Torulaspora delbrueckii Phenotypic and Metabolic Profiling towards Its Biotechnological Exploitation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Collection
2.2. Phenotypic Characterization
2.3. Individual Fermentations and Metabolites Quantification
2.4. Data Analysis Using Statistical and Data Mining Methods
3. Results and Discussion
3.1. T. delbrueckii Intra-Strain Phenotypic Heterogeneity
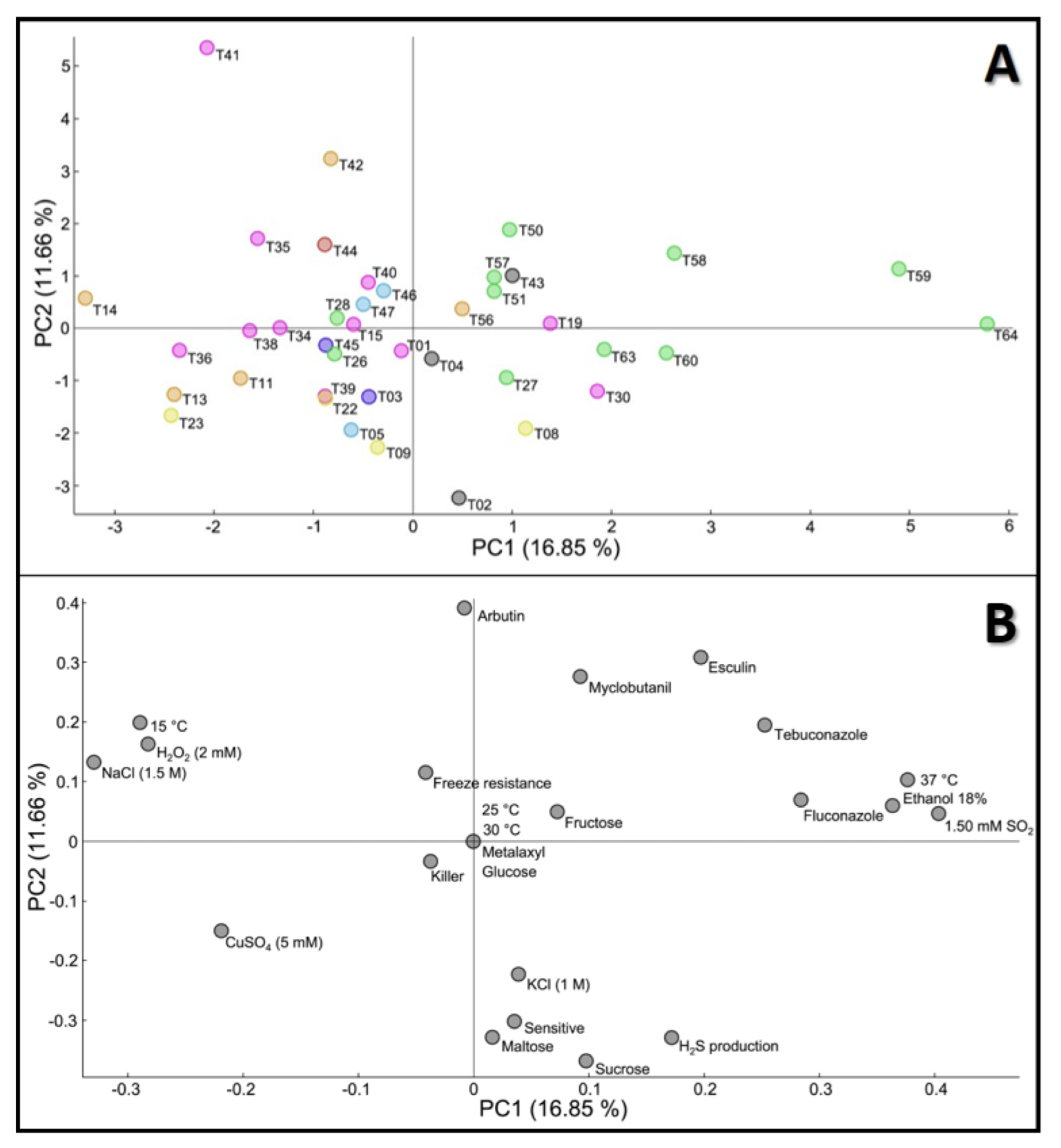
3.2. Principal Component Analysis Evaluation of Phenotypic Variation
3.3. Metabolic Characterization
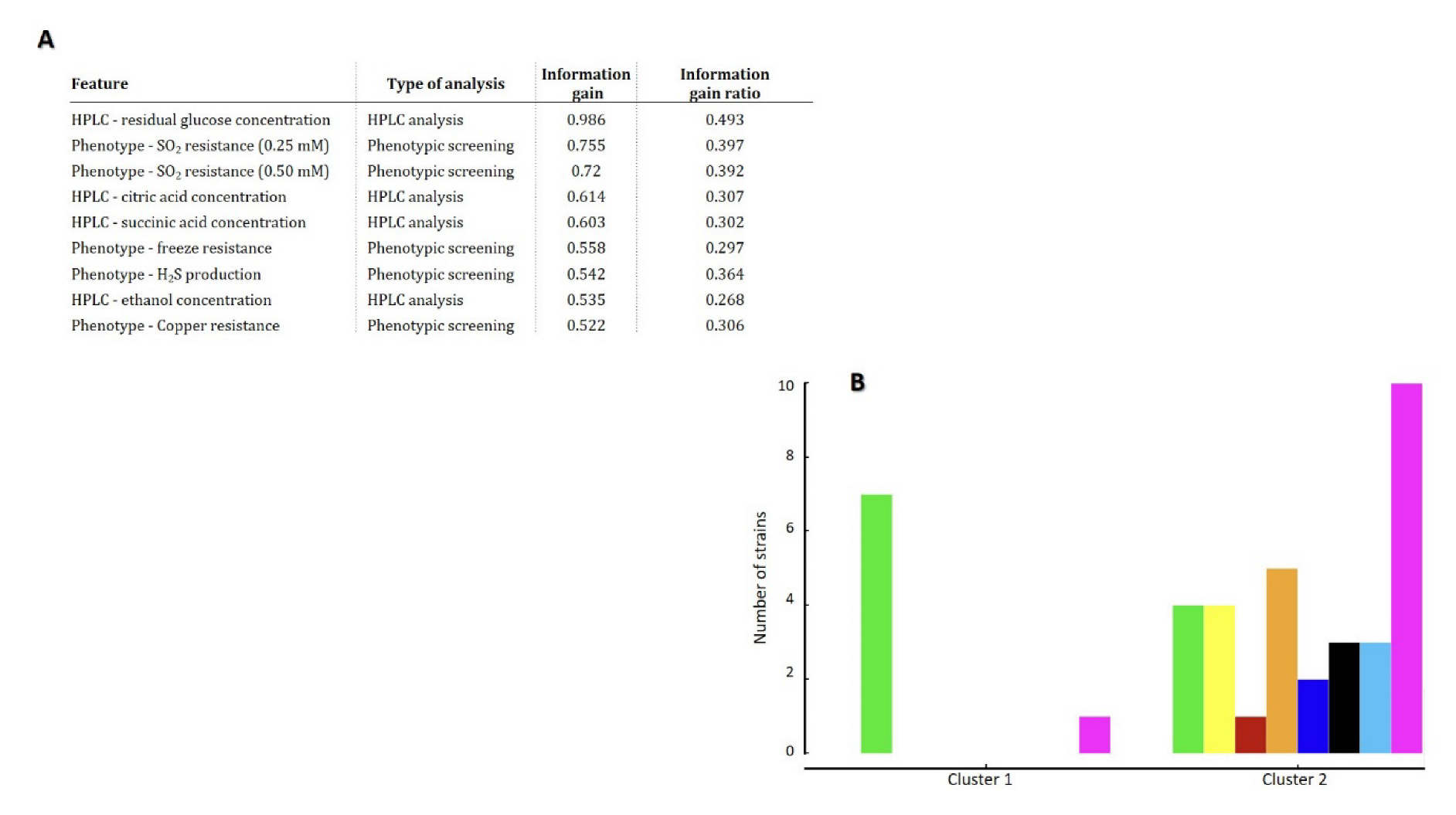
3.4. Bioinformatic Prediction of T. delbrueckii Biotechnological Potential
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Castelli, T. Les Agents de la Fermentation Vinaire. Arch. Mikrobiol. 1954, 20, 323–342. [Google Scholar] [CrossRef] [PubMed]
- Benito, S. The impact of Torulaspora delbrueckii yeast in winemaking. Appl. Microbiol. Biotechnol. 2018, 102, 3081–3094. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.; Silva-Sousa, F.; Pereira, F.; Rito, T.; Soares, P.; Franco-Duarte, R.; Sousa, M.J. Biotechnological Importance of Torulaspora delbrueckii: From the Obscurity to the Spotlight. J. Fungi 2021, 7, 712. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, I.S.; Høj, P.B. Grape and wine biotechnology: Challenges, opportunities and potential benefits. Aust. J. Grape Wine Res. 2005, 11, 83–108. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Umek, L.; Mendes, I.; Castro, C.; Fonseca, N.; Martins, R.; Ferreira, A.C.S.; Sampaio, P.; Pais, C.; Schuller, D. New integrative computational approaches unveil the Saccharomyces cerevisiae pheno-metabolomic fermentative profile and allow strain selection for winemaking. Food Chem. 2016, 211, 509–520. [Google Scholar] [CrossRef] [Green Version]
- Rainieri, S.; Pretorius, I.S. Selection and improvement of wine yeasts. Ann. Microbiol. 2000, 50, 15–31. [Google Scholar]
- Drumonde-Neves, J.; Fernandes, T.; Lima, T.; Pais, C.; Franco-Duarte, R. Learning from 80 years of studies: A comprehensive catalogue of non-Saccharomyces yeasts associated with viticulture and winemaking. FEMS Yeast Res. 2021, 21, foab017. [Google Scholar] [CrossRef]
- Ciani, M.; Beco, L.; Comitini, F. Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 2006, 108, 239–245. [Google Scholar] [CrossRef]
- Pacheco, A.; Santos, J.; Chaves, S.; Almeida, J.; Leão, C.; Sousa, M.J. The Emerging Role of the Yeast Torulaspora delbrueckii in Bread and Wine Production: Using Genetic Manipulation to Study Molecular Basis of Physiological Responses. In Structure and Function of Food Engineering; Eissa, A.A., Ed.; IntechOpen: London, UK, 2012; Chapter 13; pp. 339–370. [Google Scholar]
- Ciani, M.; Maccarelli, F. Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 1998, 14, 199–203. [Google Scholar] [CrossRef]
- Loira, I.; Vejarano, R.; Bañuelos, M.; Morata, A.; Tesfaye, W.; Uthurry, C.; Villa, A.; Cintora, I.; Suárez-Lepe, J. Influence of sequential fermentation with Torulaspora delbrueckii and Saccharomyces cerevisiae on wine quality. LWT Food Sci. Technol. 2014, 59, 915–922. [Google Scholar] [CrossRef] [Green Version]
