Intra-Species Genomic Variation in the Pine Pathogen Fusarium circinatum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolates
2.2. Genome Sequencing, Data Sets and Assembly
2.3. Identification and Annotation of SVs
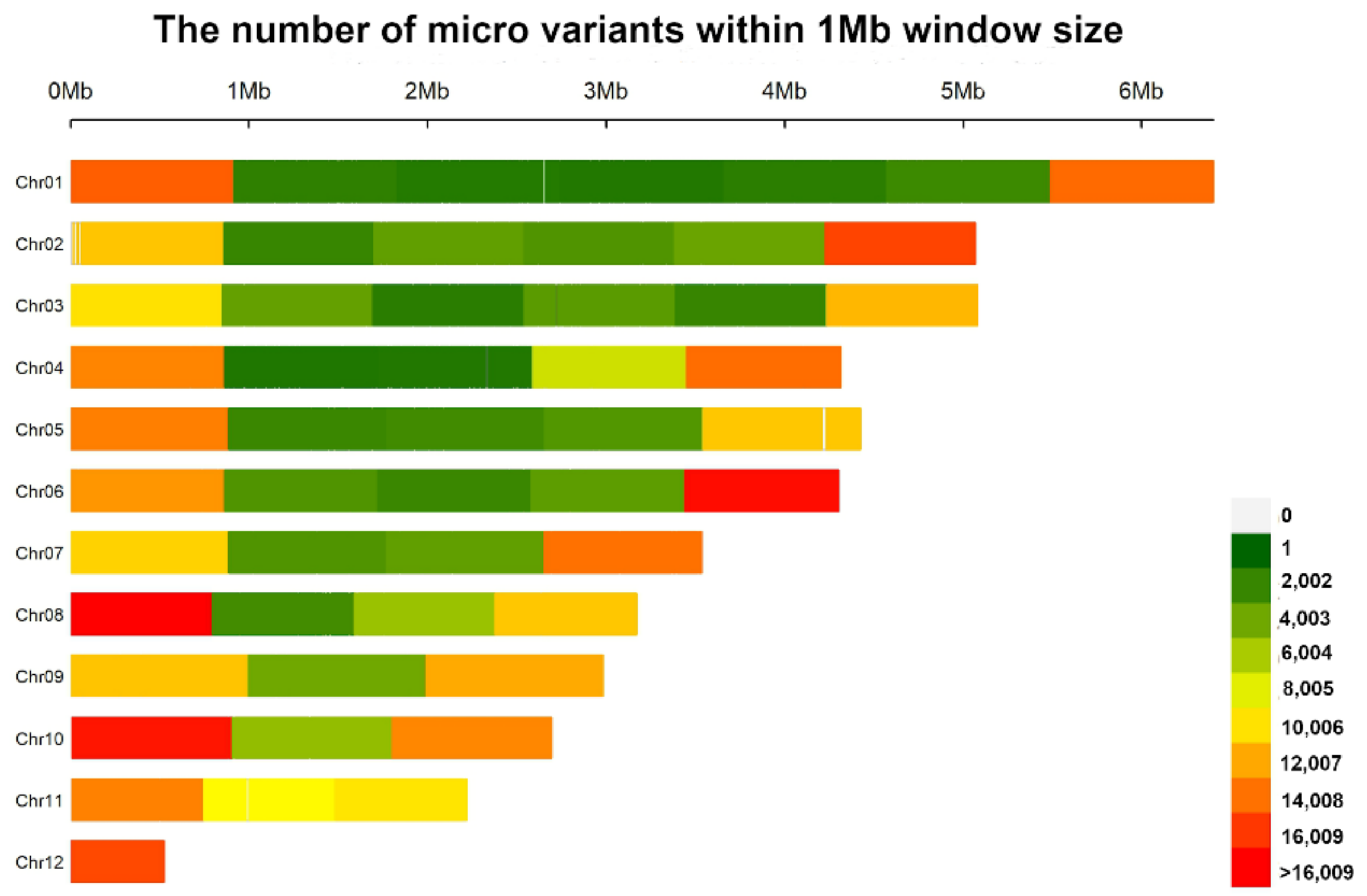
2.4. Identification and Characterization of Micro Variants
2.5. Synteny and Pangenome Analyses
3. Results
3.1. Genome Sequencing, Data Sets and Assembly
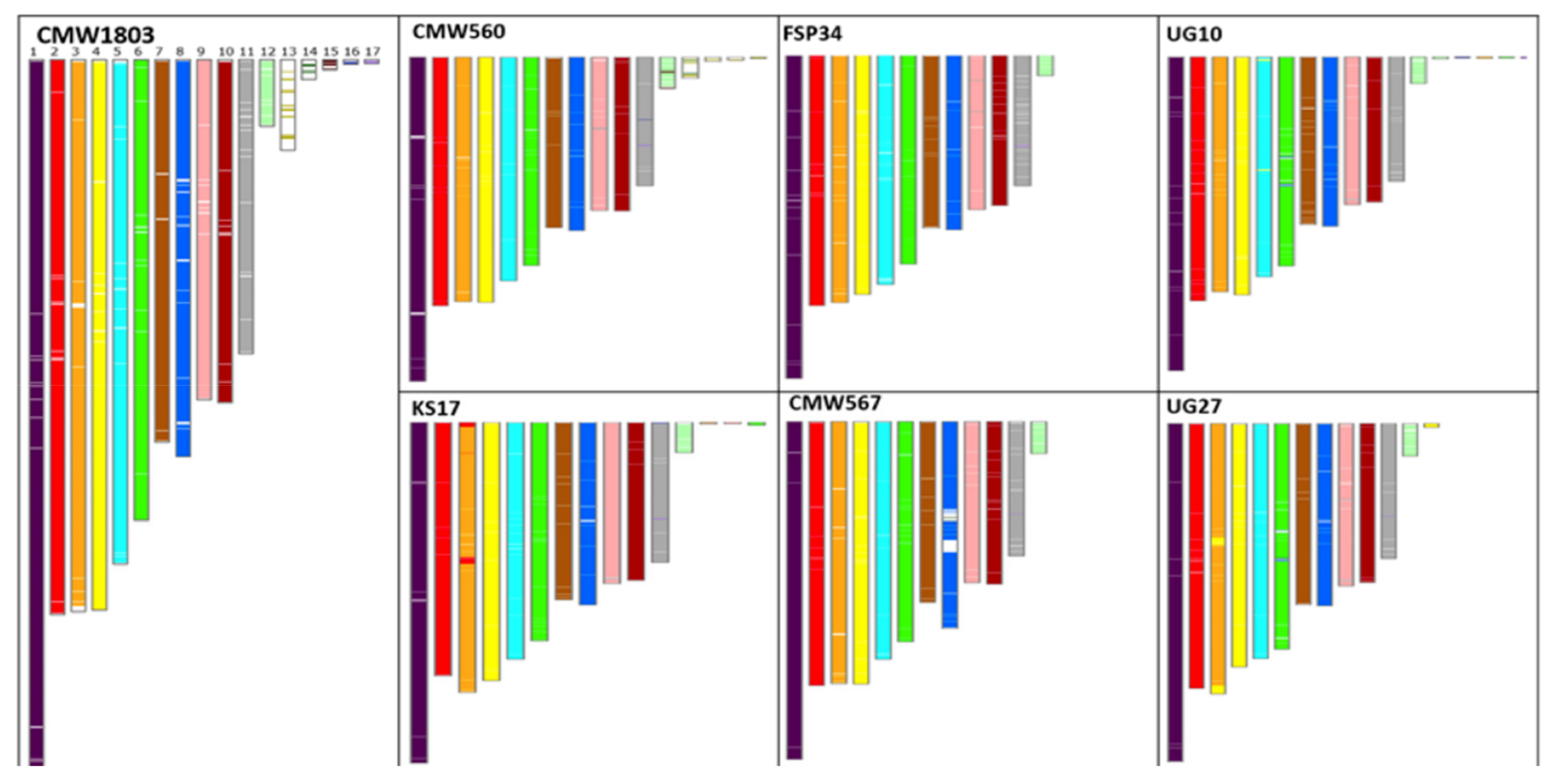
3.2. Identification and Annotation of SVs
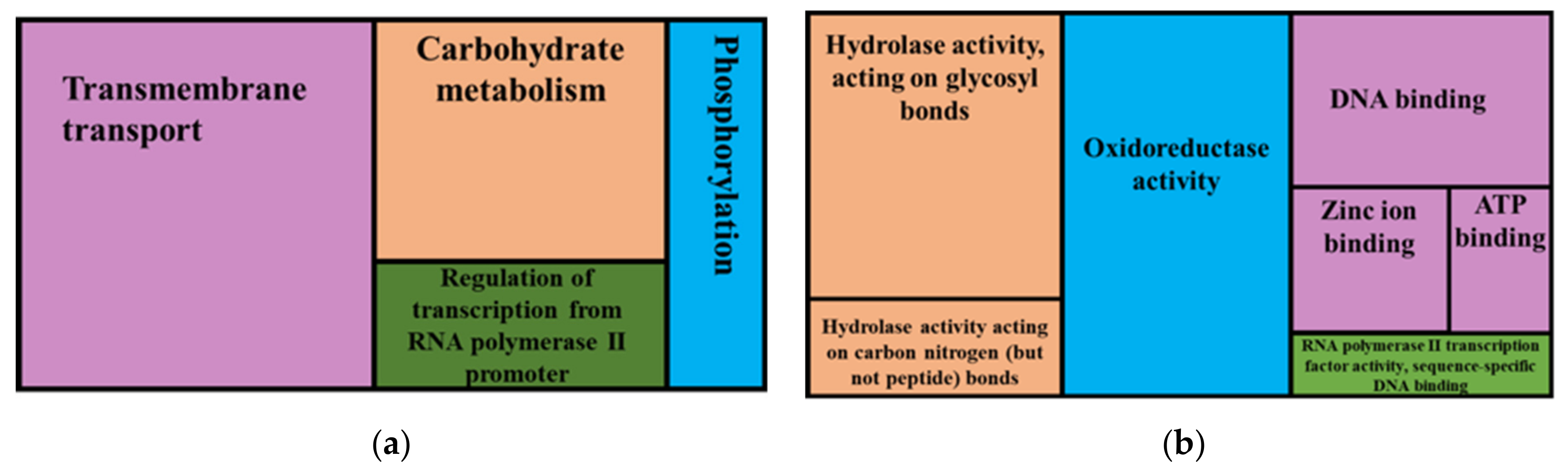
3.3. Identification and Annotation of Micro Variants
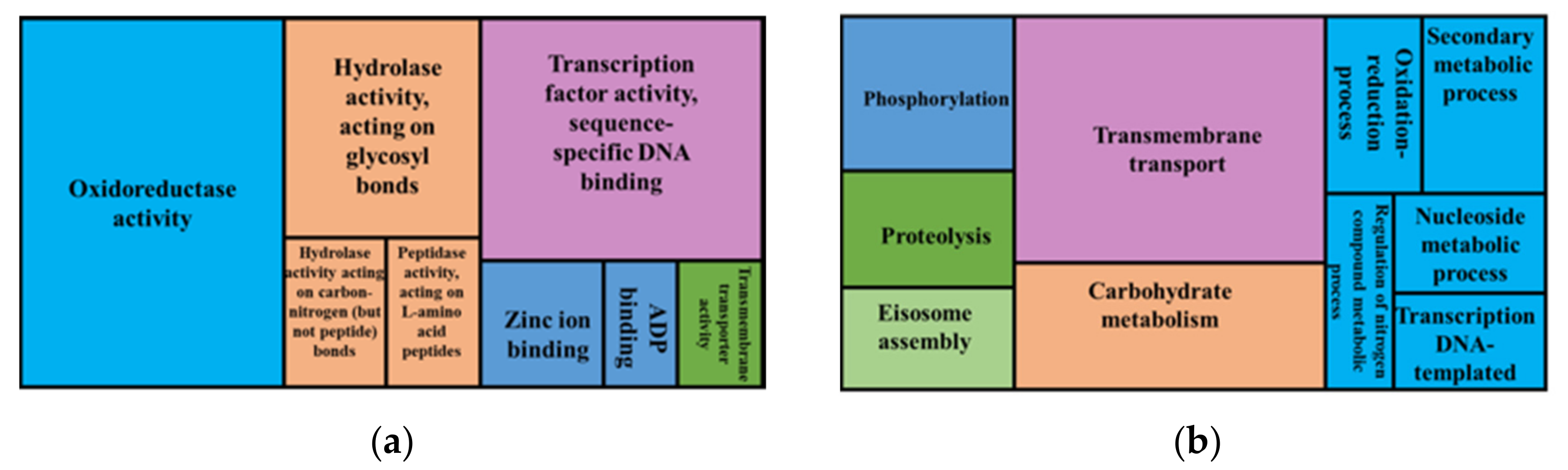
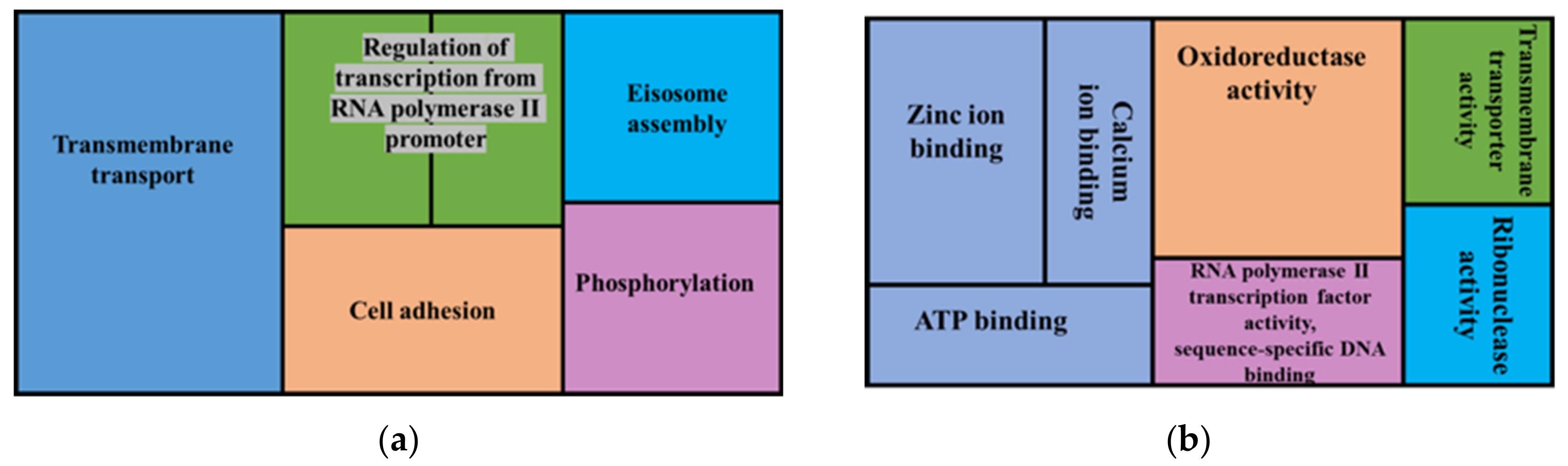
3.4. Synteny and Pangenome Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schirawski, J.; Mannhaupt, G.; Münch, K.; Brefort, T.; Schipper, K.; Doehlemann, G.; Di Stasio, M.; Rössel, N.; Mendoza-Mendoza, A.; Pester, D. Pathogenicity determinants in smut fungi revealed by genome comparison. Science 2010, 330, 1546–1548. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.M.; Panstruga, R. Pathogenomics of fungal plant parasites: What have we learnt about pathogenesis? Curr. Opin. Plant Biol. 2011, 14, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Croll, D.; McDonald, B.A. The accessory genome as a cradle for adaptive evolution in pathogens. PLoS Pathog. 2012, 8, e1002608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, D.M.; McDonald, M.C.; Covarelli, L.; Solomon, P.S.; Rusu, A.G.; Marshall, M.; Kazan, K.; Chakraborty, S.; McDonald, B.A.; Manners, J.M. Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 2012, 8, e1002952. [Google Scholar] [CrossRef] [Green Version]
- Möller, M.; Stukenbrock, E.H. Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Microbiol. 2017, 15, 756. [Google Scholar] [CrossRef]
- De Wit, P.J.; Mehrabi, R.; Van den Burg, H.A.; Stergiopoulos, I. Fungal effector proteins: Past, present and future. Mol. Plant Pathol. 2009, 10, 735–747. [Google Scholar] [CrossRef]
- Perez-Nadales, E.; Nogueira, M.F.A.; Baldin, C.; Castanheira, S.; El Ghalid, M.; Grund, E.; Lengeler, K.; Marchegiani, E.; Mehrotra, P.V.; Moretti, M. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genet. Biol. 2014, 70, 42–67. [Google Scholar] [CrossRef] [Green Version]
- Parfrey, L.W.; Lahr, D.J.; Katz, L.A. The dynamic nature of eukaryotic genomes. Mol. Biol. Evol. 2008, 25, 787–794. [Google Scholar] [CrossRef]
- McCarthy, C.G.; Fitzpatrick, D.A. Pan-genome analyses of model fungal species. Microb. Genom. 2019, 5, e000243. [Google Scholar] [CrossRef]
- Eid, J.; Fehr, A.; Gray, J.; Luong, K.; Lyle, J.; Otto, G.; Peluso, P.; Rank, D.; Baybayan, P.; Bettman, B. Real-time DNA sequencing from single polymerase molecules. Science 2009, 323, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wikler, K.; Gordon, T.R. An initial assessment of genetic relationships among populations of Fusarium circinatum in different parts of the world. Can. J. Bot. 2000, 78, 709–717. [Google Scholar] [CrossRef]
- Fru, F.F.; Steenkamp, E.T.; Wingfield, M.J.; Roux, J. High genetic diversity of Fusarium circinatum associated with the first outbreak of pitch canker on Pinus patula in South Africa. South. For. J. For. Sci. 2019, 81, 69–78. [Google Scholar] [CrossRef]
