Peroxins in Peroxisomal Receptor Export System Contribute to Development, Stress Response, and Virulence of Insect Pathogenic Fungus Beauveria bassiana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culturing Conditions
2.2. Bioinformatic Analyses of Three Pex Homologs in B. bassiana
2.3. Targeted Gene Disruption and Complementation
2.4. Assay for Fungal Vegetative Growth, Stress Response, Development, and Virulence
2.4.1. Vegetative Growth
2.4.2. Stress Response
2.4.3. Development into Conidia and Blastospores
2.4.4. Insect Bioassay
2.5. Fluorescent Microcopy
2.6. Imaging Peroxisomes with Transmission Electron Microscopy
2.7. Yeast Two-Hybrid (Y2H) Analyses
2.8. Statistical Analyses
3. Results
3.1. Bioinformatic Characterization and Molecular Manipulation
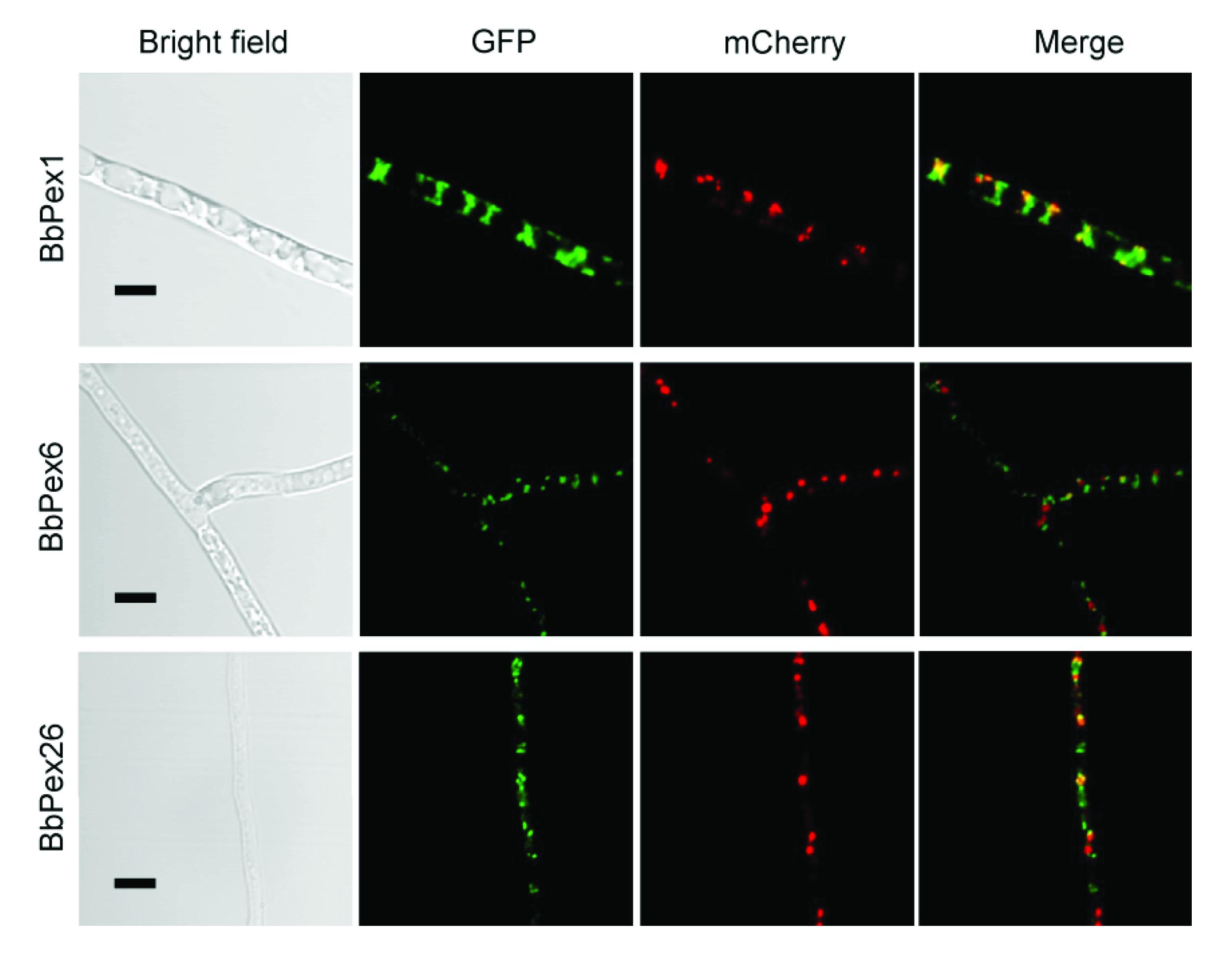
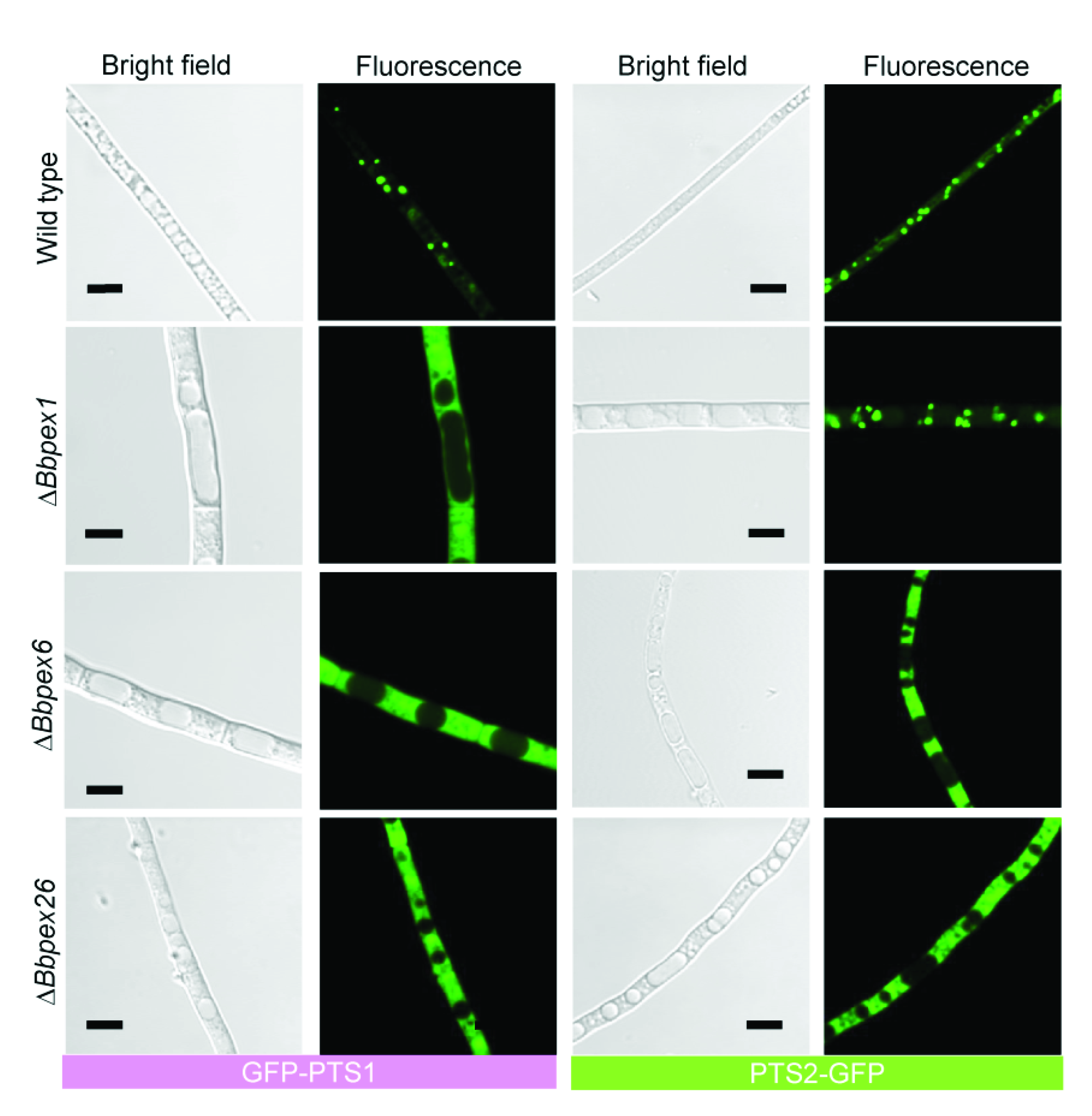
3.2. Imaging the Peroxisome Biogenesis and the Translocation of the Peroxisomal Proteins
3.3. Phenotypic Assays for Vegetative Growth and Development
3.4. Fungal Response to Stresses
3.5. Fungal Virulence against Insects
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De la Cruz, Q.R.; Roussos, S.; Hernandez, D.; Rodriguez, R.; Castillo, F.; Aguilar, C.N. Challenges and opportunities of the bio-pesticides production by solid-state fermentation: Filamentous fungi as a model. Crit. Rev. Biotechnol. 2015, 35, 326–333. [Google Scholar] [CrossRef]
- Lopez-Perez, M.; Rodriguez-Gomez, D.; Loera, O. Production of conidia of Beauveria bassiana in solid-state culture: Current status and future perspectives. Crit. Rev. Biotechnol. 2015, 35, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Wanchoo, A.; Lewis, M.W.; Keyhani, N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria bassiana. Microbiology-SGM 2009, 155, 3121–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ying, S.H.; Feng, M.G.; Keyhani, N.O. A carbon responsive G-protein coupled receptor modulates broad developmental and genetic networks in the entomopathogenic fungus, Beauveria bassiana. Environ. Microbiol. 2013, 15, 2902–2921. [Google Scholar] [CrossRef]
- He, P.H.; Dong, W.X.; Chu, X.L.; Feng, M.G.; Ying, S.H. The cellular proteome is affected by a gelsolin (BbGEL1) during morphological transitions in aerobic surface versus liquid growth in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2016, 18, 4153–4169. [Google Scholar] [CrossRef]
- Pieuchot, L.; Jedd, G. Peroxisome assembly and functional diversity in eukaryotic microorganisms. Annu. Rev. Microbiol. 2012, 66, 237–263. [Google Scholar] [CrossRef] [PubMed]