- Sauer, M.; Porro, D.; Mattanovich, D.; Branduardi, P. Microbial production of organic acids: Expanding the markets. Trends Biotechnol. 2008, 26, 100–108. [Google Scholar] [CrossRef]
- Almeida, M.J.; Pais, C. Leavening ability and freeze tolerance of yeasts isolated from traditional corn and rye bread doughs. Appl. Environ. Microbiol. 1996, 62, 4401–4404. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Lopez, M.; Prieto, J.; Randez-Gil, F. Osmotolerance and leavening ability in sweet and frozen sweet dough. Comparative analysis between Torulaspora delbrueckii and Saccharomyces cerevisiae baker’s yeast strains. Antonie Leeuwenhoek 2003, 84, 125–134. [Google Scholar] [CrossRef]
- Alves-Araújo, C.; Almeida, M.J.; Sousa, M.J.; Leão, C. Freeze tolerance of the yeast Torulaspora delbrueckii: Cellular and biochemical basis. FEMS Microbiol. Lett. 2004, 240, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Belda, I.; Navascués, E.; Marquina, D.; Santos, A.; Calderon, F.; Benito, S. Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biotechnol. 2014, 99, 1911–1922. [Google Scholar] [CrossRef]
- Ciani, M.; Picciotti, G. The growth kinetics and fermentation behaviour of some non-Saccharomyces yeasts associated with wine-making. Biotechnol. Lett. 1995, 17, 1247–1250. [Google Scholar] [CrossRef]
- King, A.J.; Dickinson, J.R. Biotransformation of hop aroma terpenoids by ale and lager yeasts. FEMS Yeast Res. 2003, 3, 53–62. [Google Scholar] [CrossRef]
- Canonico, L.; Agarbati, A.; Comitini, F.; Ciani, M. Torulaspora delbrueckii in the brewing process: A new approach to enhance bioflavour and to reduce ethanol content. Food Microbiol. 2016, 56, 45–51. [Google Scholar] [CrossRef]
- Arrizon, J.; Morel, S.; Gschaedler, A.; Monsan, P. Fructanase and fructosyltransferase activity of non-Saccharomyces yeasts isolated from fermenting musts of Mezcal. Bioresour. Technol. 2012, 110, 560–565. [Google Scholar] [CrossRef]
- De La Torre-González, F.J.; Narváez-Zapata, J.A.; Taillandier, P.; Larralde-Corona, C.P. Mezcal as a Novel Source of Mixed Yeasts Inocula for Wine Fermentation. Processes 2020, 8, 1296. [Google Scholar] [CrossRef]
- Visintin, S.; Ramos, C.; Batista, N.; Dolci, P.; Schwan, F.; Cocolin, L. Impact of Saccharomyces cerevisiae and Torulaspora delbrueckii starter cultures on cocoa beans fermentation. Int. J. Food Microbiol. 2017, 257, 31–40. [Google Scholar] [CrossRef]
- Lachance, M.-A. Yeast communities in a natural tequila fermentation. Antonie Leeuwenhoek 1995, 68, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Wang, S.; Zhang, Y.; Yuan, Y.; Yue, T. Characterization and screening of non-Saccharomyces yeasts used to produce fragrant cider. LWT 2019, 107, 191–198. [Google Scholar] [CrossRef]
- Barry, J.P.; Metz, M.S.; Hughey, J.; Quirk, A.; Bochman, M.L. Two Novel Strains of Torulaspora delbrueckii Isolated from the Honey Bee Microbiome and Their Use in Honey Fermentation. Fermentation 2018, 4, 22. [Google Scholar] [CrossRef] [Green Version]
- Welthagen, J. Yeast profile in Gouda cheese during processing and ripening. Int. J. Food Microbiol. 1998, 41, 185–194. [Google Scholar] [CrossRef]