- Wikler, K.; Gordon, T.R.; Clark, S.L.; Wingfield, M.J.; Britz, H. Potential for outcrossing in an apparently asexual population of Fusarium circinatum, the causal agent of pitch canker disease. Mycologia 2000, 92, 1085–1090. [Google Scholar] [CrossRef]
- Santana, Q.C.; Coetzee, M.P.A.; Wingfield, B.D.; Wingfield, M.J.; Steenkamp, E.T. Nursery-linked plantation outbreaks and evidence for multiple introductions of the pitch canker pathogen Fusarium circinatum into South Africa. Plant Pathol. 2016, 65, 357–368. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Sierra, A.; Landeras, E.; León, M.; Berbegal, M.; García-Jiménez, J.; Armengol, J. Characterization of Fusarium circinatum from Pinus spp. in northern Spain. Mycol. Res. 2007, 111, 832–839. [Google Scholar] [CrossRef]
- Wöstemeyer, J.; Kreibich, A. Repetitive DNA elements in fungi (Mycota): Impact on genomic architecture and evolution. Curr. Genet. 2002, 41, 189–198. [Google Scholar] [CrossRef]
- Kempken, F.; Kück, U. Transposons in filamentous fungi—Facts and perspectives. BioEssays 1998, 20, 652–659. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, E. Repeat-induced point mutation and other genome defense mechanisms in fungi. Microbiol. Spectr. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amselem, J.; Lebrun, M.-H.; Quesneville, H. Whole genome comparative analysis of transposable elements provides new insight into mechanisms of their inactivation in fungal genomes. BMC Genom. 2015, 16, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Wyk, S.; Wingfield, B.D.; De Vos, L.; van der Merwe, N.A.; Santana, Q.C.; Steenkamp, E.T. Repeat-induced point mutations drive divergence between Fusarium circinatum and its close relatives. Pathogens 2019, 8, 298. [Google Scholar] [CrossRef] [Green Version]
- Steenkamp, E.T.; Wingfield, M.J.; McTaggart, A.R.; Wingfield, B.D. Fungal species and their boundaries matter–Definitions, mechanisms and practical implications. Fungal Biol. Rev. 2018, 32, 104–116. [Google Scholar] [CrossRef]
- Van der Nest, M.A.; Beirn, L.A.; Crouch, J.A.; Demers, J.E.; De Beer, Z.W.; De Vos, L.; Gordon, T.R.; Moncalvo, J.-M.; Naidoo, K.; Sanchez-Ramirez, S. Draft genomes of Amanita jacksonii, Ceratocystis albifundus, Fusarium circinatum, Huntiella omanensis, Leptographium procerum, Rutstroemia sydowiana, and Sclerotinia echinophila. IMA Fungus 2014, 5, 472–485. [Google Scholar] [CrossRef] [Green Version]
- Slinski, S.; Kirkpatrick, S.; Gordon, T. Inheritance of virulence in Fusarium circinatum, the cause of pitch canker in pines. Plant Pathol. 2016, 65, 1292–1296. [Google Scholar] [CrossRef] [Green Version]
- Jeffares, D.C.; Jolly, C.; Hoti, M.; Speed, D.; Shaw, L.; Rallis, C.; Balloux, F.; Dessimoz, C.; Bähler, J.; Sedlazeck, F.J. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat. Commun. 2017, 8, 14061. [Google Scholar] [CrossRef] [Green Version]
- Steenwyk, J.; Rokas, A. Extensive copy number variation in fermentation-related genes among Saccharomyces cerevisiae wine strains. G3 Genes Genomes Genet. 2017, 7, 1475–1485. [Google Scholar] [CrossRef] [Green Version]