- Reverberi, M.; Punelli, M.; Smith, C.A.; Zjalic, S.; Scarpari, M.; Scala, V.; Cardinali, G.; Aspite, N.; Pinzari, F.; Payne, G.A.; et al. How peroxisomes affect aflatoxin biosynthesis in Aspergillus flavus. PLoS ONE 2012, 7, e48097. [Google Scholar] [CrossRef] [PubMed]
- Peraza-Reyes, L.; Berteaux-Lecellier, V. Peroxisomes and sexual development in fungi. Front. Physiol. 2013, 4, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, X.; Zhang, H.; Wang, X.; van der Lee, T.; Waalwijk, C.; van Diepeningen, A.; Brankovics, B.; Xu, J.; Xu, J.; Chen, W.; et al. FgPex3, a peroxisome biogenesis factor, is involved in regulating vegetative growth, conidiation, sexual development, and virulence in Fusarium graminearum. Front. Microbiol. 2019, 10, 2088. [Google Scholar] [CrossRef] [Green Version]
- Huarte-Bonnet, C.; Paixão, F.R.S.; Mascarin, G.M.; Santana, M.; Fernandes, E.K.K.; Pedrini, N. The entomopathogenic fungus Beauveria bassiana produces microsclerotia-like pellets mediated by oxidative stress and peroxisome biogenesis. Environ. Microbiol. Rep. 2019, 11, 518–524. [Google Scholar] [CrossRef]
- Pang, M.Y.; Lin, H.Y.; Hou, J.; Feng, M.G.; Ying, S.H. Different contributions of the peroxisomal import protein Pex5 and Pex7 to development, stress response and virulence of insect fungal pathogen Beauveria bassiana. J. Appl. Microbiol. 2022, 132, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Kiel, J.A.; Veenhuis, M.; van der Klei, I.J. PEX genes in fungal genomes: Common, rare or redundant. Traffic 2006, 7, 1291–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, A.; Lanyon-Hogg, T.; Warriner, S.L. Peroxisome protein import: A complex journey. Biochem. Soc. Trans. 2016, 44, 783–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, F.; Lu, Y.; Pieuchot, L.; Dhavale, T.; Jedd, G. Import oligomers induce positive feedback to promote peroxisome differentiation and control organelle abundance. Dev. Cell 2011, 21, 457–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Li, D.; Jiang, K.; Zhang, K.Q.; Yang, J. AoPEX1 and AoPEX6are required for mycelial growth, conidiation, stress response, fatty acid utilization, and trap formation in Arthrobotrys oligospora. Microbiol. Spectr. 2022, 10, e0027522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, C.; Wang, L.; Sun, S.; Liu, A.; Liang, Y.; Yu, J.; Dong, H. FgPEX1 and FgPEX10 are required for the maintenance of Woronin bodies and full virulence of Fusarium graminearum. Curr. Genet. 2019, 65, 1383–1396. [Google Scholar] [CrossRef]