- Santiago, C.; Rito, T.; Vieira, D.; Fernandes, T.; Pais, C.; Sousa, M.; Soares, P.; Franco-Duarte, R. Improvement of Torulaspora delbrueckii Genome Annotation: Towards the Exploitation of Genomic Features of a Biotechnologically Relevant Yeast. J. Fungi 2021, 7, 287. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.; Pontes, A.; Franco-Duarte, R.; Soares, P.; Sampaio, J.P.; Sousa, M.J.; Brito, P.H. A glimpse at an early stage of microbe domestication revealed in the variable genome of Torulaspora delbrueckii, an emergent industrial yeast. Mol. Ecol. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, C.; Lage, P.; Vilela, A.; Mendes-Faia, A.; Mendes-Ferreira, A. Phenotypic and metabolic traits of commercial Saccharomyces cerevisiae yeasts. AMB Express 2014, 4, 39. [Google Scholar] [CrossRef] [Green Version]
- Mendes, I.; Franco-Duarte, R.; Umek, L.; Fonseca, E.; Drumonde-Neves, J.; Dequin, S.; Zupan, B.; Schuller, D. Computational Models for Prediction of Yeast Strain Potential for Winemaking from Phenotypic Profiles. PLoS ONE 2013, 8, e66523. [Google Scholar] [CrossRef] [Green Version]
- Mendes, I.; Sanchez, I.; Franco-Duarte, R.; Camarasa, C.; Schuller, D.; Dequin, S.; Sousa, M.J. Integrating transcriptomics and metabolomics for the analysis of the aroma profiles of Saccharomyces cerevisiae strains from diverse origins. BMC Genomics 2017, 18, 455. [Google Scholar] [CrossRef] [Green Version]
- Renault, P.; Miot-Sertier, C.; Marullo, P.; Hernández-Orte, P.; Lagarrigue, L.; Lonvaud-Funel, A.; Bely, M. Genetic characterization and phenotypic variability in Torulaspora delbrueckii species: Potential applications in the wine industry. Int. J. Food Microbiol. 2009, 134, 201–210. [Google Scholar] [CrossRef]
- Escribano, R.; González-Arenzana, L.; Portu, J.; Garijo, P.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A. Wine aromatic compound production and fermentative behaviour within different non-Saccharomyces species and clones. J. Appl. Microbiol. 2018, 124, 1521–1531. [Google Scholar] [CrossRef]
- Bely, M.; Sablayrolles, J.M.; Barre, P. Description of alcoholic fermentation kinetics: Its variability and significance. Am. J. Enol. Vitic. 1990, 41, 319–324. [Google Scholar]
- Rosi, I.; Vinella, M.; Domizio, P. Characterization of β-glucosidase activity in yeasts of oenological origin. J. Appl. Bacteriol. 1994, 77, 519–527. [Google Scholar] [CrossRef]
- Bonciani, T.; De Vero, L.; Giannuzzi, E.; Verspohl, A.; Giudici, P. Qualitative and quantitative screening of the β -glucosidase activity in Saccharomyces cerevisiae and Saccharomyces uvarum strains isolated from refrigerated must. Lett. Appl. Microbiol. 2018, 67, 72–78. [Google Scholar] [CrossRef]
- Jiranek, V.; Langridge, P.; Henschke, P.A. Validation of bismuth-containing indicator media for predicting H2S-producing potential of Saccharomyces cerevisiae wine yeasts under enological conditions. Am. J. Enol. Vitic. 1995, 46, 269–273. [Google Scholar]
- Buzzini, P.; Martini, A. Large-scale screening of selected Candida maltosa, Debaryomyces hansenii and Pichia anomala killer toxin activity against pathogenic yeasts. Med. Mycol. 2001, 39, 479–482. [Google Scholar] [CrossRef] [Green Version]