- Syvänen, A.-C. Accessing genetic variation: Genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001, 2, 930–942. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Gutiérrez-Gil, B.; Klopp, C.; Tosser-Klopp, G.; Arranz, J.J. Variant discovery in the sheep milk transcriptome using RNA sequencing. BMC Genom. 2017, 18, 170. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Panitz, F.; Gregersen, V.R.; Bendixen, C.; Holm, L.-E. Deep sequencing of Danish Holstein dairy cattle for variant detection and insight into potential loss-of-function variants in protein coding genes. BMC Genom. 2015, 16, 1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daboussi, M.-J.; Capy, P. Transposable elements in filamentous fungi. Annu. Rev. Microbiol. 2003, 57, 275–299. [Google Scholar] [CrossRef] [PubMed]
- Katju, V.; Bergthorsson, U. Copy-number changes in evolution: Rates, fitness effects and adaptive significance. Front. Genet. 2013, 4, 273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuma, I.; Isobe, C.; Hotta, Y.; Ibaragi, K.; Futamata, N.; Kusaba, M.; Yoshida, K.; Terauchi, R.; Fujita, Y.; Nakayashiki, H. Multiple translocation of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011, 7, e1002147. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, M. How and why chromosome inversions evolve. PLoS Biol. 2010, 8, e1000501. [Google Scholar] [CrossRef] [Green Version]
- Kirkpatrick, M.; Barton, N. Chromosome inversions, local adaptation and speciation. Genetics 2006, 173, 419–434. [Google Scholar] [CrossRef] [Green Version]
- De Vos, L.; Myburg, A.A.; Wingfield, M.J.; Desjardins, A.E.; Gordon, T.; Wingfield, B.D. Complete genetic linkage maps from an interspecific cross between Fusarium circinatum and Fusarium subglutinans. Fungal Genet. Biol. 2007, 44, 701–714. [Google Scholar] [CrossRef] [Green Version]
- Steenkamp, E.; Makhari, O.; Coutinho, T.; Wingfield, B.; Wingfield, M. Evidence for a new introduction of the pitch canker fungus F usarium circinatum in S outh Africa. Plant Pathol. 2014, 63, 530–538. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.-S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- De Vos, L.; Steenkamp, E.T.; Martin, S.H.; Santana, Q.C.; Fourie, G.; van der Merwe, N.A.; Wingfield, M.J.; Wingfield, B.D. Genome-wide macrosynteny among Fusarium species in the Gibberella fujikuroi complex revealed by amplified fragment length polymorphisms. PLoS ONE 2014, 9, e114682. [Google Scholar] [CrossRef] [Green Version]
- Harris, R. Improved Pairwise Alignment of Genomic DNA. Ph.D. Thesis, The Pennsylvania State University, State College, PA, USA, 2007. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Wiemann, P.; Sieber, C.M.; Von Bargen, K.W.; Studt, L.; Niehaus, E.-M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D. Deciphering the cryptic genome: Genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [Green Version]
- Wingfield, B.D.; Barnes, I.; de Beer, Z.W.; De Vos, L.; Duong, T.A.; Kanzi, A.M.; Naidoo, K.; Nguyen, H.D.; Santana, Q.C.; Sayari, M. Draft genome sequences of Ceratocystis eucalypticola, Chrysoporthe cubensis, C. deuterocubensis, Davidsoniella virescens, Fusarium temperatum, Graphilbum fragrans, Penicillium nordicum, and Thielaviopsis musarum. IMA Fungus 2015, 6, 493–506. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnett, D.W.; Garrison, E.K.; Quinlan, A.R.; Strömberg, M.P.; Marth, G.T. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 2011, 27, 1691–1692. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoff, K.J.; Stanke, M. WebAUGUSTUS—A web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013, 41, W123–W128. [Google Scholar] [CrossRef]
