Establishment of CRISPR/Cas9 Genome-Editing System Based on Dual sgRNAs in Flammulina filiformis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Selection of sgRNAs of pyrG and U6 Promoters of F. filiformis
2.3. In Vitro Cas9 Cleavage Assay
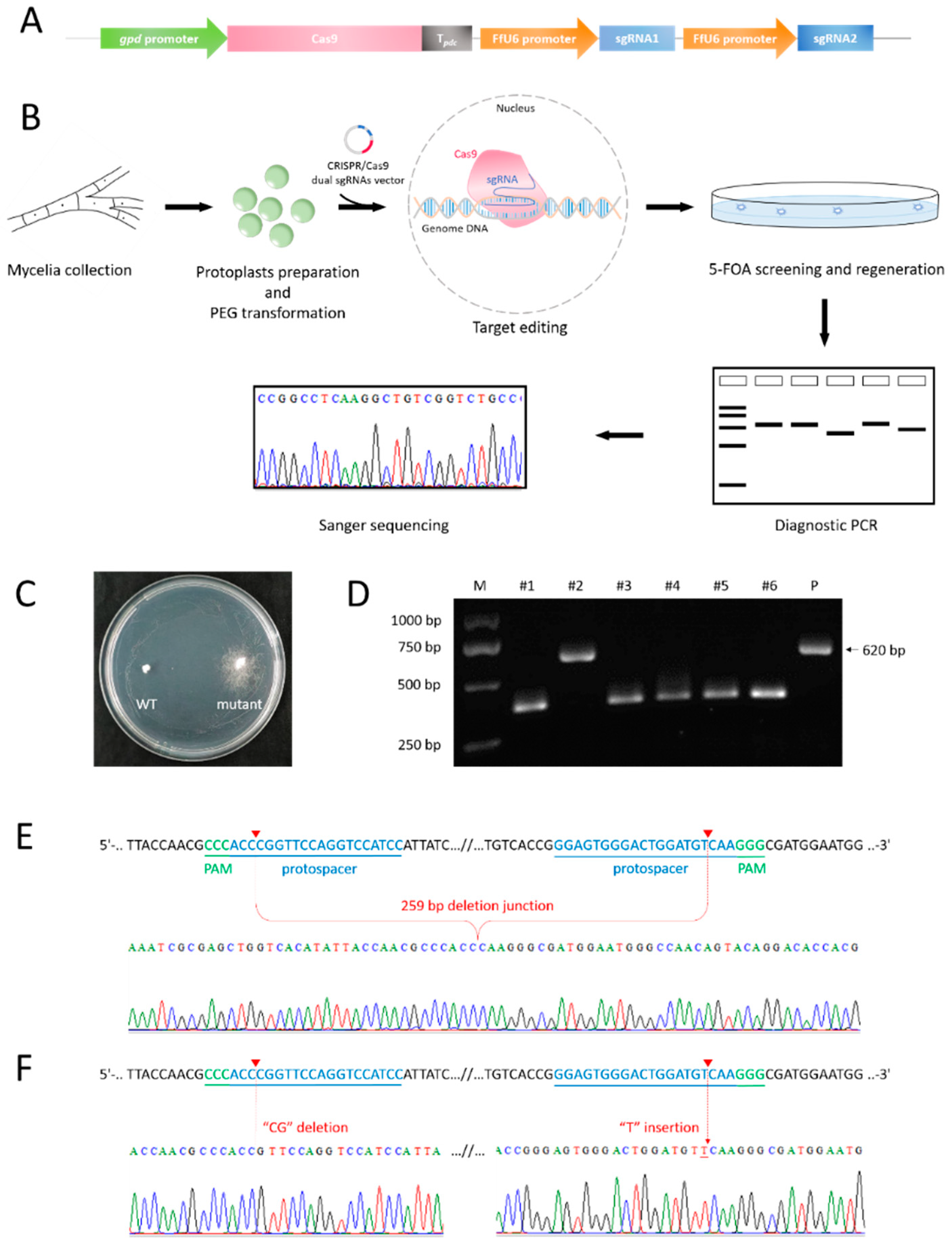
2.4. Construction of CRISPR Plasmids
2.5. PEG-Mediated Transformation of Protoplasts
2.6. Screening and Verification of Transformants
3. Results
3.1. In Vitro Cas9 Cleavage Assay
3.2. Four U6 Promoters of F. filiformis Were Identified Based on Homologous Search
3.3. Transformants Harboring pyrG Fragment Deletion or Small Indels Were Obtained
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, P.M.; Liu, X.B.; Dai, Y.C.; Horak, E.; Steffen, K.; Yang, Z.L. Phylogeny and species delimitation of Flammulina: Taxonomic status of winter mushroom in East Asia and a new European species identified using an integrated approach. Mycol. Prog. 2018, 17, 1013–1030. [Google Scholar] [CrossRef]
- Tang, C.; Hoo, P.C.-X.; Tan, L.T.-H.; Pusparajah, P.; Khan, T.M.; Lee, L.-H.; Goh, B.-H.; Chan, K.-G. Golden needle mushroom: A culinary medicine with evidenced-based biological activities and health promoting properties. Front. Pharmacol. 2016, 7, 474. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhao, Q.; Hao, X.; Wang, J.; Ma, C.; Xi, X.; Kang, W. The effect of Flammulina velutipes polysaccharide on immunization analyzed by intestinal flora and proteomics. Front. Nutr. 2022, 9, 841230. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Hu, Q.; Ma, G.; Su, A.; Xie, M.; Li, X.; Chen, G.; Zhao, L. Effects of Flammulina velutipes polysaccharide on immune response and intestinal microbiota in mice. J. Funct. Foods 2019, 56, 255–264. [Google Scholar] [CrossRef]
- Yuan, F.; Gao, Z.; Liu, W.; Li, H.; Zhang, Y.; Feng, Y.; Song, X.; Wang, W.; Zhang, J.; Huang, C.; et al. Characterization, antioxidant, anti-aging and organ protective effects of sulfated polysaccharides from Flammulina velutipes. Molecules 2019, 24, 3517. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Duan, W.; Liu, Y.; Cen, Y. Anti-inflammatory effect of the polysaccharides of golden needle mushroom in burned rats. Int. J. Biol. Macromol. 2010, 46, 100–103. [Google Scholar] [CrossRef]
- Sonnenberg, A.S.M.; Baars, J.J.P.; Gao, W.; Visser, R.G.F. Developments in breeding of Agaricus bisporus var. bisporus: Progress made and technical and legal hurdles to take. Appl. Microbiol. Biotechnol. 2017, 101, 1819–1829. [Google Scholar] [CrossRef] [Green Version]
- Tsukihara, T.; Honda, Y.; Watanabe, T.; Watanabe, T. Molecular breeding of white rot fungus Pleurotus ostreatus by homologous expression of its versatile peroxidase MnP2. Appl. Microbiol. Biotechnol. 2006, 71, 114–120. [Google Scholar] [CrossRef]
