The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
2.2. Molecular Identification
2.3. Antifungal Susceptibility Assay
2.4. Biofilm Formation
2.5. Antifungal Susceptibility of Biofilm-Forming Cells
2.6. Production of Biologically Active Extracellular Molecules
2.7. Statistics
3. Results and Discussion
3.1. Origin and Proper Identification of the C. haemulonii Clinical Isolates
3.2. Antifungal Susceptibility Assay
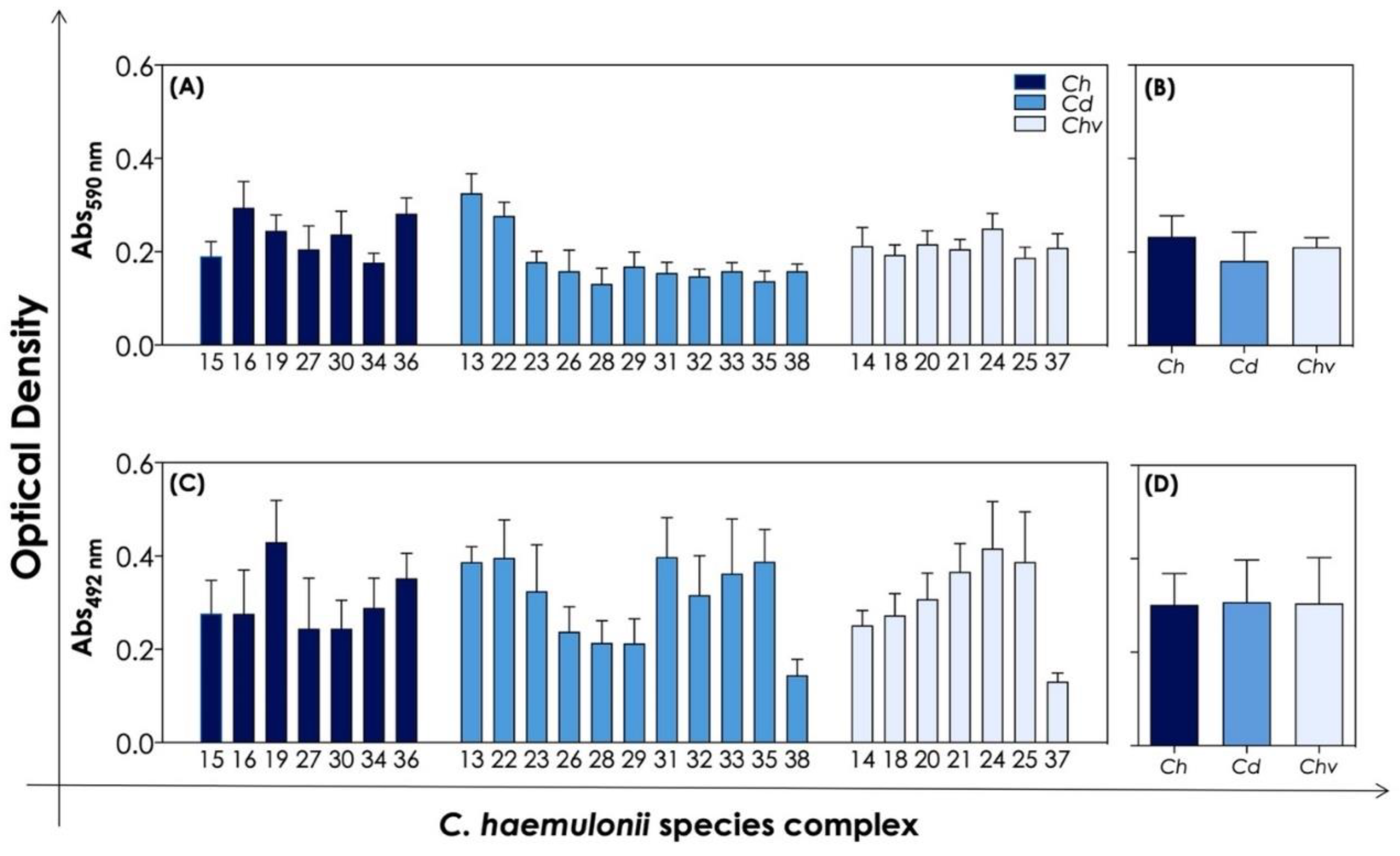
3.3. Biofilm Formation and Antifungal Susceptibility of Biofilm-Forming Cells
3.4. Production of Extracellular Biologically Active Molecules Associated with Fungal Virulence
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cendejas-Bueno, E.; Kolecka, A.; Alastruey-Izquierdo, A.; Theelen, B.; Groenewald, M.; Kostrzewa, M.; Cuenca-Estrella, M.; Gomez-Lopez, A.; Boekhout, T. Reclassification of the Candida haemulonii Complex as Candida haemulonii (C. haemulonii Group I), C. duobushaemulonii Sp. Nov. (C. haemulonii Group II), and C. haemulonii var. vulnera var. Nov.: Three Multiresistant Human Pathogenic Yeasts. J. Clin. Microbiol. 2012, 50, 3641–3651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.N.; Mello, T.P.; Ramos, L.S.; Branquinha, M.H.; Santos, A.L.S. New and Promising Chemotherapeutics for Emerging Infections Involving Drug-Resistant Non-albicans Candida Species. Curr. Top. Med. Chem. 2019, 19, 2527–2553. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.S.; Figueiredo-Carvalho, M.H.; Barbedo, L.S.; Ziccardi, M.; Chaves, A.L.; Zancope-Oliveira, R.M.; Pinto, M.R.; Sgarbi, D.B.; Dornelas-Ribeiro, M.; Branquinha, M.H.; et al. Candida haemulonii Complex: Species Identification and Antifungal Susceptibility Profiles of Clinical Isolates from Brazil. J. Antimicrob. Chemother. 2015, 70, 111–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, L.N.; Oliveira, S.S.C.; Magalhães, L.B.; Andrade Neto, V.V.; Torres-Santos, E.C.; Carvalho, M.D.C.; Pereira, M.D.; Branquinha, M.H.; Santos, A.L.S. Unmasking the Amphotericin B Resistance Mechanisms in Candida haemulonii Species Complex. ACS Infect. Dis. 2020, 6, 1273–1282. [Google Scholar] [CrossRef]
- Silva, L.N.; Ramos, L.S.; Oliveira, S.S.C.; Magalhães, L.B.; Squizani, E.D.; Kmetzsch, L.; Vainstein, M.H.; Branquinha, M.H.; Santos, A.L.S. Insights into the Multi-Azole Resistance Profile in Candida haemulonii Species Complex. J. Fungi 2020, 6, 215. [Google Scholar] [CrossRef]
- Ramos, L.S.; Mello, T.P.; Branquinha, M.H.; Santos, A.L.S. Biofilm Formed by Candida haemulonii Species Complex: Structural Analysis and Extracellular Matrix Composition. J. Fungi 2020, 6, 46. [Google Scholar] [CrossRef] [Green Version]
- Ramos, L.S.; Branquinha, M.H.; Santos, A.L.S. Different Classes of Hydrolytic Enzymes Produced by Multidrug-Resistant Yeasts Comprising the Candida haemulonii Complex. Med. Mycol. 2017, 55, 228–232. [Google Scholar] [CrossRef] [Green Version]
- Souto, X.M.; Ramos, L.S.; Branquinha, M.H.; Santos, A.L.S. Identification of Cell-Associated and Secreted Serine-Type Peptidases in Multidrug-Resistant Emergent Pathogens Belonging to the Candida haemulonii Complex. Folia Microbiol. (Praha) 2019, 64, 245–255. [Google Scholar] [CrossRef]