- Wingfield, B.D.; Steenkamp, E.T.; Santana, Q.C.; Coetzee, M.; Bam, S.; Barnes, I.; Beukes, C.W.; Yin Chan, W.; De Vos, L.; Fourie, G. First fungal genome sequence from Africa: A preliminary analysis. S. Afr. J. Sci. 2012, 108, 01–09. [Google Scholar] [CrossRef] [Green Version]
- Wingfield, B.D.; Liu, M.; Nguyen, H.D.; Lane, F.A.; Morgan, S.W.; De Vos, L.; Wilken, P.M.; Duong, T.A.; Aylward, J.; Coetzee, M.P. Nine draft genome sequences of Claviceps purpurea s. lat., including C. arundinis, C. humidiphila, and C. cf. spartinae, pseudomolecules for the pitch canker pathogen Fusarium circinatum, draft genome of Davidsoniella eucalypti, Grosmannia galeiformis, Quambalaria eucalypti, and Teratosphaeria destructans. IMA Fungus 2018, 9, 401. [Google Scholar]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24. [Google Scholar] [CrossRef] [Green Version]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [Green Version]
- Ruden, D.M.; Cingolani, P.; Patel, V.M.; Coon, M.; Nguyen, T.; Land, S.J.; Lu, X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012, 3, 35. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Drillon, G.; Carbone, A.; Fischer, G. SynChro: A fast and easy tool to reconstruct and visualize synteny blocks along eukaryotic chromosomes. PLoS ONE 2014, 9, e92621. [Google Scholar] [CrossRef]
- Ozer, E.A.; Allen, J.P.; Hauser, A.R. Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genom. 2014, 15, 737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delcher, A.L.; Phillippy, A.; Carlton, J.; Salzberg, S.L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002, 30, 2478–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5, R12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozer, E.A. ClustAGE: A tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinform. 2018, 19, 150. [Google Scholar] [CrossRef]
- Jankowsky, E. RNA helicases at work: Binding and rearranging. Trends Biochem. Sci. 2011, 36, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Strandberg, A.K.; Salter, L.A. A comparison of methods for estimating the transition: Transversion ratio from DNA sequences. Mol. Phylogenet. Evol. 2004, 32, 495–503. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2020, 19, 619–628. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walkowiak, S.; Rowland, O.; Rodrigue, N.; Subramaniam, R. Whole genome sequencing and comparative genomics of closely related Fusarium Head Blight fungi: Fusarium graminearum, F. meridionale and F. asiaticum. BMC Genom. 2016, 17, 1014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maphosa, M.N.; Steenkamp, E.T.; Wingfield, B.D. Genome-based selection and characterization of Fusarium circinatum-specific sequences. G3 Gene Genomes Genet. 2016, 6, 631–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phasha, M.M.; Wingfield, B.D.; Coetzee, M.P.; Santana, Q.C.; Fourie, G.; Steenkamp, E.T. Architecture and distribution of introns in core genes of four Fusarium species. G3 Gene Genomes Genet. 2017, 7, 3809–3820. [Google Scholar] [CrossRef] [Green Version]
- Ellison, C.E.; Hall, C.; Kowbel, D.; Welch, J.; Brem, R.B.; Glass, N.; Taylor, J.W. Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc. Natl. Acad. Sci. USA 2011, 108, 2831–2836. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, R.; Paul, J.S.; Albrechtsen, A.; Song, Y.S. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011, 12, 443–451. [Google Scholar] [CrossRef]
