 7, 636. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Huseyin, C.E.; O’Toole, P.W.; Cotter, P.D.; Scanlan, P.D. Forgotten fungi—the gut mycobiome in human health and disease. FEMS Microbiol. Rev. 2017, 41, 479–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding, C. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241. [Google Scholar] [CrossRef] [Green Version]
- Begerow, D.; Nilsson, H.; Unterseher, M.; Maier, W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 2010, 87, 99–108. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef] [Green Version]
- Ali, N.; Mac Aogain, M.; Morales, R.F.; Tiew, P.Y.; Chotirmall, S.H. Optimisation and Benchmarking of Targeted Amplicon Sequencing for Mycobiome Analysis of Respiratory Specimens. Int. J. Mol. Sci. 2019, 20, 4991. [Google Scholar] [CrossRef] [Green Version]
- Angebault, C.; Payen, M.; Woerther, P.L.; Rodriguez, C.; Botterel, F. Combined bacterial and fungal targeted amplicon sequencing of respiratory samples: Does the DNA extraction method matter? PLoS ONE 2020, 15, e0232215. [Google Scholar] [CrossRef]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Lindahl, B. Fungal identification biases in microbiome projects. Environ. Microbiol. Rep. 2016, 8, 774–779. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Miao, Q.; Ma, Y.; Wang, Q.; Pan, J.; Zhang, Y.; Jin, W.; Yao, Y.; Su, Y.; Huang, Y.; Wang, M.; et al. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin. Infect. Dis. 2018, 67, S231–S240. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, Y.; Feng, J. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm. Med. 2019, 19, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diao, Z.; Han, D.; Zhang, R.; Li, J. Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections. J. Adv. Res. 2021, in press. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Huang, J.; Jiang, E.; Yang, D.; Wei, J.; Zhao, M.; Feng, J.; Cao, J. Metagenomic Next-Generation Sequencing versus Traditional Pathogen Detection in the Diagnosis of Peripheral Pulmonary Infectious Lesions. Infect. Drug Resist. 2020, 13, 567–576. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [Green Version]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Klimek-Ochab, M.; Brzezińska-Rodak, M.; Zymańczyk-Duda, E.; Lejczak, B.; Kafarski, P. Comparative study of fungal cell disruption--scope and limitations of the methods. Folia Microbiol. 2011, 56, 469–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angebault, C.; Ghozlane, A.; Volant, S.; Botterel, F.; d’Enfert, C.; Bougnoux, M.-E. Combined bacterial and fungal intestinal microbiota analyses: Impact of storage conditions and DNA extraction protocols. PLoS ONE 2018, 13, e0201174. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum, J.; Usyk, M.; Chen, Z.; Zolnik, C.P.; Jones, H.E.; Waldron, L.; Dowd, J.B.; Thorpe, L.E.; Burk, R.D. Evaluation of Oral Cavity DNA Extraction Methods on Bacterial and Fungal Microbiota. Sci. Rep. 2019, 9, 1531. [Google Scholar] [CrossRef] [PubMed]
- Tipton, L.; Ghedin, E.; Morris, A. The lung mycobiome in the next-generation sequencing era. Virulence 2017, 8, 334–341. [Google Scholar] [CrossRef]
- Bittinger, K.; Charlson, E.S.; Loy, E.; Shirley, D.J.; Haas, A.R.; Laughlin, A.; Yi, Y.; Wu, G.D.; Lewis, J.D.; Frank, I.; et al. Improved characterization of medically relevant fungi in the human respiratory tract using next-generation sequencing. Genome Biol. 2014, 15, 487. [Google Scholar] [CrossRef]
- Whiteside, S.A.; McGinniss, J.E.; Collman, R.G. The lung microbiome: Progress and promise. J. Clin. Investig. 2021, 131, e150473. [Google Scholar] [CrossRef]
- Huseyin, C.E.; Rubio, R.C.; O’Sullivan, O.; Cotter, P.D.; Scanlan, P.D. The Fungal Frontier: A Comparative Analysis of Methods Used in the Study of the Human Gut Mycobiome. Front. Microbiol. 2017, 8, 1432. [Google Scholar] [CrossRef] [Green Version]
