Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Plant Materials, and Growth Conditions
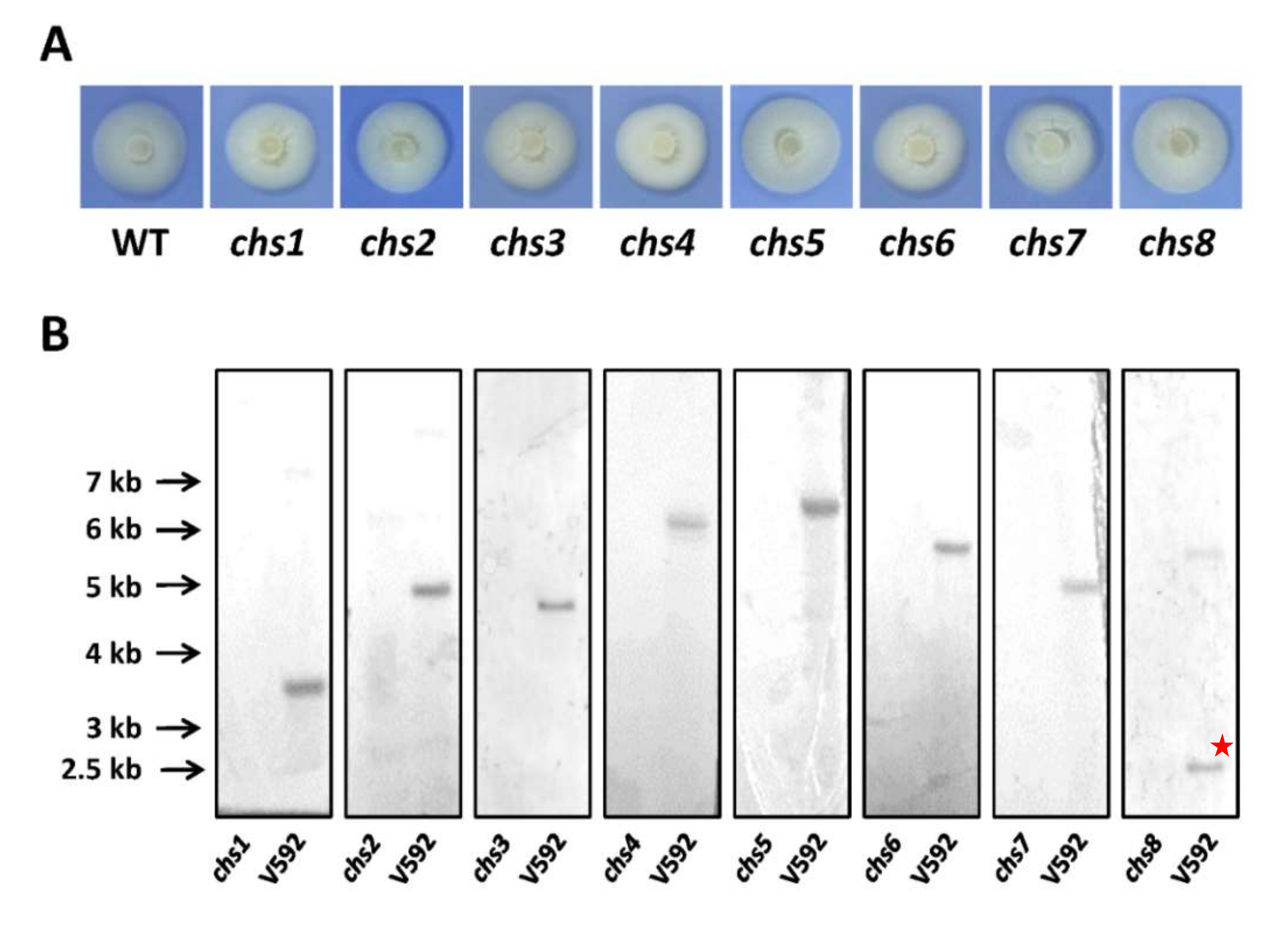
2.2. Generation of Eight CHS Deletion Mutants and Their Complementation Strains
2.3. Southern Blot Hybridizations
2.4. Plant Infection Assay
2.5. Fluorescence Microscopy
3. Results
3.1. Identification of Eight Putative Chitin Synthase Genes in Verticillium Dahliae
3.2. Generation of CHS Gene Deletion Mutants
3.3. VdCHS5 Regulates Germination and Tolerance to Hyperosmotic Stress
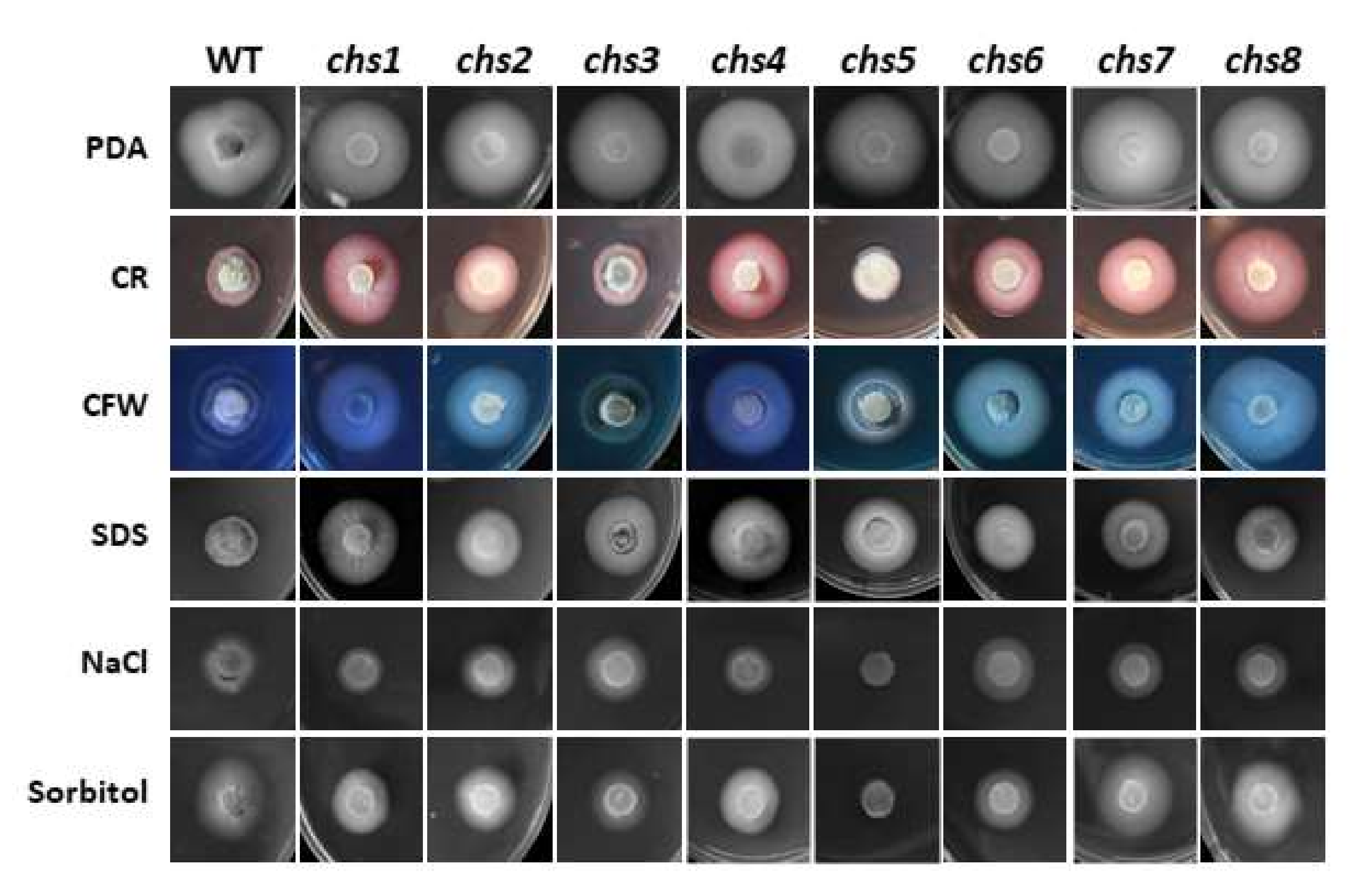
3.4. Most VdCHS Genes Function in Cell Wall Maintenance or Cell Membrane Integrity
3.5. All VdCHS Gene Deletion Mutants, except for Vdchs3, Are Compromised during Plant Infection
3.6. Four VdCHS Gene Deletion Mutants Have Impaired Penetration
3.7. The VdCHS2, VdCHS5, and VdCHS8 Genes Likely Contribute to Post-Penetration Virulence
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bulawa, C.E. Genetics and molecular-biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 1993, 47, 505–534. [Google Scholar] [CrossRef] [PubMed]
- Latgé, J.P. The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 2007, 66, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.I.; Yu, Y.M.; Rho, Y.M.; Park, B.C.; Choi, J.H.; Park, H.M.; Maeng, P.J. Differential expression of the chsE gene encoding a chitin synthase of Aspergillus nidulans in response to developmental status and growth conditions. FEMS Microbiol. Lett. 2005, 249, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munro, C.A.; Gow, N.A.R. Chitin synthesis in human pathogenic fungi. Med. Mycol. 2001, 39, 41–53. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A.R. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef] [Green Version]
- Choquer, M.; Boccara, M.; Goncalves, I.R.; Soulié, M.C.; Vidal-Cros, A. Survey of the Botrytis cinerea chitin synthase multigenic family through the analysis of six euascomycetes genomes. Eur. J. Biochem. 2004, 271, 2153–2164. [Google Scholar] [CrossRef]