- Olson, N.D.; Lund, S.P.; Colman, R.E.; Foster, J.T.; Sahl, J.W.; Schupp, J.M.; Keim, P.; Morrow, J.B.; Salit, M.L.; Zook, J.M. Best practices for evaluating single nucleotide variant calling methods for microbial genomics. Front. Genet. 2015, 6, 235. [Google Scholar] [CrossRef] [Green Version]
- Santana, Q.C.; Coetzee, M.P.; Steenkamp, E.T.; Mlonyeni, O.X.; Hammond, G.N.; Wingfield, M.J.; Wingfield, B.D. Microsatellite discovery by deep sequencing of enriched genomic libraries. Biotechniques 2009, 46, 217–223. [Google Scholar] [CrossRef]
- Wallace, M.M.; Covert, S.F. Molecular Mating Type Assay for Fusarium circinatum. Appl. Environ. Microbiol. 2000, 66, 5506–5508. [Google Scholar] [CrossRef] [Green Version]
- Fierro, F.; García-Estrada, C.; Castillo, N.I.; Rodríguez, R.; Velasco-Conde, T.; Martín, J.-F. Transcriptional and bioinformatic analysis of the 56.8 kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum. Fungal Genet. Biol. 2006, 43, 618–629. [Google Scholar] [CrossRef]
- Waalwijk, C.; Taga, M.; Zheng, S.-L.; Proctor, R.H.; Vaughan, M.M.; O’Donnell, K. Karyotype evolution in Fusarium. IMA Fungus 2018, 9, 13–33. [Google Scholar] [CrossRef] [PubMed]
- Zakian, V.A. Telomeres: Beginning to understand the end. Science 1995, 270, 1601–1607. [Google Scholar] [CrossRef] [PubMed]
- Cohn, M.; Liti, G.; Barton, D.B. Telomeres in fungi. In Comparative Genomics; Springer: Berlin/Heidelberg, Germany, 2006; pp. 101–130. [Google Scholar]
- Xu, J.-R.; Yan, K.; Dickman, M.B.; Leslie, J.F. Electrophoretic karyotypes distinguish the biological species of Gibberella fujikuroi (Fusarium section Liseola). Mol. Plant Microbe Interact. 1995, 8, 74–84. [Google Scholar] [CrossRef]
- Steenwyk, J.L.; Soghigian, J.S.; Perfect, J.R.; Gibbons, J.G. Copy number variation contributes to cryptic genetic variation in outbreak lineages of Cryptococcus gattii from the North American Pacific Northwest. BMC Genom. 2016, 17, 700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barredo, J.L.; Díez, B.; Alvarez, E.; Martín, J.F. Large amplification of a 35-kb DNA fragment carrying two penicillin biosynthetic genes in high penicillin producing strains of Penicillium chrysogenum. Curr. Genet. 1989, 16, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Bull, J.H.; Edwards, J.; Turner, G. Amplification of the isopenicillin N synthetase gene in a strain of Penicillium chrysogenum producing high levels of penicillin. Mol. Gen. Genet. 1989, 216, 492–497. [Google Scholar] [CrossRef]
- Fogel, S.; Welch, J.W. Tandem gene amplification mediates copper resistance in yeast. Proc. Natl. Acad. Sci. USA 1982, 79, 5342–5346. [Google Scholar] [CrossRef] [Green Version]
- Marcotte, E.M.; Pellegrini, M.; Ng, H.-L.; Rice, D.W.; Yeates, T.O.; Eisenberg, D. Detecting protein function and protein-protein interactions from genome sequences. Science 1999, 285, 751–753. [Google Scholar] [CrossRef] [Green Version]
- Seoighe, C.; Federspiel, N.; Jones, T.; Hansen, N.; Bivolarovic, V.; Surzycki, R.; Tamse, R.; Komp, C.; Huizar, L.; Davis, R.W. Prevalence of small inversions in yeast gene order evolution. Proc. Natl. Acad. Sci. USA 2000, 97, 14433–14437. [Google Scholar] [CrossRef] [Green Version]