- Tu, J.-L.; Bai, X.-Y.; Xu, Y.-L.; Li, N.; Xu, J.-W. Targeted gene insertion and replacement in the basidiomycete Ganoderma lucidum by inactivation of nonhomologous end joining using CRISPR/Cas9. Appl. Environ. Microbiol. 2021, 87, e0151021. [Google Scholar] [CrossRef]
- Boontawon, T.; Nakazawa, T.; Horii, M.; Tsuzuki, M.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Functional analyses of Pleurotus ostreatus pcc1 and clp1 using CRISPR/Cas9. Fungal Genet. Biol. 2021, 154, 103599. [Google Scholar] [CrossRef]
- Jin, F.-J.; Wang, B.-T.; Wang, Z.-D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.-Q.; Liu, G.-N.; Ji, R.-Y.; Shi, K.; Song, P.; Ren, L.-J.; Huang, H.; Ji, X.-J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef]
- Song, R.; Zhai, Q.; Sun, L.; Huang, E.; Zhang, Y.; Zhu, Y.; Guo, Q.; Tian, Y.; Zhao, B.; Lu, H. CRISPR/Cas9 genome editing technology in filamentous fungi: Progress and perspective. Appl. Microbiol. Biotechnol. 2019, 103, 6919–6932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Yue, S.; Jin, Y.; Wei, H.; Lu, L. Advances allowing feasible pyrG gene editing by a CRISPR-Cas9 system for the edible mushroom Pleurotus eryngii. Fungal Genet. Biol. 2021, 147, 103509. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Xiao, H.; Zou, G.; Zhou, Z.; Zhong, J.-J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 2017, 56, 57–61. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Boontawon, T.; Nakazawa, T.; Inoue, C.; Osakabe, K.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Efficient genome editing with CRISPR/Cas9 in Pleurotus ostreatus. AMB Express 2021, 11, 30. [Google Scholar] [CrossRef]
- Moon, S.; An, J.Y.; Choi, Y.-J.; Oh, Y.-L.; Ro, H.-S.; Ryu, H. Construction of a CRISPR/Cas9-mediated genome editing system in Lentinula edodes. Mycobiology 2021, 49, 599–603. [Google Scholar] [CrossRef]
- Xue, L.-J.; Tsai, C.-J. AGEseq: Analysis of genome editing by sequencing. Mol. Plant 2015, 8, 1428–1430. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, A.; Plaisier, C.L.; Carroll, D.; Drews, G.N. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 2232–2237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Kraft, K.; Geuer, S.; Will, A.J.; Chan, W.L.; Paliou, C.; Borschiwer, M.; Harabula, I.; Wittler, L.; Franke, M.; Ibrahim, D.M.; et al. Deletions, inversions, duplications: Engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 2015, 10, 833–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Sun, B.; You, H.; Tu, J.; Yu, X.; Zhao, P.; Xu, J. Dual sgRNA-directed gene deletion in basidiomycete Ganoderma lucidum using the CRISPR/Cas9 system. Microb. Biotechnol. 2020, 13, 386–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, R.; Lin, J.-F.; Guo, L.-Q.; Ye, Z.-W.; Guo, T.-F.; Yun, F. Construction of Flammulina velutipes genome editing vector by using CRISPR/Cas9 system. Sci. Technol. Food Ind. 2016, 37, 230–234. [Google Scholar] [CrossRef]