- Deng, Y.; Li, S.; Bing, J.; Liao, W.; Tao, L. Phenotypic Switching and Filamentation in Candida haemulonii, an Emerging Opportunistic Pathogen of Humans. Microbiol. Spectrum 2021, 9, E0077921. [Google Scholar] [CrossRef]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard; Clsi Document M27a3, 3rd ed.; CLSI: Wayne, PA, USA, 2008; Volume 28, p. 40. [Google Scholar]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Third Informational Supplement, CLSI Document M27-S3; CLSI: Wayne, PA, USA, 2008; 28p. [Google Scholar]
- Almeida, J.N., Jr.; Motta, A.L.; Rossi, F.; Abdala, E.; Pierrotti, L.C.; Kono, A.S.; Diz Mdel, P.; Benard, G.; Del Negro, G.M. First Report of a Clinical Isolate of Candida haemulonii in Brazil. Clinics (Sao Paulo) 2012, 67, 1229–1231. [Google Scholar] [CrossRef]
- Peeters, E.; Nelis, H.J.; Coenye, T. Comparison of Multiple Methods for Quantification of Microbial Biofilms Grown in Microtiter Plates. J. Microbiol. Meth. 2008, 72, 157–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, N.Y.; Kang, S.Y.; Kim, K.J. Artemisia Princeps Inhibits Biofilm Formation and Virulence-Factor Expression of Antibiotic-Resistant Bacteria. Biomed Res. Int. 2015, 2015, 239519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melo, A.S.; Bizerra, F.C.; Freymuller, E.; Arthington-Skaggs, B.A.; Colombo, A.L. Biofilm Production and Evaluation of Antifungal Susceptibility amongst Clinical Candida Spp. Isolates, Including Strains of the Candida parapsilosis Complex. Med. Mycol. 2011, 49, 253–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romera, D.; Aguilera-Correa, J.J.; Gadea, I.; Vinuela-Sandoval, L.; Garcia-Rodriguez, J.; Esteban, J. Candida auris: A Comparison between Planktonic and Biofilm Susceptibility to Antifungal Drugs. J. Med. Microbiol. 2019, 68, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Price, M.F.; Wilkinson, I.D.; Gentry, L.O. Plate Method for Detection of Phospholipase Activity in Candida albicans. Sabouraudia 1982, 20, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Ruchel, R.; Tegeler, R.; Trost, M. A Comparison of Secretory Proteinases from Different Strains of Candida albicans. Sabouraudia 1982, 20, 233–244. [Google Scholar] [CrossRef]
- Ziccardi, M.; Souza, L.O.; Gandra, R.M.; Galdino, A.C.; Baptista, A.R.; Nunes, A.P.; Ribeiro, M.A.; Branquinha, M.H.; Santos, A.L.S. Candida parapsilosis (sensu lato) Isolated from Hospitals Located in the Southeast of Brazil: Species Distribution, Antifungal Susceptibility and Virulence Attributes. Int. J. Med. Microbiol. 2015, 305, 848–859. [Google Scholar] [CrossRef]
- Aktas, E.; Yigit, N.; Ayyildiz, A. Esterase Activity in Various Candida Species. J. Int. Med. Res. 2002, 30, 322–324. [Google Scholar] [CrossRef]
- Tsang, P.W. Differential Phytate Utilization in Candida Species. Mycopathologia 2011, 172, 473–479. [Google Scholar] [CrossRef]
- Luo, G.; Samaranayake, L.P.; Yau, J.Y. Candida Species Exhibit Differential in vitro Hemolytic Activities. J. Clin. Microbiol. 2001, 39, 2971–2974. [Google Scholar] [CrossRef] [Green Version]
- Lakshmanan, V.; Shantharaj, D.; Li, G.; Seyfferth, A.L.; Janine Sherrier, D.; Bais, H.P. A Natural Rice Rhizospheric Bacterium Abates Arsenic Accumulation in Rice (Oryza sativa L.). Planta 2015, 242, 1037–1050. [Google Scholar] [CrossRef] [PubMed]
- Ambaraghassi, G.; Dufresne, P.J.; Dufresne, S.F.; Vallières, É.; Muñoz, J.F.; Cuomo, C.A.; Berkow, E.L.; Lockhart, S.R.; Luong, M.L. Identification of Candida auris by Use of the Updated Vitek 2 Yeast Identification System, Version 8.01: A Multilaboratory Evaluation Study. J. Clin. Microbiol. 2019, 57, e00884-19. [Google Scholar] [CrossRef] [PubMed]