- Strauss, B.S. Frameshift mutation, microsatellites and mismatch repair. Mutat. Res. Rev. Mutat. Res. 1999, 437, 195–203. [Google Scholar] [CrossRef]
- Simmonds, J.; Scott, P.; Brinton, J.; Mestre, T.C.; Bush, M.; Del Blanco, A.; Dubcovsky, J.; Uauy, C. A splice acceptor site mutation in TaGW2-A1 increases thousand grain weight in tetraploid and hexaploid wheat through wider and longer grains. Theor. Appl. Genet. 2016, 129, 1099–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenn, J.; Boursnell, M.; Hitti, R.J.; Jenkins, C.A.; Terry, R.L.; Priestnall, S.L.; Kenny, P.J.; Mellersh, C.S.; Forman, O.P. Genome sequencing reveals a splice donor site mutation in the SNX14 gene associated with a novel cerebellar cortical degeneration in the Hungarian Vizsla dog breed. BMC Genet. 2016, 17, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golicz, A.A.; Bayer, P.E.; Bhalla, P.L.; Batley, J.; Edwards, D. Pangenomics comes of age: From bacteria to plant and animal applications. Trends Genet. 2020, 36, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Domazet-Lošo, T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011, 12, 692–702. [Google Scholar] [CrossRef] [PubMed]




| Isolate | CMWF560 | CMWF567 | CMW1803 | UG10 | UG27 |
|---|---|---|---|---|---|
| Accession number | JAEHFI 000000000 | JADZLS 000000000 | JAEHFH 000000000 | JAGJRQ 000000000 | JAELVK 000000000 |
| Genome size (bp) | 46,691,343 | 45,984,420 | 46,810,763 | 44,774,968 | 45,546,500 |
| Genome coverage 1 | 35 (58) | 65 (56) | 54 (56) | 31 (60) | 43 (43) |
| N50 | 4,436,154 bp | 4,431,017 bp | 4,492,802 bp | 4,380,615 bp | 4,358,900 bp |
| N75 | 3,211,240 bp | 3,566,220 bp | 3,263,251 bp | 3,014,022 bp | 3,202,209 bp |
| L50 | 5 | 5 | 5 | 5 | 5 |
| L75 | 8 | 8 | 8 | 8 | 8 |
| % G+C | 46.78 | 46.87 | 47.05 | 47.50 | 46.83 |
| BUSCO (%) | 99.0 | 99.0 | 99.1 | 98.9 | 99.1 |
| Number of chromosomes | 12 | 12 | 12 | 12 | 12 |
| Uncharacterised contigs | 37 | 12 | 7 | 16 | 35 |
| ORF | 14,170 | 14,116 | 14,382 | 14,094 | 13,987 |
| Scaffold 1 | F. circinatum Isolate 2 | ||||||
|---|---|---|---|---|---|---|---|
| FSP34 | CMWF560 | CMWF567 | CMWF1803 | UG10 | UG27 | KS17 | |
| Chr01 | 6,407,689 | 6,519,590 | 6,423,820 | 6,503,389 | 6,400,822 | 6,430,758 | 6,397,914 |
| Chr02 | 5,066,197 | 5,005,606 | 5,040,005 | 5,011,558 | 4,859,649 | 4,976,076 | 4,709,326 |
| Chr03 | 5,081,888 | 5,037,826 | 5,141,289 | 5,085,556 | 4,913,444 | 5,257,461 | 5,148,568 |
| Chr04 | 4,313,168 | 4,556,945 | 4,479,672 | 4,551,627 | 4,383,147 | 4,236,059 | 4,401,926 |
| Chr05 | 4,432,553 | 4,436,323 | 4,431,017 | 4,492,934 | 4,408,860 | 4,425,559 | 4,304,443 |
| Chr06 | 4,301,895 | 4,284,508 | 4,281,318 | 4,282,150 | 4,261,236 | 4,358,679 | 4,219,930 |
| Chr07 | 3,541,054 | 3,578,508 | 3,565,977 | 3,555,373 | 3,412,693 | 3,591,987 | 3,312,103 |
| Chr08 | 3,172,915 | 3,210,569 | 3,718,348 | 3,263,924 | 3,015,014 | 3,202,361 | 3,066,990 |
| Chr09 | 2,981,544 | 2,920,641 | 2,844,992 | 2,857,338 | 2,289,925 | 2,952,169 | 2,282,005 |
| Chr10 | 2,698,820 | 2,714,737 | 2,642,782 | 2,649,870 | 2,413,675 | 2,564,176 | 2,483,521 |
| Chr11 | 2,228,420 | 2,291,757 | 2,249799 | 2,266,931 | 2,087,508 | 2,247,110 | 2,291,537 |
| Chr12 | 525,065 | 969,164 | 857,395 | 771,183 | 680,337 | 978,035 | 870,680 |