- van Burik, J.A.; Schreckhise, R.W.; White, T.C.; Bowden, R.A.; Myerson, D. Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 1998, 36, 299–303. [Google Scholar] [CrossRef]
- Goig, G.A.; Blanco, S.; Garcia-Basteiro, A.L.; Comas, I. Contaminant DNA in bacterial sequencing experiments is a major source of false genetic variability. BMC Biol. 2020, 18, 24. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef] [Green Version]
- Ronholm, J.; Nasheri, N.; Petronella, N.; Pagotto, F. Navigating Microbiological Food Safety in the Era of Whole-Genome Sequencing. Clin. Microbiol. Rev. 2016, 29, 837–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mac Aogáin, M.; Chaturvedi, V.; Chotirmall, S.H. MycopathologiaGENOMES: The New ‘Home’ for the Publication of Fungal Genomes. Mycopathologia 2019, 184, 551–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallen-Adams, H.E.; Kachman, S.D.; Kim, J.; Legge, R.M.; Martínez, I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015, 15, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Donovan, P.D.; Gonzalez, G.; Higgins, D.G.; Butler, G.; Ito, K. Identification of fungi in shotgun metagenomics datasets. PLoS ONE 2018, 13, e0192898. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [Green Version]
- Rick, E.M.; Woolnough, K.F.; Seear, P.J.; Fairs, A.; Satchwell, J.; Richardson, M.; Monteiro, W.R.; Craner, M.; Bourne, M.; Wardlaw, A.J. The airway fungal microbiome in asthma. Clin. Exp. Allergy 2020, 50, 1325–1341. [Google Scholar] [CrossRef]
- Charlson, E.S.; Diamond, J.M.; Bittinger, K.; Fitzgerald, A.S.; Yadav, A.; Haas, A.R.; Bushman, F.D.; Collman, R.G. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012, 186, 536–545. [Google Scholar] [CrossRef] [Green Version]
- Carpagnano, G.E.; Susca, A.; Scioscia, G.; Lacedonia, D.; Cotugno, G.; Soccio, P.; Santamaria, S.; Resta, O.; Logrieco, G.; Foschino Barbaro, M.P. A survey of fungal microbiota in airways of healthy volunteer subjects from Puglia (Apulia), Italy. BMC Infect. Dis. 2019, 19, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Ghannoum, M.A.; Jurevic, R.J.; Mukherjee, P.K.; Cui, F.; Sikaroodi, M.; Naqvi, A.; Gillevet, P.M. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010, 6, e1000713. [Google Scholar] [CrossRef] [Green Version]
- Krause, R.; Halwachs, B.; Thallinger, G.G.; Klymiuk, I.; Gorkiewicz, G.; Hoenigl, M.; Prattes, J.; Valentin, T.; Heidrich, K.; Buzina, W. Characterisation of Candida within the mycobiome/microbiome of the lower respiratory tract of ICU patients. PLoS ONE 2016, 11, e0155033. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Murphy, T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 2008, 359, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- Bafadhel, M.; McKenna, S.; Agbetile, J.; Fairs, A.; Desai, D.; Mistry, V.; Morley, J.P.; Pancholi, M.; Pavord, I.D.; Wardlaw, A.J.; et al. Aspergillus fumigatus during stable state and exacerbations of COPD. Eur. Respir. J. 2014, 43, 64. [Google Scholar] [CrossRef] [PubMed]
- Guinea, J.; Torres-Narbona, M.; Gijón, P.; Muñoz, P.; Pozo, F.; Peláez, T.; de Miguel, J.; Bouza, E. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: Incidence, risk factors, and outcome. Clin. Microbiol. Infect. 2010, 16, 870–877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connell, D.; Shah, A. The contribution of Aspergillus fumigatus to COPD exacerbations: A “sensitive” topic. Eur. Respiratory Soc. 2020, 56, 2002223. [Google Scholar] [CrossRef]
- Martinsen, E.M.; Eagan, T.M.; Leiten, E.O.; Haaland, I.; Husebø, G.R.; Knudsen, K.S.; Drengenes, C.; Sanseverino, W.; Paytuví-Gallart, A.; Nielsen, R. The pulmonary mycobiome—A study of subjects with and without chronic obstructive pulmonary disease. PLoS ONE 2021, 16, e0248967. [Google Scholar] [CrossRef]