- Mandel, M.A.; Galgiani, J.N.; Kroken, S.; Orbach, M.J. Coccidioides posadasii contains single chitin synthase genes corresponding to classes I to VII. Fungal Genet. Biol. 2006, 43, 775–788. [Google Scholar] [CrossRef]
- Cabib, E.; Roh, D.H.; Schmidt, M.; Crotti, L.B.; Varma, A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 2001, 276, 19679–19682. [Google Scholar] [CrossRef] [Green Version]
- Ford, R.A.; Shaw, J.A.; Cabib, E. Yeast chitin synthases 1 and 2 consist of a non-homologous and dispensable N-terminal region and of a homologous moiety essential for function. Mol. Gen. Genet. 1996, 252, 420–428. [Google Scholar] [CrossRef]
- Klis, F.M.; Boorsma, A.; De Groot, P.W.J. Cell wall construction in Saccharomyces cerevisiae. Yeast 2006, 23, 185–202. [Google Scholar] [CrossRef]
- Cabib, E.; Sburlati, A.; Bowers, B.; Silverman, S.J. Chitin synthase-1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1989, 108, 1665–1672. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S.J.; Sburlati, A.; Slater, M.L.; Cabib, E. Chitin synthase 2 is essential for septum formation and cell division in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1988, 85, 4735–4739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, M. Survival and cytokinesis of Saccharomyces cerevisiae in the absence of chitin. Microbiology 2004, 150, 3253–3260. [Google Scholar] [CrossRef]
- Shaw, J.A.; Mol, P.C.; Bowers, B.; Silverman, S.J.; Valdivieso, M.H.; Duran, A.; Cabib, E. The function of chitin synthase 2 and synthase 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 1991, 114, 111–123. [Google Scholar] [CrossRef] [Green Version]
- Matsuo, Y.; Tanaka, K.; Nakagawa, T.; Matsuda, H.; Kawamukai, M. Genetic analysis of chs1+ and chs2+ encoding chitin synthases from Schizosaccharomyces pombe. Biosci. Biotechnol. Biochem. 2004, 68, 1489–1499. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Winter, K.; Buchan, A.; Henry, K.; Becker, J.M.; Brown, A.J.; Bulawa, C.E.; Gow, N.A. Chs1 of Candida albicans is an essential chitin synthase required for synthesis of the septum and for cell integrity. Mol. Microbiol. 2001, 39, 1414–1426. [Google Scholar] [CrossRef]