- García-Martos, P.; Domínguez, I.; Marín, P.; García-Agudo, R.; Aoufi, S.; Mira, J. Antifungal Susceptibility of Emerging Yeast Pathogens. Enferm. Infecc. Microbiol. Clin. 2001, 19, 249–256. [Google Scholar] [CrossRef]
- Rodero, L.; Cuenca-Estrella, M.; Córdoba, S.; Cahn, P.; Davel, G.; Kaufman, S.; Guelfand, L.; Rodríguez-Tudela, J.L. Transient Fungemia Caused by an Amphotericin B-Resistant Isolate of Candida haemulonii. J. Clin. Microbiol. 2002, 40, 2266–2269. [Google Scholar] [CrossRef] [Green Version]
- Khan, Z.U.; Al-Sweih, N.A.; Ahmad, S.; Al-Kazemi, N.; Khan, S.; Joseph, L.; Chandy, R. Outbreak Of Fungemia Among Neonates Caused By Candida haemulonii Resistant to Amphotericin B, Itraconazole, and Fluconazole. J. Clin. Microbiol. 2007, 45, 2025–2027. [Google Scholar] [CrossRef] [Green Version]
- Pfaller, M.A.; Diekema, D.J.; Gibbs, D.L.; Newell, V.A.; Ellis, D.; Tullio, V.; Rodloff, A.; Fu, W.; Ling, T.A. Results from the Artemis Disk Global Antifungal Surveillance Study, 1997 to 2007: A 10.5-Year Analysis of Susceptibilities of Candida Species to Fluconazole and Voriconazole as Determined by CLSI Standardized Disk Diffusion. J. Clin. Microbiol. 2010, 48, 1366–1377. [Google Scholar] [CrossRef] [Green Version]
- Ruan, S.Y.; Kuo, Y.W.; Huang, C.T.; Hsiue, H.C.; Hsueh, P.R. Infections Due to Candida haemulonii: Species Identification, Antifungal Susceptibility and Outcomes. Int. J. Antimicrob. Agents 2010, 35, 85–88. [Google Scholar] [CrossRef]
- Gomez-Lopez, A.; Buitrago, M.J.; Rodriguez-Tudela, J.L.; Cuenca-Estrella, M. In Vitro Antifungal Susceptibility Pattern and Ergosterol Content in Clinical Yeast Strains. Rev. Iberoam. Micol. 2011, 28, 100–103. [Google Scholar] [CrossRef]
- Kim, S.; Ko, K.S.; Moon, S.Y.; Lee, M.S.; Lee, M.Y.; Son, J.S. Catheter-Related Candidemia Caused by Candida haemulonii in a Patient in Long-Term Hospital Care. J. Korean Med. Sci. 2011, 26, 297–300. [Google Scholar] [CrossRef] [Green Version]
- Oberoi, J.K.; Wattal, C.; Goel, N.; Raveendran, R.; Datta, S.; Prasad, K. Non-albicans Candida Species in Blood Stream Infections in a Tertiary Care Hospital at New Delhi, India. Indian J. Med. Res. 2012, 136, 997–1003. [Google Scholar]
- Hou, X.; Xiao, M.; Chen, S.C.; Wang, H.; Cheng, J.W.; Chen, X.X.; Xu, Z.P.; Fan, X.; Kong, F.; Xu, Y.C. Identification and Antifungal Susceptibility Profiles of Candida haemulonii Species Complex Clinical Isolates from a Multicenter Study in China. J. Clin. Microbiol. 2016, 54, 2676–2680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pagani, D.M.; Heidrich, D.; Paulino, G.V.; De Oliveira Alves, K.; Dalbem, P.T.; De Oliveira, C.F.; Andrade, Z.M.; Silva, C.; Correia, M.D.; Scroferneker, M.L.; et al. Susceptibility to Antifungal Agents and Enzymatic Activity of Candida haemulonii and Cutaneotrichosporon dermatis Isolated from Soft Corals on the Brazilian Reefs. Arch. Microbiol. 2016, 198, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Sotto, A.; Picard, E.; Lachaud, L.; Bourgeois, N. A Case of Candida haemulonii Osteitis: Clinical Features, Biochemical Characteristics, and Antifungal Resistance Profile. Clin. Microbiol. Infect. 2011, 17, 1068–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Almeida, J.N., Jr.; Assy, J.G.; Levin, A.S.; Del Negro, G.M.; Giudice, M.C.; Tringoni, M.P.; Thomaz, D.Y.; Motta, A.L.; Abdala, E.; Pierroti, L.C.; et al. Candida haemulonii Complex Species, Brazil, January 2010–March 2015. Emerg. Infect. Dis. 2016, 22, 561–563. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Prakash, A.; Singh, A.; Kumar, H.; Hagen, F.; Meis, J.F.; Chowdhary, A. Candida haemulonii Species Complex: An Emerging Species in India and Its Genetic Diversity Assessed with Multilocus Sequence and Amplified Fragment-Length Polymorphism Analyses. Emerg. Microbes Infect. 2016, 5, E49. [Google Scholar] [CrossRef] [Green Version]