- Yang, R.; Zhang, Q.; Ren, Z.; Li, H.; Ma, Q. Different Airway Inflammatory Phenotypes Correlate with Specific Fungal and Bacterial Microbiota in Asthma and Chronic Obstructive Pulmonary Disease. J. Immunol. Res. 2022, 2022, 2177884. [Google Scholar] [CrossRef]
- Flume, P.A.; Chalmers, J.D.; Olivier, K.N. Advances in bronchiectasis: Endotyping, genetics, microbiome, and disease heterogeneity. Lancet 2018, 392, 880–890. [Google Scholar] [CrossRef] [Green Version]
- Chotirmall, S.H.; Chalmers, J.D. Bronchiectasis: An emerging global epidemic. BMC Pulm. Med. 2018, 18, 76. [Google Scholar] [CrossRef]
- Richardson, H.; Dicker, A.J.; Barclay, H.; Chalmers, J.D. The microbiome in bronchiectasis. Eur. Respir. Rev. 2019, 28, 190048. [Google Scholar] [CrossRef]
- Dicker, A.J.; Lonergan, M.; Keir, H.R.; Smith, A.H.; Pollock, J.; Finch, S.; Cassidy, A.J.; Huang, J.T.J.; Chalmers, J.D. The sputum microbiome and clinical outcomes in patients with bronchiectasis: A prospective observational study. Lancet Respir. Med. 2021, 9, 885–896. [Google Scholar] [CrossRef]
- Al Shakirchi, M.; Sorjonen, K.; Klingspor, L.; Bergman, P.; Hjelte, L.; de Monestrol, I. The Effects of Aspergillus fumigatus Colonization on Lung Function in Patients with Cystic Fibrosis. J. Fungi 2021, 7, 944. [Google Scholar] [CrossRef] [PubMed]
- Chotirmall, S.H.; Branagan, P.; Gunaratnam, C.; McElvaney, N.G. Aspergillus/allergic bronchopulmonary aspergillosis in an Irish cystic fibrosis population: A diagnostically challenging entity. Respir. Care 2008, 53, 1035–1041. [Google Scholar] [PubMed]
- Chotirmall, S.H. Candida albicans in cystic fibrosis: “Opening statements presented, let the trial begin”. Pediatr. Pulmonol. 2016, 51, 445–446. [Google Scholar] [CrossRef] [PubMed]
- Chotirmall, S.H.; McElvaney, N.G. Fungi in the cystic fibrosis lung: Bystanders or pathogens? Int. J. Biochem. Cell Biol. 2014, 52, 161–173. [Google Scholar] [CrossRef]
- Cuthbertson, L.; Felton, I.; James, P.; Cox, M.J.; Bilton, D.; Schelenz, S.; Loebinger, M.R.; Cookson, W.O.C.; Simmonds, N.J.; Moffatt, M.F. The fungal airway microbiome in cystic fibrosis and non-cystic fibrosis bronchiectasis. J. Cyst. Fibros. 2021, 20, 295–302. [Google Scholar] [CrossRef]
- Reese, T.A.; Liang, H.-E.; Tager, A.M.; Luster, A.D.; Van Rooijen, N.; Voehringer, D.; Locksley, R.M. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 2007, 447, 92–96. [Google Scholar] [CrossRef] [Green Version]
- Livingston, B.; Crimi, C.; Newman, M.; Higashimoto, Y.; Appella, E.; Sidney, J.; Sette, A. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J. Immunol. 2002, 168, 5499–5506. [Google Scholar] [CrossRef]
- Neveu, W.A.; Bernardo, E.; Allard, J.L.; Nagaleekar, V.; Wargo, M.J.; Davis, R.J.; Iwakura, Y.; Whittaker, L.A.; Rincon, M. Fungal allergen β-glucans trigger p38 mitogen-activated protein kinase–mediated IL-6 translation in lung epithelial cells. Am. J. Respir. Cell Mol. Biol. 2011, 45, 1133–1141. [Google Scholar] [CrossRef] [Green Version]
- Amitani, R.; Taylor, G.; Elezis, E.-N.; Llewellyn-Jones, C.; Mitchell, J.; Kuze, F.; Cole, P.J.; Wilson, R. Purification and characterization of factors produced by Aspergillus fumigatus which affect human ciliated respiratory epithelium. Infect. Immun. 1995, 63, 3266–3271. [Google Scholar] [CrossRef] [Green Version]