- Bulawa, C.E.; Miller, D.W.; Henry, L.K.; Becker, J.M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc. Natl. Acad. Sci. USA 1995, 92, 10570–10574. [Google Scholar] [CrossRef] [Green Version]
- Gow, N.A.R.; Robbins, P.W.; Lester, J.W.; Brown, A.J.P.; Fonzi, W.A.; Chapman, T.; Kinsman, O.S. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 1994, 91, 6216–6220. [Google Scholar] [CrossRef] [Green Version]
- Mio, T.; Yabe, T.; Sudoh, M.; Satoh, Y.; Nakajima, T.; Arisawa, M.; Yamadaokabe, H. Role of three chitin synthase genes in the growth of Candida albicans. J. Bacteriol. 1996, 178, 2416–2419. [Google Scholar] [CrossRef] [Green Version]
- Munro, C.A.; Schofield, D.A.; Gooday, G.W.; Gow, N.A.R. Regulation of chitin synthesis during dimorphic growth of Candida albicans. Microbiology 1998, 144, 391–401. [Google Scholar] [CrossRef] [Green Version]
- Munro, C.A.; Whitton, R.K.; Hughes, H.B.; Rella, M.; Selvaggini, S.; Gow, N.A. CHS8-a fourth chitin synthase gene of Candida albicans contributes to in vitro chitin synthase activity, but is dispensable for growth. Fungal Genet. Biol. 2003, 40, 146–158. [Google Scholar] [CrossRef]
- Sánchez-León, E.; Verdin, J.; Freitag, M.; Roberson, R.W.; Bartnicki-Garcia, S.; Riquelme, M. Traffic of chitin synthase 1 (CHS-1) to the Spitzenkörper and developing septa in hyphae of Neurospora crassa: Actin dependence and evidence of distinct microvesicle populations. Eukaryot. Cell 2011, 10, 683–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarden, O.; Yanofsky, C. Chitin synthase 1 plays a major role in cell wall biogenesis in Neurospora crassa. Genes Dev. 1991, 5, 2420–2430. [Google Scholar] [CrossRef] [PubMed]
- Beth-Din, A.; Yarden, O. The Neurospora crassa chs3 gene encodes an essential class I chitin synthase. Mycologia 2000, 92, 65–73. [Google Scholar] [CrossRef]
- Din, A.B.; Yarden, O. The Neurospora crassa chs-2 gene encodes a non-essential chitin synthase. Microbiology 1994, 140, 2189–2197. [Google Scholar] [CrossRef] [Green Version]