- Muro, M.D.; Motta Fde, A.; Burger, M.; Melo, A.S.; Dalla-Costa, L.M. Echinocandin Resistance in Two Candida haemulonii Isolates from Pediatric Patients. J. Clin. Microbiol. 2012, 50, 3783–3785. [Google Scholar] [CrossRef] [Green Version]
- Lima, S.L.; Francisco, E.C.; De Almeida Júnior, J.N.; Santos, D.W.C.L.; Carlesse, F.; Queiroz-Telles, F.; Melo, A.S.A.; Colombo, A.L. Increasing Prevalence of Multidrug-Resistant Candida haemulonii Species Complex Among All Yeast Cultures Collected by a Reference Laboratory over the Past 11 Years. J. Fungi 2020, 6, 110. [Google Scholar] [CrossRef]
- Oh, B.J.; Shin, J.H.; Kim, M.N.; Sung, H.; Lee, K.; Joo, M.Y.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Biofilm Formation and Genotyping of Candida haemulonii, Candida pseudohaemulonii, and a Proposed New Species (Candida auris) Isolates from Korea. Med. Mycol. 2011, 49, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Ramos, L.S.; Oliveira, S.S.C.; Souto, X.M.; Branquinha, M.H.; Santos, A.L.S. Planktonic Growth and Biofilm Formation Profiles in Candida haemulonii Species Complex. Med. Mycol. 2017, 55, 785–789. [Google Scholar] [CrossRef] [Green Version]
- Lima, S.L.; Rossato, L.; Salles De Azevedo Melo, A. Evaluation of the Potential Virulence of Candida haemulonii Species Complex and Candida auris Isolates in Caenorhabditis elegans as an in vivo Model and Correlation to Their Biofilm Production Capacity. Microb. Pathog. 2020, 148, 104461. [Google Scholar] [CrossRef]
- Santos, A.L.S.; Mello, T.P.; Ramos, L.S.; Branquinha, M.H. Biofilm: A Robust and Efficient Barrier to Antifungal Chemotherapy. J. Antimicrob. 2015, 1, E101. [Google Scholar] [CrossRef]
- Mello, T.P.; Ramos, L.S.; Braga-Silva, L.A.; Branquinha, M.H.; Santos, A.L.S. Fungal Biofilm—A Real Obstacle against an Efficient Therapy: Lessons from Candida. Curr. Top. Med. Chem. 2017, 17, 1987–2004. [Google Scholar] [CrossRef]
- Ramos, L.S.; Silva, L.N.; Branquinha, M.H.; Santos, A.L.S. Susceptibility of the Candida haemulonii Complex to Echinocandins: Focus on both Planktonic and Biofilm Life Styles and a Literature Review. J. Fungi 2020, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; George, T.; Chandra, J.; Mukherjee, P.K.; Ghannoum, M.A. Antifungal Susceptibility of Candida Biofilms: Unique Efficacy of Amphotericin B Lipid Formulations and Echinocandins. Antimicrob. Agents Chemother. 2002, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.W.; Shin, J.H.; Jung, S.I.; Park, K.H.; Cho, D.; Kee, S.J.; Shin, M.G.; Suh, S.P.; Ryang, D.W. Species-Specific Differences in the Susceptibilities of Biofilms Formed by Candida Bloodstream Isolates to Echinocandin Antifungals. Antimicrob. Agents Chemother. 2007, 51, 1520–1523. [Google Scholar] [CrossRef] [Green Version]
- Schaller, M.; Borelli, C.; Korting, H.C.; Hube, B. Hydrolytic Enzymes as Virulence Factors of Candida albicans. Mycoses 2005, 48, 365–377. [Google Scholar] [CrossRef]