- Hino, A.; Mihara, K.; Nakashima, K.; Takano, H. Trehalose levels and survival ratio of freeze-tolerant versus freeze-sensitive yeasts. Appl. Environ. Microbiol. 1990, 56, 1386–1391. [Google Scholar] [CrossRef] [Green Version]
- Demšar, J.; Zupan, B.; Leban, G.; Curk, T. Orange: From experimental machine learning to interactive data mining. In Proceedings of the European Conference on Principles of Data Mining and Knowledge Discovery, Pisa, Italy, 20–24 September 2004; pp. 537–539. [Google Scholar]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Walsh, R.M.; Martin, P.A. Growth Of Saccharomyces Cerevisiae And Saccharomyces Uvarum In A Temperature Gradient Incubator. J. Inst. Brew. 1977, 83, 169–172. [Google Scholar] [CrossRef]
- Salvadó, Z.; Arroyo-López, F.N.; Guillamón, J.M.; Salazar, G.; Querol, A.; Barrio, E. Temperature Adaptation Markedly Determines Evolution within the Genus Saccharomyces. Appl. Environ. Microbiol. 2011, 77, 2292–2302. [Google Scholar] [CrossRef] [Green Version]
- García-Ríos, E.; Ramos-Alonso, L.; Guillamón, J.M. Correlation between Low Temperature Adaptation and Oxidative Stress in Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Vargas, S.; Estruch, F.; Randez-Gil, F. Gene Expression Analysis of Cold and Freeze Stress in Baker’s Yeast. Appl. Environ. Microbiol. 2002, 68, 3024–3030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Franco-Duarte, R.; Umek, L.; Zupan, B.; Schuller, D. Computational approaches for the genetic and phenotypic characterization of a Saccharomyces cerevisiaewine yeast collection. Yeast 2009, 26, 675–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batistote, M.; da Cruz, S.H.; Ernandes, J.R. Altered Patterns of Maltose and Glucose Fermentation by Brewing and Wine Yeasts Influenced by the Complexity of Nitrogen Source. J. Inst. Brew. 2006, 112, 84–91. [Google Scholar] [CrossRef]
- Hackenberg, S.; Leitner, T.; Jekle, M.; Becker, T. Maltose formation in wheat dough depending on mechanical starch modification and dough hydration. Carbohydr. Polym. 2018, 185, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts a Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Alves-Araújo, C.; Hernandez-Lopez, M.J.; Prieto, J.A.; Randez-Gil, F.; Sousa, M.J. Isolation and characterization of the LGT1 gene encoding a low-affinity glucose transporter from Torulaspora delbrueckii. Yeast 2005, 22, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Donzella, L.; Hernandez-Lopez, M.J.; Almeida, M.J.; Prieto, J.A.; Randez-Gil, F.; Morrissey, J.P.; Sousa, M.J. Hexose transport in Torulaspora delbrueckii: Identification of Igt1, a new dual-affinity transporter. FEMS Yeast Res. 2020, 20, foaa004. [Google Scholar] [CrossRef]
- Tronchoni, J.; Gamero, A.; Arroyo-López, F.N.; Barrio, E.; Querol, A. Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int. J. Food Microbiol. 2009, 134, 237–243. [Google Scholar] [CrossRef] [Green Version]
- Marques, W.L.; Raghavendran, V.; Stambuk, B.U.; Gombert, A.K. Sucrose and Saccharomyces cerevisiae: A relationship most sweet. FEMS Yeast Res. 2015, 16, fov107. [Google Scholar] [CrossRef] [Green Version]