- Din, A.B.; Specht, C.A.; Robbins, P.W.; Yarden, O. chs-4, a class IV chitin synthase gene from Neurospora crassa. Mol. Gen. Genet. 1996, 250, 214–222. [Google Scholar] [CrossRef]
- Takeshita, N.; Yamashita, S.; Ohta, A.; Horiuchi, H. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 2006, 59, 1380–1394. [Google Scholar] [CrossRef]
- Tsuizaki, M.; Takeshita, N.; Ohta, A.; Horiuchi, H. Myosin motor-like domain of the class VI chitin synthase CsmB is essential to its functions in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2009, 73, 1163–1167. [Google Scholar] [CrossRef] [Green Version]
- Horiuchi, H.; Fujiwara, M.; Yamashita, S.; Ohta, A.; Takagi, M. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 1999, 181, 3721–3729. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, K.; Yamada, K.; Deoka, K.; Yamashita, S.; Ohta, A.; Horiuchi, H. Class III chitin synthase ChsB of Aspergillus nidulans localizes at the sites of polarized cell wall synthesis and is required for conidial development. Eukaryot. Cell 2009, 8, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, M.; Ichinomiya, M.; Motoyama, T.; Horiuchi, H.; Ohta, A.; Takagi, M. Evidence that the Aspergillus nidulans class I and class II chitin synthase genes, chsC and chsA, share critical roles in hyphal wall integrity and conidiophore development. J. Biochem. 2000, 127, 359–366. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Atallah, Z.K.; Vallad, G.E.; Subbarao, K.V. Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 2009, 47, 39–62. [Google Scholar] [CrossRef] [Green Version]
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by Verticillium dahliae and Verticillium albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.J.; Liu, Y.J.; Zhu, H.Q.; Shang, W.J.; Yang, J.R.; Hu, X.P. Verticillium wilt of redbud in China caused by Verticillium dahliae. Plant Dis. 2013, 97, 1513. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Tian, C. Quantitative detection of pathogen DNA of Verticillium wilt on smoke tree Cotinus coggygria. Plant Dis. 2013, 97, 1645–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, S. Longevity of the Verticillium wilt fungus in the laboratory and field. Phytopathology 1955, 45, 180–181. [Google Scholar]