- Lin, J.-D.; Yang, X.-Q.; Wei, T.; Guo, L.-Q.; Lin, J.-F.; Chen, Y.-S.; Huang, S.-S. Construction and transformation of CRISPR/Cas9 genome editing vector of Flammulina filiformis G protein-coupled receptor gene. Mycosystema 2019, 38, 349–361. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.; Herrera-Carrillo, E.; Berkhout, B. RNA polymerase II activity of type 3 Pol III promoters. Mol. Ther.-Nucleic Acids 2018, 12, 135–145. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Zhang, C.; Meng, X.; Wei, X.; Lu, L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 2016, 86, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Xiao, M.; Chai, S.; Zhu, Z.; Wang, Y.; Zhou, Z. Efficient genome editing in filamentous fungi via an improved CRISPR-Cas9 ribonucleoprotein method facilitated by chemical reagents. Microb. Biotechnol. 2021, 14, 2343–2355. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terns, M.P. CRISPR-based technologies: Impact of RNA-targeting systems. Mol. Cell 2018, 72, 404–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.Y.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Zheng, X.; Qi, Y.; Zhang, D.; Cheng, Y.; Tang, A.; Voytas, D.F.; Zhang, Y. A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol. Plant 2016, 9, 1088–1091. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhao, L.; Gao, Y.; Xu, J.; Han, R. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res. 2016, 45, e28. [Google Scholar] [CrossRef]
- Wang, P.-A.; Xiao, H.; Zhong, J.-J. CRISPR-Cas9 assisted functional gene editing in the mushroom Ganoderma lucidum. Appl. Microbiol. Biotechnol. 2020, 104, 1661–1671. [Google Scholar] [CrossRef]
- Sugano, S.S.; Suzuki, H.; Shimokita, E.; Chiba, H.; Noji, S.; Osakabe, Y.; Osakabe, K. Genome editing in the mushroom-forming basidiomycete Coprinopsis cinerea, optimized by a high-throughput transformation system. Sci. Rep. 2017, 7, 1260. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; An, C.; Shu, X.; Meng, X.; Yao, Y.; Zhang, J.; Chen, F.; Xiang, H.; Yang, S.; Gao, X.; et al. A dual-plasmid CRISPR/Cas system for mycotoxin elimination in polykaryotic industrial fungi. ACS Synth. Biol. 2020, 9, 2087–2095. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, J. Genome editing technology and its application potentials in the industrial filamentous fungus Aspergillus oryzae. J. Fungi 2021, 7, 638. [Google Scholar] [CrossRef] [PubMed]