- Alves-Araújo, C.; Pacheco, A.; Almeida, M.J.; Spencer-Martins, I.; Leão, C.; Sousa, M.J. Sugar utilization patterns and respiro-fermentative metabolism in the baker’s yeast Torulaspora delbrueckii. Microbiology 2007, 153, 898–904. [Google Scholar] [CrossRef] [Green Version]
- Diezmann, S.; Dietrich, F.S. Saccharomyces cerevisiae: Population Divergence and Resistance to Oxidative Stress in Clinical, Domesticated and Wild Isolates. PLoS ONE 2009, 4, e5317. [Google Scholar] [CrossRef] [Green Version]
- Kvitek, D.J.; Will, J.L.; Gasch, A.P. Variations in Stress Sensitivity and Genomic Expression in Diverse S. cerevisiae Isolates. PLoS Genet. 2008, 4, e1000223. [Google Scholar] [CrossRef] [Green Version]
- Gibson, B.R.; Lawrence, S.; LeClaire, J.P.R.; Powell, C.; Smart, K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef] [Green Version]
- Budroni, M.; Zara, S.; Zara, G.; Pirino, G.; Mannazzu, I. Peculiarities of flor strains adapted to Sardinian sherry-like wine ageing conditions. FEMS Yeast Res. 2005, 5, 951–958. [Google Scholar] [CrossRef] [Green Version]
- Capece, A.; Siesto, G.; Poeta, C.; Pietrafesa, R.; Romano, P. Indigenous yeast population from Georgian aged wines produced by traditional “Kakhetian” method. Food Microbiol. 2013, 36, 447–455. [Google Scholar] [CrossRef]
- Calhelha, R.C.; Andrade, J.V.; Ferreira, I.C.; Estevinho, L.M. Toxicity effects of fungicide residues on the wine-producing process. Food Microbiol. 2006, 23, 393–398. [Google Scholar] [CrossRef]
- Russo, P.; Berbegal, C.; De Ceglie, C.; Grieco, F.; Spano, G.; Capozzi, V. Pesticide Residues and Stuck Fermentation in Wine: New Evidences Indicate the Urgent Need of Tailored Regulations. Fermentation 2019, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Zhou, T.; Zhu, L.; Wang, X.; Wang, J.; Wang, J.; Du, Z.; Li, B. Effects of successive metalaxyl application on soil microorganisms and the residue dynamics. Ecol. Indic. 2019, 103, 194–201. [Google Scholar] [CrossRef]
- Francois, I.; Cammue, B.; Borgers, M.; Ausma, J.; Dispersyn, G.; Thevissen, K. Azoles: Mode of Antifungal Action and Resistance Development. Effect of Miconazole on Endogenous Reactive Oxygen Species Production in Candida albicans. Anti Infect. Agents Med. Chem. 2006, 5, 3–13. [Google Scholar] [CrossRef]
- Claus, H.; Mojsov, K. Enzymes for Wine Fermentation: Current and Perspective Applications. Fermentation 2018, 4, 52. [Google Scholar] [CrossRef] [Green Version]
- Andrade, R.P.; Oliveira, D.R.; Lopes, A.C.A.; de Abreu, L.R.; Duarte, W.F. Survival of Kluyveromyces lactis and Torulaspora delbrueckii to simulated gastrointestinal conditions and their use as single and mixed inoculum for cheese production. Food Res. Int. 2019, 125, 108620. [Google Scholar] [CrossRef] [PubMed]
- Borelli, B.M.; Ferreira, E.G.; Lacerda, I.C.; Franco, G.R.; Rosa, C.A. Yeast populations associated with the artisanal cheese produced in the region of Serra da Canastra, Brazil. World J. Microbiol. Biotechnol. 2006, 22, 1115–1119. [Google Scholar] [CrossRef]