| UC > 500 Kb | - | 536,197 | - | 1,045,802 | - | - | - |
| UC < 500 Kb | 257,344 | 631,872 | 308,006 | 473,128 | 560,738 | 328,470 | 339,343 |
| Sequencing Platform | Isolate | Total Number of Quality-Filtered Reads | Number of Reads Mapped to the FSP34 Reference Assembly (%) |
|---|---|---|---|
| Illumina | FSP34 | 7,840,006 | 7,762,015 (99.0) |
| CMWF560 | 9,471,099 | 8,931,113 (94.0) | |
| CMWF567 | 9,482,308 | 9,012,583 (95.1) | |
| CMWF1803 | 9,860,143 | 9,028,628 (91.6) | |
| UG10 | 9,737,181 | 9,394,412 (96.5) | |
| UG27 | 9,743,693 | 8,934,966 (91.7) | |
| KS17 | 7,995,046 | 7,228,461 (90.4) | |
| PacBio | CMWF560 | 300,889 | 281,873 (93.7) |
| CMWF567 | 187,327 | 181,372 (96.8) | |
| CMWF1803 | 194,164 | 183,210 (94.4) | |
| UG10 | 256,808 | 247,772 (96.5)) | |
| UG27 | 357,305 | 339,326 (95.0) | |
| MinION | KS17 | 95,510 | 92,336 (96.7) |
| FSP34 | 165,477 | 164,130 (99.2) |
| Chromosome | Micro Variants 1 | SVs 2 | ||||
|---|---|---|---|---|---|---|
| Number of Variants | Variant Rate 3 | Number of Deletions | Number of Duplications | Number of Inversions | Number of Insertions | |
| Chr01 | 41,809 | 153 | 99 | 1 | 10 | 83 |
| Chr02 | 44,429 | 114 | 104 | 2 | 14 | 72 |
| Chr03 | 35,947 | 141 | 97 | 0 | 18 | 69 |
| Chr04 | 42,594 | 101 | 80 | 1 | 4 | 71 |
| Chr05 | 36,398 | 121 | 81 | 0 | 8 | 67 |
| Chr06 | 44,223 | 97 | 112 | 0 | 11 | 68 |
| Chr07 | 36,258 | 97 | 75 | 3 | 8 | 63 |
| Chr08 | 43,130 | 73 | 97 | 0 | 4 | 61 |
| Chr09 | 31,971 | 93 | 60 | 0 | 6 | 44 |
| Chr10 | 43,870 | 61 | 80 | 0 | 8 | 39 |
| Chr11 | 36,755 | 60 | 68 | 1 | 10 | 49 |
| Chr12 | 17,314 | 30 | 33 | 0 | 2 | 31 |
| Total | 46,1683 | 97 | 986 | 8 | 103 | 717 |
| Effect Classification | Fields | SnpEff Count | Percentage (%) |
|---|---|---|---|
| Number of effects by impact | High | 420,017 | 88.2 |
| Low | 1 | 0.0 | |
| Moderate | 49,895 | 10.5 | |
| Modifier | 6316 | 1.3 | |
| Number of effects by type | Bidirectional gene fusion | 222,121 | 46.6 |
| Chromosome number variation | 10 | 0.002 | |
| Conservative in-frame deletion | 103 | 0.02 | |
| Disruptive in-frame deletion | 57 | 0.01 | |
| Downstream gene variant | 2499 | 0.5 | |
| Duplication | 1 | 0.0 | |
| Exon loss variant | 100 | 0.02 | |
| Exon region | 2 | 0.0 | |
| Feature ablation | 7724 | 1.6 | |
| Frame shift variant | 149 | 0.03 | |
| Gene fusion | 180,468 | 37.9 | |
| Intergenic region | 608 | 0.13 | |
| Intragenic variant | 367 | 0.08 | |
| Intron variant | 49 | 0.01 | |
| Inversion | 49,863 | 10.5 | |
| Non-coding transcript variant | 310 | 0.07 | |
| Splice acceptor variant | 16 | 0.003 | |
| Splice donor variant | 19 | 0.004 | |
| Splice region variant | 67 | 0.01 | |
| Splice site region | 1 | 0.0 | |
| Start lost | 45 | 0.01 | |
| Stop gained | 1 | 0.0 | |
| Stop lost | 39 | 0.008 | |
| Transcript ablation | 9316 | 2.0 | |
| Upstream gene variant | 2515 | 0.5 | |
| Number of effects by region | Chromosome | 49 | 0.01 |
| Downstream | 2 499 | 0.53 | |
| Exon | 449 | 0.1 | |
| Gene | 438,343 | 92.1 | |
| Intergenic | 608 | 0.1 | |
| Intron | 15 | 0.003 | |
| Splice site acceptor | 5 | 0.001 | |
| Splice site donor | 6 | 0.001 | |
| Splice site region | 1 | 0.0 | |
| Transcript | 31,739 | 6.7 | |
| Upstream | 2515 | 0.5 |
| Effect Classification | Fields | SnpEff Count | Percentage (%) |
|---|---|---|---|