- Naglik, J.R.; Challacombe, S.J.; Hube, B. Candida albicans Secreted Aspartyl Proteinases in Virulence and Pathogenesis. Microbiol. Mol. Biol. Rev. 2003, 67, 400–428. [Google Scholar] [CrossRef] [Green Version]
- Ramos, L.S.; Oliveira, S.S.C.; Braga-Silva, L.A.; Branquinha, M.H.; Santos, A.L.S. Secreted Aspartyl Peptidases by the Emerging, Opportunistic and Multidrug-Resistant Fungal Pathogens Comprising the Candida haemulonii Complex. Fungal Biol. 2020, 124, 700–707. [Google Scholar] [CrossRef]
- Parra-Ortega, B.; Cruz-Torres, H.; Villa-Tanaca, L.; Hernandez-Rodriguez, C. Phylogeny and Evolution of the Aspartyl Protease Family from Clinically Relevant Candida Species. Mem. Inst. Oswaldo Cruz 2009, 104, 505–512. [Google Scholar] [CrossRef] [Green Version]
- Portela, M.B.; Souza, I.P.; Abreu, C.M.; Bertolini, M.; Holandino, C.; Alviano, C.S.; Santos, A.L.S.; Soares, R.M.A. Effect of Serine-Type Protease of Candida spp. Isolated from Linear Gingival Erythema of HIV-Positive Children: Critical Factors in the Colonization. J. Oral Pathol. Med. 2010, 39, 753–760. [Google Scholar] [CrossRef]
- Santos, A.L.S.; De Carvalho, I.M.; Da Silva, B.A.; Portela, M.B.; Alviano, C.S.; Soares, R.M.A. Secretion of Serine Peptidase by a Clinical Strain of Candida albicans: Influence of Growth Conditions and Cleavage of Human Serum Proteins and Extracellular Matrix Components. FEMS Immunol. Med. Microbiol. 2006, 46, 209–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, A.L.S.; Soares, R.M.A. Candida guilliermondii Isolated from HIV-Infected Human Secretes a 50 kDa Serine Proteinase that Cleaves a Broad Spectrum of Proteinaceous Substrates. Fems. Immunol. Med. Microbiol. 2005, 43, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Souto, X.M.; Branquinha, M.H.; Santos, A.L.S. Chymotrypsin- and Trypsin-Like Activities Secreted by the Multidrug-Resistant Yeasts Forming the Candida haemulonii Complex. An. Acad. Bras. Cienc. 2019, 91, E20180735. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Do, E.; Jung, W.H. Lipolytic Enzymes Involved in the Virulence of Human Pathogenic Fungi. Mycobiology 2013, 41, 67–72. [Google Scholar] [CrossRef] [Green Version]
- Mroczyńska, M.; Brillowska-Dąbrowska, A. Virulence of Clinical Candida Isolates. Pathogens 2021, 10, 466. [Google Scholar] [CrossRef]
- Niewerth, M.; Korting, H.C. Phospholipases of Candida albicans. Mycoses 2001, 44, 361–367. [Google Scholar] [CrossRef]
- Costa, C.R.; Passos, X.S.; E Souza, L.K.; Lucena Pde, A.; Fernandes Ode, F.; Silva Mdo, R. Differences in Exoenzyme Production and Adherence Ability of Candida spp. Isolates from Catheter, Blood and Oral Cavity. Rev. Inst. Med. Trop. Sao Paulo 2010, 52, 139–143. [Google Scholar] [CrossRef]
- Treviño-Rangel Rde, J.; González, J.G.; González, G.M. Aspartyl Proteinase, Phospholipase, Esterase and Hemolysin Activities of Clinical Isolates of the Candida parapsilosis Species Complex. Med. Mycol. 2013, 51, 331–335. [Google Scholar] [CrossRef] [Green Version]
- Abi-chacra, E.A.; Souza, L.O.; Cruz, L.P.; Braga-Silva, L.A.; Goncalves, D.S.; Sodre, C.L.; Ribeiro, M.D.; Seabra, S.H.; Figueiredo-Carvalho, M.H.; Barbedo, L.S.; et al. Phenotypical Properties Associated with Virulence from Clinical Isolates Belonging to the Candida parapsilosis Complex. FEMS Yeast Res. 2013, 13, 831–848. [Google Scholar] [CrossRef] [Green Version]