- Imazaki, A.; Tanaka, A.; Harimoto, Y.; Yamamoto, M.; Akimitsu, K.; Park, P.; Tsuge, T. Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen Alternaria alternata. Eukaryot. Cell 2010, 9, 682–694. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Zhu, W.; Cheng, J.; Xie, J.; Li, B.; Jiang, D.; Li, G.; Yi, X.; Fu, Y. CmPEX6, a gene involved in peroxisome biogenesis, is essential for parasitism and conidiation by the sclerotial parasite Coniothyrium minitans. Appl. Environ. Microbiol. 2013, 79, 3658–3666. [Google Scholar] [CrossRef] [Green Version]
- Xiao, G.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.; Xie, X.Q.; Shang, Y.; Zheng, H.; Zhou, Y.; St Leger, R.J.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolu tionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, J.L.; Hou, J.; Li, X.H.; Feng, M.G.; Ying, S.H. Transcription activator Swi6 interacts with Mbp1 in MluI cell cycle box-binding complex and regulates hyphal differentiation and virulence in Beauveria bassiana. J. Fungi 2021, 7, 411. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Peng, Y.J.; Feng, M.G.; Ying, S.H. Functional analysis of the mitochondrial gene mitofilin in the filamentous entomopathogenic fungus Beauveria bassiana. Fungal Genet. Biol. 2019, 132, 103250. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.L.; Peng, Y.J.; Chu, X.L.; Feng, M.G.; Ying, S.H. Autophagy-related gene BbATG11 is indispensable for pexophagy and mitophagy, and contributes to stress response, conidiation and virulence in the insect mycopathogen Beauveria bassiana. Environ. Microbiol. 2018, 20, 3309–3324. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Peng, Y.J.; Ding, J.L.; Feng, M.G.; Ying, S.H. Mitochondrial fission is necessary for mitophagy, development and virulence of the insect pathogenic fungus Beauveria bassiana. J. Appl. Microbiol. 2020, 129, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Ding, J.L.; Peng, Y.J.; Feng, M.G.; Ying, S.H. Proteomic and phosphoryproteomic investigations reveal that autophagy-related protein 1, a protein kinase for autophagy initiation, synchronously deploys phospho-regulation on ubiquitin-like conjugation system in mycopathogen Beauveria bassiana. mSystems 2022, 7, e01463-21. [Google Scholar] [CrossRef]
- Matsumoto, N.; Tamura, S.; Fujiki, Y. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat. Cell Biol. 2003, 5, 454–460. [Google Scholar] [CrossRef]