- Fitzell, R.; Evans, G.; Fahy, P.C. Studies on the colonization of plant roots by Verticillium dahliae Klebahn with use of immunofluorescent staining. Aust. J. Bot. 1980, 28, 357–368. [Google Scholar] [CrossRef]
- Vallad, G.E.; Subbarao, K.V. Colonization of resistant and susceptible lettuce cultivars by a green fluorescent protein-tagged isolate of Verticillium dahliae. Phytopathology 2008, 98, 871–885. [Google Scholar] [CrossRef] [Green Version]
- Klimes, A.; Dobinson, K.F.; Thomma, B.P.H.J.; Klosterman, S.J. Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu. Rev. Phytopathol. 2015, 53, 181–198. [Google Scholar] [CrossRef]
- Yadeta, K.; Thomma, B.P.H.J. The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 2013, 4, 97. [Google Scholar] [CrossRef] [Green Version]
- Gao, F.; Zhou, B.J.; Li, G.Y.; Jia, P.S.; Li, H.; Zhao, Y.L.; Zhao, P.; Xia, G.X.; Guo, H.-S. A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS ONE 2010, 5, e15319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.-L.; Zhou, T.-T.; Guo, H.-S. Hyphopodium-specific VdNoxB/VdPls1-dependent ROS-Ca2+ signaling is required for plant infection by Verticillium dahliae. PLoS Pathog. 2016, 12, e1005793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, T.-T.; Zhao, Y.-L.; Guo, H.-S. Secretory proteins are delivered to the septin-organized penetration interface during root infection by Verticillium dahliae. PLoS Pathog. 2017, 13, e1006275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Xing, H.Y.; Hua, C.L.; Guo, H.-S.; Zhang, J. An improved single-step cloning strategy simplifies the Agrobacterium tumefaciens-mediated transformation (ATMT)-based gene-disruption method for Verticillium dahliae. Phytopathology 2016, 106, 645–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Wang, K.; Sun, L.; Xing, H.; Wang, S.; Li, L.; Chen, S.; Guo, H.