- de Jesus Carrion, S.; Leal, S.M.; Ghannoum, M.A.; Aimanianda, V.; Latgé, J.-P.; Pearlman, E. The rodA hydrophobin on Aspergillus fumigatus spores masks dectin-1–and dectin-2–dependent responses and enhances fungal survival in vivo. J. Immunol. 2013, 191, 2581–2588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oosthuizen, J.L.; Gomez, P.; Ruan, J.; Hackett, T.L.; Moore, M.M.; Knight, D.A.; Tebbutt, S.J. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS ONE 2011, 6, e20527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Máiz, L.; Nieto, R.; Cantón, R.; Gómez, G.; Martinez-García, M.Á. Fungi in bronchiectasis: A concise review. Int. J. Mol. Sci. 2018, 19, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poh, T.Y.; Ali, N.; Chan, L.L.Y.; Tiew, P.Y.; Chotirmall, S.H. Evaluation of Droplet Digital Polymerase Chain Reaction (ddPCR) for the Absolute Quantification of Aspergillus species in the Human Airway. Int. J. Mol. Sci. 2020, 21, 3043. [Google Scholar] [CrossRef]
- Rogers, G.B.; Zain, N.M.M.; Bruce, K.D.; Burr, L.D.; Chen, A.C.; Rivett, D.W.; McGuckin, M.A.; Serisier, D.J. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann. Am. Thorac. Soc. 2014, 11, 496–503. [Google Scholar] [CrossRef]
- Tunney, M.M.; Einarsson, G.G.; Wei, L.; Drain, M.; Klem, E.R.; Cardwell, C.; Ennis, M.; Boucher, R.C.; Wolfgang, M.C.; Elborn, J.S. Lung microbiota and bacterial abundance in patients with bronchiectasis when clinically stable and during exacerbation. Am. J. Respir. Crit. Care Med. 2013, 187, 1118–1126. [Google Scholar] [CrossRef] [Green Version]
- Rogers, G.B.; Van Der Gast, C.J.; Cuthbertson, L.; Thomson, S.K.; Bruce, K.D.; Martin, M.L.; Serisier, D.J. Clinical measures of disease in adult non-CF bronchiectasis correlate with airway microbiota composition. Thorax 2013, 68, 731–737. [Google Scholar] [CrossRef] [Green Version]
- Maiz, L.; Vendrell, M.; Olveira, C.; Giron, R.; Nieto, R.; Martinez-Garcia, M.A. Prevalence and factors associated with isolation of Aspergillus and Candida from sputum in patients with non-cystic fibrosis bronchiectasis. Respiration 2015, 89, 396–403. [Google Scholar] [CrossRef]
- Skalski, J.H.; Limon, J.J.; Sharma, P.; Gargus, M.D.; Nguyen, C.; Tang, J.; Coelho, A.L.; Hogaboam, C.M.; Crother, T.R.; Underhill, D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018, 14, e1007260. [Google Scholar] [CrossRef]
- Li, X.; Leonardi, I.; Semon, A.; Doron, I.; Gao, I.H.; Putzel, G.G.; Kim, Y.; Kabata, H.; Artis, D.; Fiers, W.D.; et al. Response to Fungal Dysbiosis by Gut-Resident CX3CR1(+) Mononuclear Phagocytes Aggravates Allergic Airway Disease. Cell Host Microbe 2018, 24, 847–856. [Google Scholar] [CrossRef] [Green Version]
- Noverr, M.C.; Noggle, R.M.; Toews, G.B.; Huffnagle, G.B. Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 2004, 72, 4996–5003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Stage | Challenges |
|---|---|
| Sample processing | Contamination DNA degradation with lysis of the fungal cell walls Variation in the DNA yield and quality between different commercial kits and extraction methods |
| Targeted amplicon sequencing | Primer bias Amplification bias Target accuracy Data reproducibility |
| Shotgun metagenomic sequencing | Low overall fungal abundance relative to bacteria High levels of host DNA impedes fungal detection High costs Data reproducibility |
| Bioinformatics analyses | Lack of consensus on best practices Limited established bioinformatics pipelines Poorly curated fungal reference databases Large numbers of unidentified taxa Poor species-level resolution |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiew, P.Y.; Thng, K.X.; Chotirmall, S.H. Clinical Aspergillus Signatures in COPD and Bronchiectasis. J. Fungi 2022, 8, 480. https://doi.org/10.3390/jof8050480
Tiew PY, Thng KX, Chotirmall SH. Clinical Aspergillus Signatures in COPD and Bronchiectasis. Journal of Fungi. 2022; 8(5):480. https://doi.org/10.3390/jof8050480
Chicago/Turabian StyleTiew, Pei Yee, Kai Xian Thng, and Sanjay H. Chotirmall. 2022. "Clinical Aspergillus Signatures in COPD and Bronchiectasis" Journal of Fungi 8, no. 5: 480. https://doi.org/10.3390/jof8050480