- Fatahinia, M.; Poormohamadi, F.; Zarei Mahmoudabadi, A. Comparative Study of Esterase and Hemolytic Activities in Clinically Important Candida Species, Isolated from Oral Cavity of Diabetic and Non-Diabetic Individuals. Jundishapur J. Microbiol. 2015, 8, E20893. [Google Scholar] [CrossRef] [Green Version]
- Sasani, E.; Yadegari, M.H.; Khodavaisy, S.; Rezaie, S.; Salehi, M.; Getso, M.I. Virulence Factors and Azole-Resistant Mechanism of Candida tropicalis Isolated from Candidemia. Mycopathologia 2021, 186, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Kauffman, S.; Reynolds, T.B. Candida albicans Uses Multiple Mechanisms to Acquire the Essential Metabolite Inositol During Infection. Infect. Immun. 2008, 76, 2793–2801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, V.K.; Foong, K.J.; Maha, A.; Rusliza, B.; Norhafizah, M.; Ng, K.P.; Chong, P.P. Candida albicans Isolates from a Malaysian Hospital Exhibit More Potent Phospholipase and Haemolysin Activities than Non-albicans Candida Isolates. Trop. Biomed. 2013, 30, 654–662. [Google Scholar] [PubMed]
- Riceto, E.B.; Menezes Rde, P.; Penatti, M.P.; Pedroso Rdos, S. Enzymatic and Hemolytic Activity in Different Candida Species. Rev. Iberoam. Micol. 2015, 32, 79–82. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.S.; Wilson, D.; Hube, B. Candida albicans Iron Acquisition within the Host. FEMS Yeast Res. 2009, 9, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
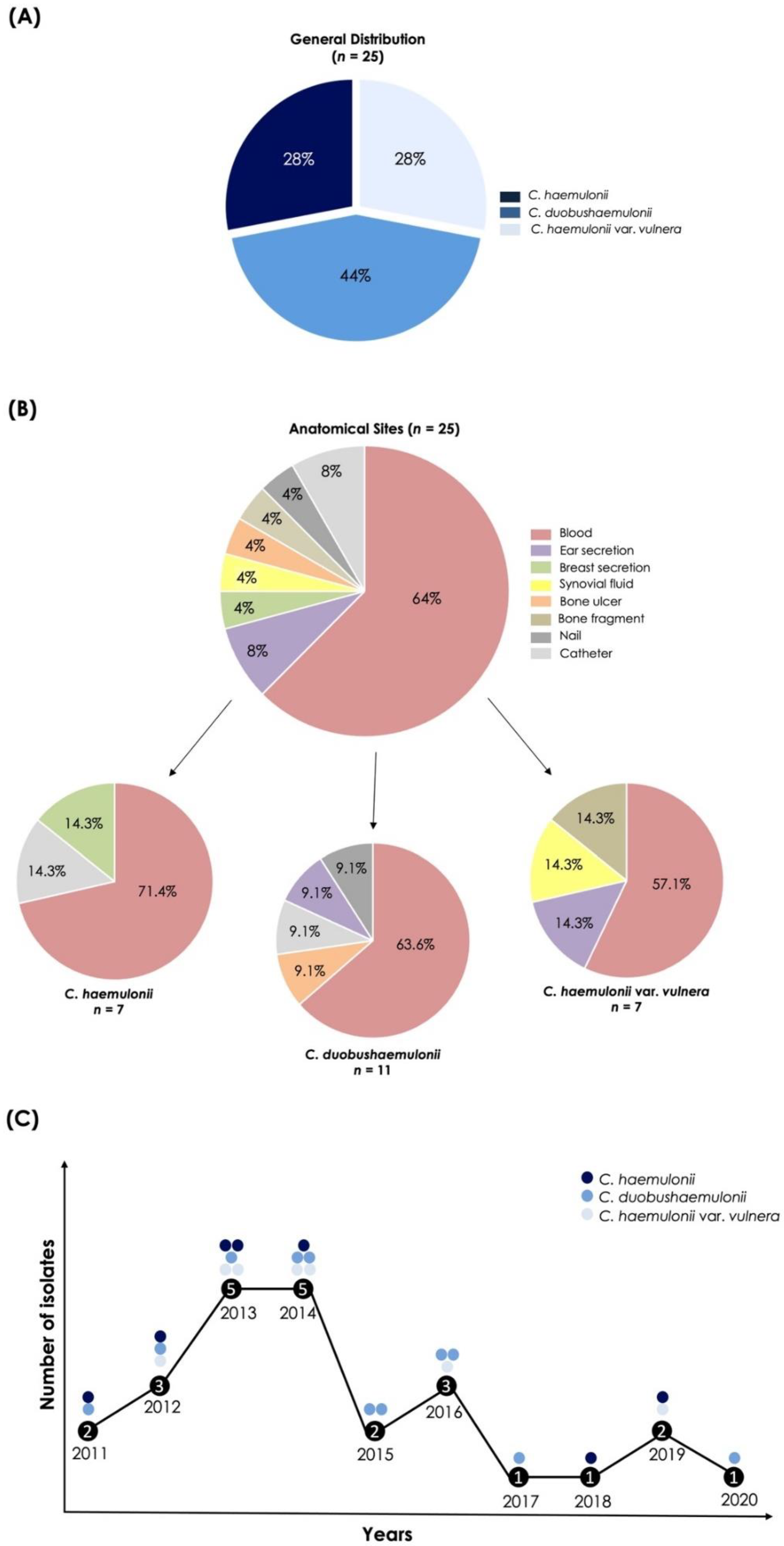

| Code | Source of Isolates | Isolation Year | Identification by Vitek® 2 YST System (Probability of Identity) | Molecular Identification (ITS1-5.8S-ITS2 Gene) | GenBank Accession Number |
|---|---|---|---|---|---|
| LIPCh13 | Blood | 2012 | C. haemulonii (90%) | C. duobushaemulonii | OM575048 |
| LIPCh14 | Blood | 2012 | C. haemulonii (97%) | C. haemulonii var. vulnera | OM575049 |
| LIPCh15 | Blood | 2012 | C. haemulonii/Kodamaea ohmeri | C. haemulonii | OM575050 |
| LIPCh16 | Catheter tip | 2013 | C. haemulonii (97%) | C. haemulonii | OM575051 |
| LIPCh18 | Blood | 2014 | C. haemulonii (99%) | C. haemulonii var. vulnera | OM575052 |
| LIPCh19 | Blood | 2011 | C. haemulonii/Kodamaea ohmeri | C. haemulonii | OM575053 |
| LIPCh20 | Blood | 2013 | C. haemulonii/Kodamaea ohmeri | C. haemulonii var. vulnera | OM575054 |
| LIPCh21 | Ear secretion | 2016 | C. haemulonii/Kodamaea ohmeri | C. haemulonii var. vulnera | OM575055 |
| LIPCh22 | Blood | 2015 | C. auris/C. duobushaemulonii | C. duobushaemulonii | OM575056 |
| LIPCh23 | Blood | 2011 | C. duobushaemulonii (93%) | C. duobushaemulonii | OM575057 |
| LIPCh24 | Blood | 2014 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii var. vulnera | OM575058 |
| LIPCh25 | Synovial fluid | 2013 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii var. vulnera | OM575059 |
| LIPCh26 | Blood | 2016 | C. duobushaemulonii (92%) | C. duobushaemulonii | OM575060 |
| LIPCh27 | Blood | 2018 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii | OM575061 |
| LIPCh28 | Bone ulcer | 2017 | C. duobushaemulonii (92%) | C. duobushaemulonii | OM575062 |
| LIPCh29 | Catheter tip | 2016 | C. duobushaemulonii (90%) | C. duobushaemulonii | OM575063 |
| LIPCh30 | Breast secretion | 2014 | C. haemulonii (95%) | C. haemulonii | OM575064 |
| LIPCh31 | Ear secretion | 2014 | C. duobushaemulonii (90%) | C. duobushaemulonii | OM575065 |
| LIPCh32 | Blood | 2015 | C. haemulonii/C. neoformans/C. duobushaemulonii | C. duobushaemulonii | OM575066 |
| LIPCh33 | Blood | 2013 | C. duobushaemulonii (90%) | C. duobushaemulonii | OM575067 |