-S.; Zhang, J. The plant-specific transcription factors CBP60g and SARD1 are targeted by a Verticillium secretory protein VdSCP41 to modulate immunity. eLife 2018, 7, 34902. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Subbarao, K.V.; Kang, S.; Veronese, P.; Gold, S.E.; Thomma, B.P.H.J.; Chen, Z.; Henrissat, B.; Lee, Y.-H.; Park, J.; et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 2011, 7, e1002137. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Z.; Chen, Q.; Liu, C.H.; Liu, Y.B.; Yi, P.; Niu, K.X.; Wang, Y.Q.; Wang, A.Q.; Yu, H.Y.; Pu, Z.E.; et al. Chitin synthase gene FgCHS8 affects virulence and fungal cell wall sensitivity to environmental stress in Fusarium graminearum. Fungal Biol. 2016, 120, 764–774. [Google Scholar] [CrossRef]
- Kong, L.A.; Yang, J.; Li, G.T.; Qi, L.L.; Zhang, Y.J.; Wang, C.F.; Zhao, W.S.; Xu, J.R.; Peng, Y.L. Different chitin synthase genes are required for various developmental and plant infection processes in the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2012, 8, e1002526. [Google Scholar] [CrossRef] [Green Version]
- Cheng, W.; Song, X.S.; Li, H.P.; Cao, L.H.; Sun, K.; Qiu, X.L.; Xu, Y.B.; Yang, P.; Huang, T.; Zhang, J.B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef]
- Kim, J.E.; Lee, H.J.; Lee, J.; Kim, K.W.; Yun, S.H.; Shim, W.B.; Lee, Y.W. Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 2009, 55, 449–459. [Google Scholar] [CrossRef]
- Odenbach, D.; Thines, E.; Anke, H.; Foster, A.J. The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Mol. Plant Pathol. 2009, 10, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Wang, Y.; Lei, N.; Wang, K.; Zhu, T. Botrytis cinerea chitin synthase BcChsVI is required for normal growth and pathogenicity. Curr. Genet. 2013, 59, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Soulié, M.-C.; Perino, C.; Piffeteau, A.; Choquer, M.; Malfatti, P.; Cimerman, A.; Kunz, C.; Boccara, M.; Vidal-Cros, A. Botrytis cinerea virulence is drastically reduced after disruption of chitin synthase class III gene (Bcchs3a). Cell Microbiol. 2006, 8, 1310–1321. [Google Scholar] [CrossRef] [PubMed]