- Huang, C.-W.; Walker, M.E.; Fedrizzi, B.; Gardner, R.C.; Jiranek, V. Hydrogen sulfide and its roles in Saccharomyces cerevisiae in a winemaking context. FEMS Yeast Res. 2017, 17, fox058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divol, B.; du Toit, M.; Duckitt, E. Surviving in the presence of sulphur dioxide: Strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 2012, 95, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Simonin, S.; Alexandre, H.; Nikolantonaki, M.; Coelho, C.; Tourdot-Maréchal, R. Inoculation of Torulaspora delbrueckii as a bio-protection agent in winemaking. Food Res. Int. 2018, 107, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Sangorrín, M.P.; Lopes, C.A.; Giraudo, M.R.; Caballero, A.C. Diversity and killer behaviour of indigenous yeasts isolated from the fermentation vat surfaces in four Patagonian wineries. Int. J. Food Microbiol. 2007, 119, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Velázquez, R.; Zamora, E.; Álvarez, M.L.; Hernández, L.M.; Ramírez, M. Effects of new Torulaspora delbrueckii killer yeasts on the must fermentation kinetics and aroma compounds of white table wine. Front. Microbiol. 2015, 6, 1222. [Google Scholar] [CrossRef] [Green Version]
- Ramírez, M.; Velázquez, R.; Maqueda, M.; López-Piñeiro, A.; Ribas, J.C. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 2015, 6, 983. [Google Scholar] [CrossRef] [Green Version]
- Franco-Duarte, R.; Bigey, F.; Carreto, L.; Mendes, I.; Dequin, S.; Santos, M.A.; Pais, C.; Schuller, D. Intrastrain genomic and phenotypic variability of the commercial Saccharomyces cerevisiae strain Zymaflore VL1 reveals microevolutionary adaptation to vineyard environments. FEMS Yeast Res. 2015, 15, fov063. [Google Scholar] [CrossRef] [Green Version]
- González, M.L.; Valero, E.; Chimeno, S.V.; Garrido-Fernández, A.; Rodríguez-Gómez, F.; Rojo, M.C.; Paolinelli, M.; Arroyo-López, F.N.; Combina, M.; Mercado, L.A. Growth response of Saccharomyces cerevisiae strains to stressors associated to the vine cycle. LWT 2022, 158, 113157. [Google Scholar] [CrossRef]
- Drumonde-Neves, J.; Franco-Duarte, R.; Vieira, E.; Mendes, I.; Lima, T.; Schuller, D.; Pais, C. Differentiation of Saccharomyces cerevisiae populations from vineyards of the Azores Archipelago: Geography vs Ecology. Food Microbiol. 2018, 74, 151–162. [Google Scholar] [CrossRef]
- Ramírez, M.; Velázquez, R. The Yeast Torulaspora delbrueckii: An Interesting But Difficult-To-Use Tool for Winemaking. Fermentation 2018, 4, 94. [Google Scholar] [CrossRef] [Green Version]
- Canonico, L.; Comitini, F.; Ciani, M. Torulaspora delbrueckii for secondary fermentation in sparkling wine production. Food Microbiol. 2018, 74, 100–106. [Google Scholar] [CrossRef]
- Catrileo, D.; Acuña-Fontecilla, A.; Godoy, L. Adaptive Laboratory Evolution of Native Torulaspora delbrueckii YCPUC10 With Enhanced Ethanol Resistance and Evaluation in Co-inoculated Fermentation. Front. Microbiol. 2020, 11, 3318. [Google Scholar] [CrossRef]
- Mores, S.; Vandenberghe, L.P.D.S.; Júnior, A.I.M.; de Carvalho, J.C.; de Mello, A.F.M.; Pandey, A.; Soccol, C.R. Citric acid bioproduction and downstream processing: Status, opportunities, and challenges. Bioresour. Technol. 2020, 320, 124426. [Google Scholar] [CrossRef]