| LIPCh34 | Blood | 2013 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii | OM575068 |
| LIPCh35 | Blood | 2014 | C. duobushaemulonii (90%) | C. duobushaemulonii | OM575069 |
| LIPCh36 | Blood | 2019 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii | OM575070 |
| LIPCh37 | Bone fragment | 2019 | C. haemulonii/C. haemulonii var. vulnera | C. haemulonii var. vulnera | OM575071 |
| LIPCh38 | Nail | 2020 | C. duobushaemulonii (97%) | C. duobushaemulonii | OM575072 |
| Clinical Isolates | MIC (mg/L) a | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLC b | ITC | VRC | PSC | AMB | TRB | 5-FC | CSF | MCF | ANF | |
| Ch | ||||||||||
| LIPCh15 | >64 | >16 | >16 | >16 | >16 | 8 | 0.5 | 1 | 0.125 | 0.25 |
| LIPCh16 | >64 | >16 | >16 | >16 | 4 | 8 | 0.5 | 1 | 0.06 | 0.125 |
| LIPCh19 | >64 | >16 | >16 | >16 | 2 | 16 | 0.25 | 1 | 0.125 | 0.25 |
| LIPCh27 | >64 | >16 | >16 | >16 | 2 | >16 | 0.5 | 1 | 0.125 | 0.125 |
| LIPCh30 | >64 | >16 | >16 | >16 | 8 | >16 | 0.5 | 1 | 0.125 | 0.125 |
| LIPCh34 | >64 | >16 | >16 | >16 | 4 | >16 | 0.125 | 1 | 0.06 | 0.06 |
| LIPCh36 | 64 | 0.25 | 1 | 0.25 | 16 | >16 | 0.25 | 0.25 | <0.015 | 0.125 |
| Cd | ||||||||||
| LIPCh13 | 8 | 0.125 | 0.12 | 0.25 | 2 | 1 | >64 | >8 | 0.125 | 2 |
| LIPCh22 | >64 | 8 | >16 | >16 | 16 | 16 | 0.5 | 0.5 | 0.06 | 0.125 |
| LIPCh23 | 64 | 8 | 16 | 16 | >16 | >16 | 0.25 | 0.5 | 0.125 | 0.06 |
| LIPCh26 | >64 | >16 | >16 | >16 | >16 | >16 | 1 | 0.5 | 0.06 | 0.06 |
| LIPCh28 | >64 | >16 | 8 | >16 | 4 | >16 | 0.25 | 0.125 | 0.06 | 0.06 |
| LIPCh29 | 64 | >16 | 16 | >16 | >16 | >16 | 0.25 | 0.5 | 0.125 | 0.06 |
| LIPCh31 | >64 | 0.5 | 0.5 | 0.125 | 4 | >16 | 1 | 0.125 | 0.06 | 0.06 |
| LIPCh32 | >64 | >16 | >16 | >16 | >16 | >16 | 0.125 | 0.5 | 0.125 | 0.06 |
| LIPCh33 | 64 | >16 | >16 | >16 | 4 | >16 | 0.125 | 0.25 | 0.125 | 0.06 |
| LIPCh35 | 32 | 0.5 | 0.5 | 0.06 | 4 | 8 | 0.5 | 0.25 | 0.125 | 0.06 |
| LIPCh38 | 32 | 0.5 | >16 | >16 | >16 | >16 | >64 | 0.25 | 0.06 | 0.125 |
| Chv | ||||||||||
| LIPCh14 | >64 | >16 | >16 | >16 | 2 | 8 | 0.25 | 0.25 | 0.06 | 0.125 |
| LIPCh18 | >64 | >16 | >16 | >16 | 4 | 8 | 0.25 | 1 | 0.125 | 0.125 |
| LIPCh20 | >64 | >16 | >16 | >16 | 8 | 16 | 0.25 | 1 | 0.125 | 0.25 |
| LIPCh21 | >64 | >16 | >16 | >16 | 4 | 8 | 0.25 | 1 | 0.125 | 0.06 |
| LIPCh24 | 64 | 0.25 | 0.25 | 0.125 | 16 | >16 | 0.25 | 1 | 0.125 | 0.06 |
| LIPCh25 | >64 | >16 | >16 | >16 | 4 | >16 | 0.25 | 0.5 | 0.06 | 0.06 |
| LIPCh37 | >64 | >16 | >16 | >16 | 8 | >16 | <0.125 | 0.5 | 0.06 | 0.06 |
| Antifungals | Ch (n = 7) | Cd (n = 11) | Chv (n = 7) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MIC Range | MIC50 | MIC90 | MIC Range | MIC50 | MIC90 | MIC Range | MIC50 | MIC90 | |
| AMB | 2–>16 | 4 | 16 | 2–>16 | 16 | >16 | 2–16 | 4 | 8 |
| FLC | 64–>64 | >64 | >64 | 8–>64 | 64 | >64 | 64–>64 | >64 | >64 |
| ITC | 0.25–>16 | >16 | >16 | 0.125–>16 | 8 | >16 | 0.25–>16 | >16 | >16 |
| VRC | 1–>16 | >16 | >16 | 0.125–>16 | 16 | >16 | 0.25–>16 | >16 | >16 |
| PSC | 0.25–>16 | >16 | >16 | 0.06–>16 | >16 | >16 | 0.125–>16 | >16 | >16 |
| CSF | 0.25–1 | 1 | 1 | 0.125–>8 | 0.5 | 0.5 | 0.25–1 | 1 | 1 |
| MCF | <0.015–0.125 | 0.125 | 0.125 | 0.06–0.125 | 0.125 | 0.125 | 0.06–0.125 | 0.125 | 0.125 |
| ANF | 0.06–0.25 | 0.125 | 0.25 | 0.06–2 | 0.06 | 0.125 | 0.06–0.25 | 0.06 | 0.125 |
| 5-FC | 0.125–0.5 | 0.5 | 0.5 | 0.125–>64 | 0.5 | >64 | <0.125–0.25 | 0.25 | 0.25 |