- Soulié, M.C.; Piffeteau, A.; Choquer, M.; Boccara, M.; Vidal-Cros, A. Disruption of Botrytis cinerea class I chitin synthase gene Bcchs1 results in cell wall weakening and reduced virulence. Fungal Genet. Biol. 2003, 40, 38–46. [Google Scholar] [CrossRef]
- Cui, Z.; Ding, Z.; Yang, X.; Wang, K.; Zhu, T. Gene disruption and characterization of a class V chitin synthase in Botrytis cinerea. Can. J. Microbiol. 2009, 55, 1267–1274. [Google Scholar] [CrossRef]
- Morcx, S.; Kunz, C.; Choquer, M.; Assie, S.; Blondet, E.; Simond-Côte, E.; Gajek, K.; Chapeland-Leclerc, F.; Expert, D.; Soulié, M.-C. Disruption of Bcchs4, Bcchs6 or Bcchs7 chitin synthase genes in Botrytis cinerea and the essential role of class VI chitin synthase (Bcchs6). Fungal Genet. Biol. 2013, 52, 1–8. [Google Scholar] [CrossRef]
- Xu, Y.B.; Li, H.P.; Zhang, J.B.; Song, B.; Chen, F.F.; Duan, X.J.; Xu, H.Q.; Liao, Y.C. Disruption of the chitin synthase gene CHS1 from Fusarium asiaticum results in an altered structure of cell walls and reduced virulence. Fungal Genet. Biol. 2010, 47, 205–215. [Google Scholar] [CrossRef]
- Free, S.J. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013, 81, 33–82. [Google Scholar] [CrossRef]


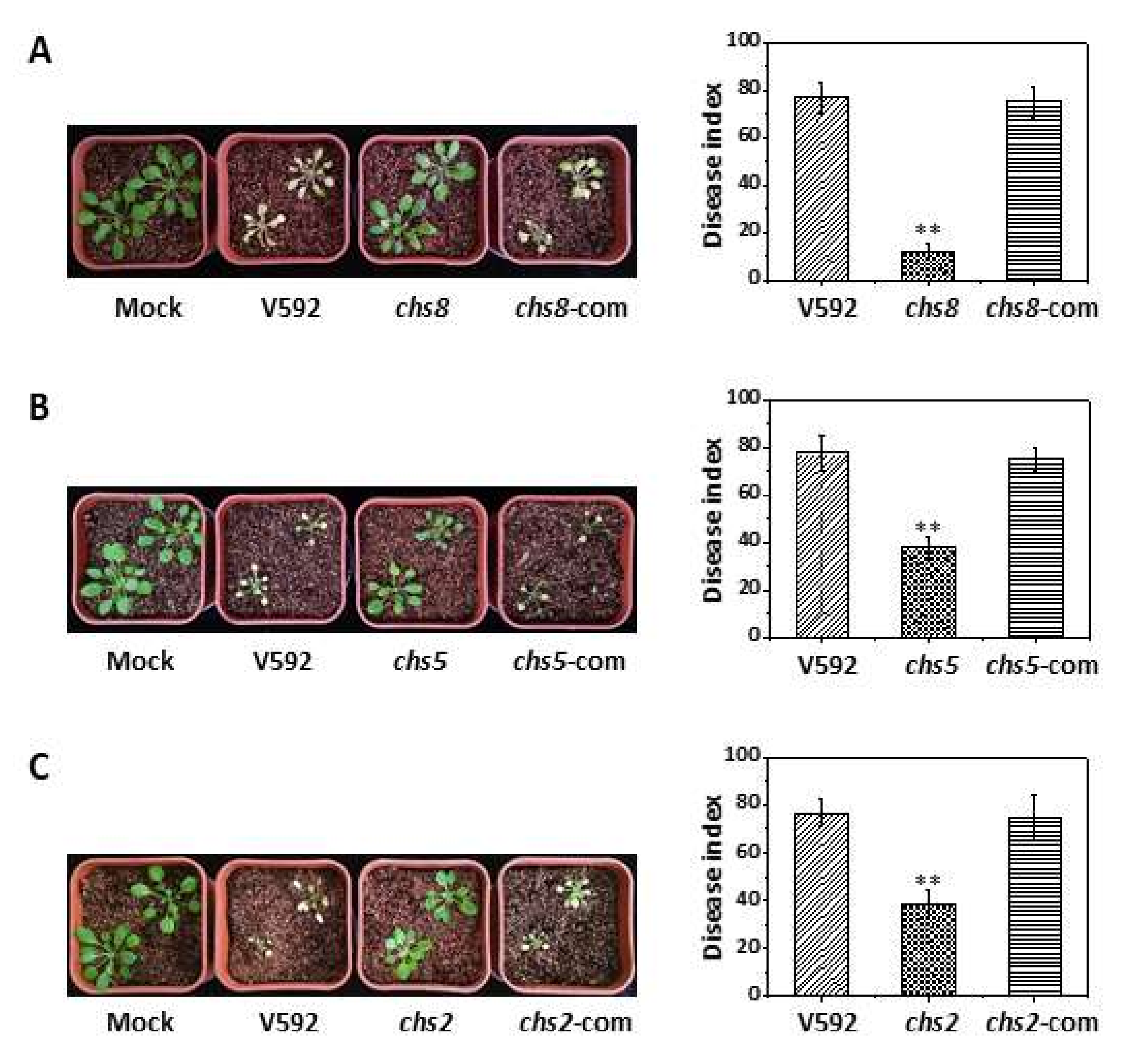
| Growth Rate (mm/Day) 1 | Germination (%) 2 | Conidiation (106 Spores/ Plate) 3 | Disease Index on Arabidopsis 4 | Disease Index on Cotton 5 | |
|---|---|---|---|---|---|
| V592 (WT) | 1.9 ± 0.1 a,* | 97.0 ± 4.5 a | 1117.5 ± 502.6 b | 67.7 ± 5.5 a | 60.5 ± 3.3 b |
| chs1 | 1.8 ± 0.1 a | 93.0 ± 6.1 a | 0.4 ± 0.4 e | 26.0 ± 2.8 de | 17.1 ± 2.0 f |
| chs2 | 1.8 ± 0.1 a | 98.0 ± 2.6 a | 535 ± 487.7 c | 16.7 ± 3.8 f | 3.9 ± 1.9 h |
| chs3 | 1.8 ± 0.1 a | 95.5 ± 5.2 a | 476.7 ± 77.7 cd | 59.4 ± 5.5 b | 66.1 ± 5.0 a |
| chs4 | 1.8 ± 0.1 a | 90.5 ± 7.4 a | 38.2 ± 32.2 de | 16.3 ± 3.2 f | 22.7 ± 3.5 e |
| chs5 | 1.8 ± 0.1 a | 44.0 ± 6.1 b | 1823.3 ± 402.5 a | 42.7 ± 5.8 c | 31.2 ± 2.8 d |