- Liu, S.; Laaksonen, O.; Kortesniemi, M.; Kalpio, M.; Yang, B. Chemical composition of bilberry wine fermented with non-Saccharomyces yeasts (Torulaspora delbrueckii and Schizosaccharomyces pombe) and Saccharomyces cerevisiae in pure, sequential and mixed fermentations. Food Chem. 2018, 266, 262–274. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Bessa, D.; Gonçalves, F.; Martins, R.; Ferreira, A.C.S.; Schuller, D.; Sampaio, P.; Pais, C. Genomic and transcriptomic analysis of Saccharomyces cerevisiae isolates with focus in succinic acid production. FEMS Yeast Res. 2017, 17, fox057. [Google Scholar] [CrossRef] [Green Version]
- Puertas, B.; Jiménez, M.; Cantos-Villar, E.; Cantoral, J.; Rodríguez, M. Use of Torulaspora delbrueckii and Saccharomyces cerevisiaein semi-industrial sequential inoculation to improve quality of Palomino and Chardonnay wines in warm climates. J. Appl. Microbiol. 2017, 122, 733–746. [Google Scholar] [CrossRef]
- Contreras, A.; Hidalgo, C.; Henschke, P.A.; Chambers, P.J.; Curtin, C.; Varela, C. Evaluation of Non-Saccharomyces Yeasts for the Reduction of Alcohol Content in Wine. Appl. Environ. Microbiol. 2014, 80, 1670–1678. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.C.; Hynes, S.H.; Ingledew, W.M. Influence of medium buffering capacity on inhibition of Saccharomyces cerevisiae growth by acetic and lactic acids. Appl. Environ. Microbiol. 2002, 68, 1616–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pais, C.; Franco-Duarte, R.; Sampaio, P.; Wildner, J.; Carolas, A.; Figueira, D.; Ferreira, B. Production of Dicarboxylic Acid Platform Chemicals Using Yeasts: Focus on Succinic Acid. Biotransform. Agric. Waste By-Prod. 2016, 237–269. [Google Scholar] [CrossRef]
- Paraggio, M.; Fiore, C. Screening of Saccharomyces cerevisiae wine strains for the production of acetic acid. World J. Microbiol. Biotechnol. 2004, 20, 743–747. [Google Scholar] [CrossRef]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii–Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Benito, S. The Influence of Non-Saccharomyces Species on Wine Fermentation Quality Parameters. Fermentation 2019, 5, 54. [Google Scholar] [CrossRef] [Green Version]
- Likas, A.; Vlassis, N.; Verbeek, J.J. The global k-means clustering algorithm. Pattern Recognit. 2003, 36, 451–461. [Google Scholar] [CrossRef] [Green Version]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef] [Green Version]
- Franco-Duarte, R.; Mendes, I.; Umek, L.; Drumonde-Neves, J.; Zupan, B.; Schuller, D. Computational models reveal genotype-phenotype associations in Saccharomyces cerevisiae. Yeast 2014, 31, 265–277. [Google Scholar] [CrossRef] [Green Version]

| Phenotypic Test | Type of Medium | Phenotypic Classes | |||
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | ||
| 15 °C | Liquid (synthetic must) | 3 | 3 | 26 | 8 |
| 25 °C | Liquid (synthetic must) | 0 | 0 | 0 | 40 |
| 30 °C | Liquid (synthetic must) | 0 | 0 | 0 | 40 |
| 37 °C | Liquid (synthetic must) | 35 | 1 | 2 | 2 |
| Ethanol 5% (v/v) | Liquid (synthetic must) | 1 | 1 | 2 | 36 |
| Ethanol 10% (v/v) | Liquid (synthetic must) | 21 | 11 | 5 | 3 |
| Ethanol 14% (v/v) | Liquid (synthetic must) | 37 | 3 | 0 | 2 |
| Ethanol 18% (v/v) | Liquid (synthetic must) | 38 | 2 | 0 | 0 |