| TRB | 8–>16 | >16 | >16 | 1–>64 | >16 | >16 | 8–>16 | 16 | >16 |
| Species | % of Resistant Strains | |||||||
|---|---|---|---|---|---|---|---|---|
| FLC CLSI/CDC | ITC CLSI/CDC | VRC CLSI/CDC | AMB CLSI/CDC | 5-FC CLSI/CDC | CSF CLSI/CDC | MCF CLSI/CDC | ANF CLSI/CDC | |
| Ch | 100%/100% | 85.7%/ND | 85.7%/ND | 100%/100% | 0%/ND | 0%/0% | 0%/0% | 0%/0% |
| Cd | 72.7%/90.9% | 72.7%/ND | 72.7%/ND | 100%/100% | 18.2%/ND | 9.1%/9.1% | 0%/0% | 0%/0% |
| Chv | 100%/100% | 85.7%/ND | 85.7%/ND | 100%/100% | 0%/ND | 0%/0% | 0%/0% | 0%/0% |
| Clinical Isolates | MIC for 5-FC | MBEC for 5-FC | Variation (MBEC/MIC) | MIC for AND | MBEC for AND | Variation (MBEC/MIC) |
|---|---|---|---|---|---|---|
| Ch | ||||||
| LIPCh15 | 0.5 (S) | 2 (S) | 4× | 0.25 (S) | 2 (S) | 8× |
| LIPCh16 | 0.5 (S) | >64 (R) | >128× | 0.125 (S) | >64 (R) | >512× |
| LIPCh19 | 0.25 (S) | 0.25 (S) | - | 0.25 (S) | 0.5 (S) | 2× |
| LIPCh27 | 0.5 (S) | 8 (I) | 16× | 0.125 (S) | >8 (R) | >64× |
| LIPCh30 | 0.5 (S) | 64 (R) | 128× | 0.125 (S) | 0.25 (S) | 2× |
| LIPCh34 | 0.125 (S) | >64 (R) | >512× | 0.06 (S) | 2 (S) | 32× |
| LIPCh36 | 0.25 (S) | >64 (R) | >256× | 0.125 (S) | >8 (R) | >64× |
| Cd | ||||||
| LIPCh13 | >64 (R) | >64 (R) | - | 2 (S) | >8 (R) | 4× |
| LIPCh22 | 0.5 (S) | 0.5 (S) | - | 0.125 (S) | 0.25 (S) | 2× |
| LIPCh23 | 0.25 (S) | 1 (S) | 4× | 0.06 (S) | 0.06 (S) | - |
| LIPCh26 | 1 (S) | >64 (R) | >64× | 0.06 (S) | 4 (R) | 64× |
| LIPCh28 | 0.25 (S) | 0.5 (S) | 2× | 0.06 (S) | 0.06 (S) | - |
| LIPCh29 | 0.25 (S) | 2 (S) | 8× | 0.06 (S) | 0.25 (S) | 4× |
| LIPCh31 | 1 (S) | 1 (S) | - | 0.06 (S) | 0.125 (S) | 2× |
| LIPCh32 | 0.125 (S) | 0.5 (S) | 4× | 0.06 (S) | 0.125 (S) | 2× |
| LIPCh33 | 0.125 (S) | 1 (S) | 8× | 0.06 (S) | 0.125 (S) | 2× |
| LIPCh35 | 0.5 (S) | 1 (S) | 2× | 0.06 (S) | 0.125 (S) | 2× |
| LIPCh38 | >64 (R) | >64 (R) | - | 0.125 (S) | >8 (R) | >64× |
| Chv | ||||||
| LIPCh14 | 0.25 (S) | 4 (S) | 16× | 0.125 (S) | 0.125 (S) | - |
| LIPCh18 | 0.25 (S) | 8 (I) | 32× | 0.125 (S) | 0.25 (S) | 2× |
| LIPCh20 | 0.25 (S) | 8 (I) | 32× | 0.25 (S) | 1 (S) | 4× |
| LIPCh21 | 0.25 (S) | 4 (S) | 16× | 0.06 (S) | 0.5 (S) | 8× |
| LIPCh24 | 0.25 (S) | 32 (R) | 128× | 0.06 (S) | 1 (S) | 16× |
| LIPCh25 | 0.25 (S) | 8 (I) | 32× | 0.06 (S) | 1 (S) | 16× |
| LIPCh37 | <0.125 (S) | 4 (S) | 32× | 0.06 (S) | 0.25 (S) | 4× |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramos, L.S.; Figueiredo-Carvalho, M.H.G.; Silva, L.N.; Siqueira, N.L.M.; Lima, J.C.; Oliveira, S.S.; Almeida-Paes, R.; Zancopé-Oliveira, R.M.; Azevedo, F.S.; Ferreira, A.L.P.; et al. The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes. J. Fungi 2022, 8, 574. https://doi.org/10.3390/jof8060574
Ramos LS, Figueiredo-Carvalho MHG, Silva LN, Siqueira NLM, Lima JC, Oliveira SS, Almeida-Paes R, Zancopé-Oliveira RM, Azevedo FS, Ferreira ALP, et al. The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes. Journal of Fungi. 2022; 8(6):574. https://doi.org/10.3390/jof8060574
Chicago/Turabian StyleRamos, Lívia S., Maria Helena G. Figueiredo-Carvalho, Laura N. Silva, Nahyara L. M. Siqueira, Joice C. Lima, Samuel S. Oliveira, Rodrigo Almeida-Paes, Rosely M. Zancopé-Oliveira, Fabio S. Azevedo, Adriana L. P. Ferreira, and et al. 2022. "The Threat Called Candida haemulonii Species Complex in Rio de Janeiro State, Brazil: Focus on Antifungal Resistance and Virulence Attributes" Journal of Fungi 8, no. 6: 574. https://doi.org/10.3390/jof8060574