- Rinaldi, M.A.; Fleming, W.A.; Gonzalez, K.L.; Park, J.; Ventura, M.J.; Patel, A.B.; Bartel, B. The PEX1 ATPase stabilizes PEX6 and plays essential roles in peroxisome biology. Plant Physiol. 2017, 174, 2231–2247. [Google Scholar] [CrossRef] [Green Version]
- Gardner, B.M.; Chowdhury, S.; Lander, G.C.; Martin, A. The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J. Mol. Biol. 2015, 427, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Buentzel, J.; Vilardi, F.; Lotz-Havla, A.; Gärtner, J.; Thoms, S. Conserved targeting information in mammalian and fungal peroxisomal tail-anchored proteins. Sci. Rep. 2015, 5, 17420. [Google Scholar] [CrossRef] [Green Version]
- Ortiz-Urquiza, A.; Keyhani, N.O. Molecular genetics of Beauveria bassianainfection of insects. Adv. Genet. 2016, 94, 165–249. [Google Scholar] [PubMed]
- Kazek, M.; Kaczmarek, A.; Wrońska, A.K.; Boguś, M.I. Diet influences the bacterial and free fatty acid profiles of the cuticle of Galleria mellonella larvae. PLoS ONE 2019, 14, e0211697. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Urquiza, A.; Keyhani, N.O. Action on the surface: Entomopathogenic fungi versus the insect cuticle. Insects 2013, 4, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Huarte-Bonnet, C.; Paixão, F.R.S.; Ponce, J.C.; Santana, M.; Prieto, E.D.; Pedrini, N. Alkane-grown Beauveria bassiana produce mycelial pellets displaying peroxisome proliferation, oxidative stress, and cell surface alterations. Fungal Biol. 2018, 122, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Freitag, J.; Ast, J.; Bölker, M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 2012, 485, 522–525. [Google Scholar] [CrossRef]
- Ding, J.L.; Hou, J.; Feng, M.G.; Ying, S.H. Transcriptomic analyses reveal comprehensive responses of insect hemocytes to mycopathogen Beauveria bassiana, and fungal virulence-related cell wall protein assists pathogen to evade host cellular defense. Virulence 2020, 11, 1352–1365. [Google Scholar] [CrossRef]
- Bergin, D.; Reeves, E.P.; Renwick, J.; Wientjes, F.B.; Kavanagh, K. Superoxide production in Galleria mellonella hemocytes: Identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 2005, 73, 4161–4170. [Google Scholar] [CrossRef] [Green Version]
- Paixão, F.; Huarte-Bonnet, C.; Ribeiro-Silva, C.D.; Mascarin, G.M.; Fernandes, E.K.K.; Pedrini, N. Tolerance to abiotic factors of microsclerotia and mycelial pellets from Metarhizium robertsii, and molecular and ultrastructural changes during microsclerotial differentiation. Front. Fungal Biol. 2021, 2, 654737. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, J.; Fang, W.; Zhang, J.; Luo, Z.; Zhang, M.; Fan, Y.; Pei, Y. Mitogen-activated protein kinase hog1 in the entomopathogenic fungus Beauveria bassiana regulates environmental stress responses and virulence to insects. Appl. Environ. Microbiol. 2009, 75, 3787–3795. [Google Scholar] [CrossRef] [Green Version]