| Glucose (200 g/L) | Liquid (synthetic must) | 0 | 0 | 0 | 40 |
| Fructose (200 g/L) | Liquid (synthetic must) | 0 | 0 | 2 | 38 |
| Sucrose (200 g/L) | Liquid (synthetic must) | 0 | 0 | 1 | 39 |
| Maltose (200 g/L) | Liquid (synthetic must) | 0 | 8 | 25 | 7 |
| NaCl (1.5 M) | Liquid (synthetic must) | 2 | 8 | 26 | 4 |
| KCl (1 M) | Liquid (synthetic must) | 0 | 0 | 1 | 39 |
| H2O2 (2 mM) | Liquid (synthetic must) | 13 | 0 | 1 | 26 |
| CuSO4 (5 mM) | Liquid (synthetic must) | 19 | 11 | 8 | 2 |
| Fluconazole (0.5 mg/mL) | Liquid (synthetic must) | 0 | 0 | 9 | 31 |
| Myclobutanil (0.5 mg/mL) | Liquid (synthetic must) | 0 | 4 | 26 | 10 |
| Metalaxyl (0.5 mg/mL) | Liquid (synthetic must) | 0 | 0 | 0 | 40 |
| Tebuconazole (0.5 mg/mL) | Liquid (synthetic must) | 0 | 2 | 19 | 19 |
| β-glucosidase activity | Solid (Arbutin Agar) | 0 | 19 | 18 | 3 |
| Solid (Bile Esculin Agar) | 2 | 12 | 24 | 2 | |
| H2S production | Solid (BiGGY Agar) | 4 | 2 | 10 | 24 |
| Ethanol 12% (v/v) | Solid (MEA) | 3 | 15 | 13 | 9 |
| Ethanol 12% (v/v) + 0.25 mM SO2 | Solid (MEA) | 15 | 11 | 7 | 7 |
| Ethanol 12% (v/v) + 0.5 mM SO2 | Solid (MEA) | 18 | 10 | 5 | 7 |
| Ethanol 12% (v/v) + 1.0 mM SO2 | Solid (MEA) | 23 | 8 | 5 | 4 |
| Ethanol 12% (v/v) + 1.5 mM SO2 | Solid (MEA) | 28 | 3 | 6 | 3 |
| Killer activity—killer phenotype | Solid (YPD-MB Agar) | 39 | 1 | - | - |
| Killer activity—sensitive phenotype | Solid (YPD-MB Agar) | 37 | 3 | - | - |
| Freeze resistance | Liquid (LF) | 7 | 6 | 10 | 17 |
| Predicted Technological Group | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arboreal/Soil | Bread | Clinical | Food | Other Beverages | Unknown | Water | Wine | Total | ||
| Real technological group | Arboreal/Soil | 6 | 0 | 0 | 0 | 1 | 0 | 0 | 4 | 11 |
| Bread | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | |
| Clinical | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| Food | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 5 | |
| Other beverages | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | |
| Unknown | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | |
| Water | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 | |
| Wine | 4 | 0 | 0 | 0 | 0 | 0 | 1 | 6 | 11 | |
| Total | 11 | 5 | 0 | 3 | 2 | 1 | 3 | 15 | 40 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Sousa, F.; Fernandes, T.; Pereira, F.; Rodrigues, D.; Rito, T.; Camarasa, C.; Franco-Duarte, R.; Sousa, M.J. Torulaspora delbrueckii Phenotypic and Metabolic Profiling towards Its Biotechnological Exploitation. J. Fungi 2022, 8, 569. https://doi.org/10.3390/jof8060569
Silva-Sousa F, Fernandes T, Pereira F, Rodrigues D, Rito T, Camarasa C, Franco-Duarte R, Sousa MJ. Torulaspora delbrueckii Phenotypic and Metabolic Profiling towards Its Biotechnological Exploitation. Journal of Fungi. 2022; 8(6):569. https://doi.org/10.3390/jof8060569
Chicago/Turabian StyleSilva-Sousa, Flávia, Ticiana Fernandes, Fábio Pereira, Diana Rodrigues, Teresa Rito, Carole Camarasa, Ricardo Franco-Duarte, and Maria João Sousa. 2022. "Torulaspora delbrueckii Phenotypic and Metabolic Profiling towards Its Biotechnological Exploitation" Journal of Fungi 8, no. 6: 569. https://doi.org/10.3390/jof8060569