| Number of effects by impact | High | 18,047 | 0.7 |
| Moderate | 63,033 | 2.3 | |
| Low | 74,845 | 2.8 | |
| Modifier | 2,563,288 | 94.3 | |
| Number of effects by functional class | Missense | 46,372 | 42.0 |
| Nonsense | 1027 | 0.9 | |
| Silent | 62,991 | 57.1 | |
| Number of effects by type | Conservative in-frame deletion | 404 | 0.02 |
| Conservative in-frame insertion | 396 | 0.02 | |
| Disruptive in-frame deletion | 354 | 0.01 | |
| Disruptive in-frame insertion | 216 | 0.01 | |
| Downstream gene variant | 977,631 | 35.9 | |
| Frameshift variant | 1881 | 0.07 | |
| Gene fusion | 13.881 | 0.5 | |
| Initiator codon variant | 17 | 0.001 | |
| Intergenic region | 277.047 | 10.2 | |
| Intragenic variant | 168.282 | 6.2 | |
| Intron variant | 23.572 | 0.9 | |
| Missense variant | 62.233 | 2.3 | |
| Non-coding transcript variant | 133.146 | 4.9 | |
| Splice acceptor variant | 287 | 0.01 | |
| Splice donor variant | 325 | 0.01 | |
| Splice region variant | 5689 | 0.2 | |
| Start lost | 110 | 0.004 | |
| Stop gained | 1434 | 0.05 | |
| Stop lost | 269 | 0.01 | |
| Stop retained variant | 170 | 0.006 | |
| Synonymous variant | 71,054 | 2.6 | |
| Upstream gene variant | 988,027 | 36.2 | |
| Number of effects by region | Downstream | 977,631 | 36.0 |
| Exon | 136,959 | 5.0 | |
| Gene | 13,881 | 0.5 | |
| Intergenic | 277,047 | 10.2 | |
| Intron | 19,155 | 0.7 | |
| Splice site acceptor | 266 | 0.01 | |
| Splice site donor | 301 | 0.01 | |
| Splice site region | 4518 | 0.2 | |
| Transcript | 301,428 | 0.0 | |
| Upstream | 988,027 | 36.3 |
| Isolate | Source | Total bp | GC% |
|---|---|---|---|
| FSP34 | Accessory | 2,669,713 | 44.19 |
| Core | 42,260,189 | 47.20 | |
| CMWF560 | Accessory | 3,831,939 | 45.05 |
| Core | 42,633,795 | 47.02 | |
| CMWF567 | Accessory | 3,290,975 | 45.14 |
| Core | 42,465,505 | 47.06 | |
| CMWF1803 | Accessory | 4,359,907 | 45.27 |
| Core | 42,501,744 | 47.22 | |
| KS17 | Accessory | 2,369,668 | 44.57 |
| Core | 42,008,433 | 47.42 | |
| UG10 | Accessory | 2,733,552 | 45.07 |
| Core | 41,905,489 | 47.72 | |
| UG27 | Accessory | 2,919,797 | 44.25 |
| Core | 42,449,440 | 47.06 | |
| Backbone | 42,260,189 | 47.20 | |
| Pangenome | 50,076,541 | 46.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maphosa, M.N.; Steenkamp, E.T.; Kanzi, A.M.; van Wyk, S.; De Vos, L.; Santana, Q.C.; Duong, T.A.; Wingfield, B.D. Intra-Species Genomic Variation in the Pine Pathogen Fusarium circinatum. J. Fungi 2022, 8, 657. https://doi.org/10.3390/jof8070657
Maphosa MN, Steenkamp ET, Kanzi AM, van Wyk S, De Vos L, Santana QC, Duong TA, Wingfield BD. Intra-Species Genomic Variation in the Pine Pathogen Fusarium circinatum. Journal of Fungi. 2022; 8(7):657. https://doi.org/10.3390/jof8070657
Chicago/Turabian StyleMaphosa, Mkhululi N., Emma T. Steenkamp, Aquillah M. Kanzi, Stephanie van Wyk, Lieschen De Vos, Quentin C. Santana, Tuan A. Duong, and Brenda D. Wingfield. 2022. "Intra-Species Genomic Variation in the Pine Pathogen Fusarium circinatum" Journal of Fungi 8, no. 7: 657. https://doi.org/10.3390/jof8070657
APA StyleMaphosa, M. N., Steenkamp, E. T., Kanzi, A. M., van Wyk, S., De Vos, L., Santana, Q. C., Duong, T. A., & Wingfield, B. D. (2022). Intra-Species Genomic Variation in the Pine Pathogen Fusarium circinatum. Journal of Fungi, 8(7), 657. https://doi.org/10.3390/jof8070657