- Guan, H.; Guo, Y.; Zhu, L.; Jiao, Y.; Liu, X. Peroxisome deficiency dysregulates fatty acid oxidization and exacerbates lipotoxicity in β cells. Oxid. Med. Cell. Longev. 2021, 2021, 7726058. [Google Scholar] [CrossRef]
- Lu, Y.X.; Zhang, Q.; Xu, W.H. Global metabolomic analyses of the hemolymph and brain during the initiation, maintenance, and termination of pupal diapause in the cotton bollworm, Helicoverpa armigera. PLoS ONE 2014, 9, e99948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.X.; He, P.H.; Feng, M.G.; Ying, S.H. BbSNF1 contributes to cell differentiation, extracellular acidification, and virulence in Beauveria bassiana, a filamentous entomopathogenic fungus. Appl. Microbiol. Biotechnol. 2014, 98, 8657–8673. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Villaseñor, N.; Sánchez-Arreguín, J.A.; Herrera-Estrella, A.H. Trichoderma: Sensing the environment for survival and dispersal. Microbiology-SGM 2012, 158, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Hull, C.M. Sporulation: How to survive on planet Earth (and beyond). Curr. Genet. 2017, 63, 831–838. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Lin, H.; Ding, J.; Feng, M.; Ying, S. Peroxins in Peroxisomal Receptor Export System Contribute to Development, Stress Response, and Virulence of Insect Pathogenic Fungus Beauveria bassiana. J. Fungi 2022, 8, 622. https://doi.org/10.3390/jof8060622
Hou J, Lin H, Ding J, Feng M, Ying S. Peroxins in Peroxisomal Receptor Export System Contribute to Development, Stress Response, and Virulence of Insect Pathogenic Fungus Beauveria bassiana. Journal of Fungi. 2022; 8(6):622. https://doi.org/10.3390/jof8060622
Chicago/Turabian StyleHou, Jia, Haiyan Lin, Jinli Ding, Mingguang Feng, and Shenghua Ying. 2022. "Peroxins in Peroxisomal Receptor Export System Contribute to Development, Stress Response, and Virulence of Insect Pathogenic Fungus Beauveria bassiana" Journal of Fungi 8, no. 6: 622. https://doi.org/10.3390/jof8060622
APA StyleHou, J., Lin, H., Ding, J., Feng, M., & Ying, S. (2022). Peroxins in Peroxisomal Receptor Export System Contribute to Development, Stress Response, and Virulence of Insect Pathogenic Fungus Beauveria bassiana. Journal of Fungi, 8(6), 622. https://doi.org/10.3390/jof8060622