- Boontawon, T.; Nakazawa, T.; Xu, H.; Kawauchi, M.; Sakamoto, M.; Honda, Y. Gene targeting using pre-assembled Cas9 ribonucleoprotein and split-marker recombination in Pleurotus ostreatus. FEMS Microbiol. Lett. 2021, 368, fnab080. [Google Scholar] [CrossRef]
- Vonk, P.J.; Escobar, N.; Wösten, H.A.B.; Lugones, L.G.; Ohm, R.A. High-throughput targeted gene deletion in the model mushroom Schizophyllum commune using pre-assembled Cas9 ribonucleoproteins. Sci. Rep. 2019, 9, 7632. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Name | Sequence (5′–3′) | Descriptions |
|---|---|---|
| FfpyrG-vitro-F1 | ATGGCACATATCCTCAACTTTG | Amplification of pyrG for in vitro cleavage assay |
| FfpyrG-vitro-R1 | TCATGCTGTTCTCTCCAAGTATG | |
| FfpyrG-vitro-F2 | ACGCCGCTCGCGCCTCAAAACATTC | Amplification of pyrG for in vitro cleavage assay |
| FfpyrG-vitro-R2 | TGGCCCATTCCATCGCCCTTGAC | |
| FfU6-1-F1 | TCGGGAAGAGCAGAGCGGGCAAGTTATCAACAAGCGTG | Amplification of FfU6-1 promoter |
| FfU6-1-R1 | TTGACATCCAGTCCCACTCCATTAAACGATGAAAAGGAGACATC | |
| pyrG-sgRNA1-F | GGAGTGGGACTGGATGTCAAGTTTTAGAGCTAGAAATAGCAAG | Amplification of sgRNA1 |
| pyrG-sgRNA1-R | TCTAAAACAAAAAAGCACCGACTCGGTG | |
| FfU6-1-F2 | GTCGGTGCTTTTTTGTTTTAGAGGGCAAGTTATCAACAAGCGTG | Amplification of FfU6-1 promoter |
| FfU6-1-R2 | ACCCGGTTCCAGGTCCATCCATTAAACGATGAAAAGGAGACATC | |
| pyrG-sgRNA2-F | GGATGGACCTGGAACCGGGTGTTTTAGAGCTAGAAATAGCAAG | Amplification of sgRNA2 |
| pyrG-sgRNA2-R | GGATCCTCTAGAGATGCGGCCGCTCTAAAACAAAAAAGCACCGAC | |
| FfU6-3-F1 | TCGGGAAGAGCAGAGCGTATACGGCTTAATAGCCCTCTT | Amplification of FfU6-3 promoter |
| FfU6-3-R1 | TTGACATCCAGTCCCACTCCATAAGCCTGTGGAAGAGAGGC | |
| FfU6-3-F2 | GTCGGTGCTTTTTTGTTTTAGACGTATACGGCTTAATAGCCCTCTT | Amplification of FfU6-3 promoter |
| FfU6-3-R2 | ACCCGGTTCCAGGTCCATCCATAAGCCTGTGGAAGAGAGGC | |
| FfpyrG-chk-F | CAACTTTGCGAGCATAGACCC | Amplification of pyrG for sequencing |
| FfpyrG-chk-R | AATAATCCGTCCCATACACCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Dong, J.; Liao, J.; Tian, L.; Qiu, H.; Wu, T.; Ge, F.; Zhu, J.; Shi, L.; Jiang, A.; et al. Establishment of CRISPR/Cas9 Genome-Editing System Based on Dual sgRNAs in Flammulina filiformis. J. Fungi 2022, 8, 693. https://doi.org/10.3390/jof8070693
Liu X, Dong J, Liao J, Tian L, Qiu H, Wu T, Ge F, Zhu J, Shi L, Jiang A, et al. Establishment of CRISPR/Cas9 Genome-Editing System Based on Dual sgRNAs in Flammulina filiformis. Journal of Fungi. 2022; 8(7):693. https://doi.org/10.3390/jof8070693
Chicago/Turabian StyleLiu, Xiaotian, Jianghan Dong, Jian Liao, Li Tian, Hao Qiu, Tao Wu, Feng Ge, Jing Zhu, Liang Shi, Ailiang Jiang, and et al. 2022. "Establishment of CRISPR/Cas9 Genome-Editing System Based on Dual sgRNAs in Flammulina filiformis" Journal of Fungi 8, no. 7: 693. https://doi.org/10.3390/jof8070693