| chs6 | 1.9 ± 0.1 a | 94.5 ± 5.2 a | 210 ± 166.4 cde | 13.9 ± 3.3 f | 11 ± 3.4 g |
| chs7 | 1.9 ± 0.1 a | 96.5 ± 3.8 a | 137 ± 40.8 cde | 32.6 ± 6.1 d | 36.3 ± 2.6 cd |
| chs8 | 1.9 ± 0.1 a | 99.0 ± 2.0 a | 0.3 ± 0.2 e | 18.4 ± 5.0 ef | 37.9 ± 3.5 c |
| 0.7 M NaCl | 1.2 M Sorbitol | 0.01% SDS | 50 μg/mL CFW | 0.2 mg/mL Congo Red | |
|---|---|---|---|---|---|
| V592 (WT) | 0.39 ± 0.04 bc,* | 0.69 ± 0.06 ab | 0.55 ± 0.02 c | 0.71 ± 0.05 e | 0.40 ± 0.03 f |
| chs1 | 0.25 ± 0.02 d | 0.48 ± 0.01 de | 0.83 ± 0.04 a | 0.77 ± 0.07 cde | 0.87 ± 0.04 b |
| chs2 | 0.36 ± 0.02 c | 0.52 ± 0.07 cde | 0.77 ± 0.06 ab | 0.85 ± 0.02 c | 0.69 ± 0.01 cd |
| chs3 | 0.45 ± 0.03 a | 0.42 ± 0.04 ef | 0.73 ± 0.13 ab | 0.68 ± 0.10 e | 0.49 ± 0.07 e |
| chs4 | 0.27 ± 0.06 d | 0.54 ± 0.02 cd | 0.78 ± 0.03 ab | 0.82 ± 0.02 cd | 0.83 ± 0.03 b |
| chs5 | 0.00 ± 0.00 e | 0.11 ± 0.02 g | 0.86 ± 0.03 a | 0.74 ± 0.02 de | 0.35 ± 0.02 g |
| chs6 | 0.42 ± 0.03 ab | 0.37 ± 0.03 f | 0.68 ± 0.02 b | 0.98 ± 0.07 b | 0.71 ± 0.02 c |
| chs7 | 0.35 ± 0.01 c | 0.73 ± 0.05 a | 0.75 ± 0.05 ab | 0.75 ± 0.08 de | 0.63 ± 0.05 d |
| chs8 | 0.34 ± 0.03 c | 0.60 ± 0.12 bc | 0.66 ± 0.01 b | 1.10 ± 0.03 a | 0.98 ± 0.02 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, J.; Zhao, P.; Ye, Z.; Sun, L.; Hu, X.; Zhang, J. Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae. J. Fungi 2022, 8, 681. https://doi.org/10.3390/jof8070681
Qin J, Zhao P, Ye Z, Sun L, Hu X, Zhang J. Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae. Journal of Fungi. 2022; 8(7):681. https://doi.org/10.3390/jof8070681
Chicago/Turabian StyleQin, Jun, Peichen Zhao, Ziqin Ye, Lifan Sun, Xiaoping Hu, and Jie Zhang. 2022. "Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae" Journal of Fungi 8, no. 7: 681. https://doi.org/10.3390/jof8070681
APA StyleQin, J., Zhao, P., Ye, Z., Sun, L., Hu, X., & Zhang, J. (2022). Chitin Synthase Genes Are Differentially Required for Growth, Stress Response, and Virulence in Verticillium dahliae. Journal of Fungi, 8(7), 681. https://doi.org/10.3390/jof8070681








