Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Candida albicans Sample Preparation and Quantification
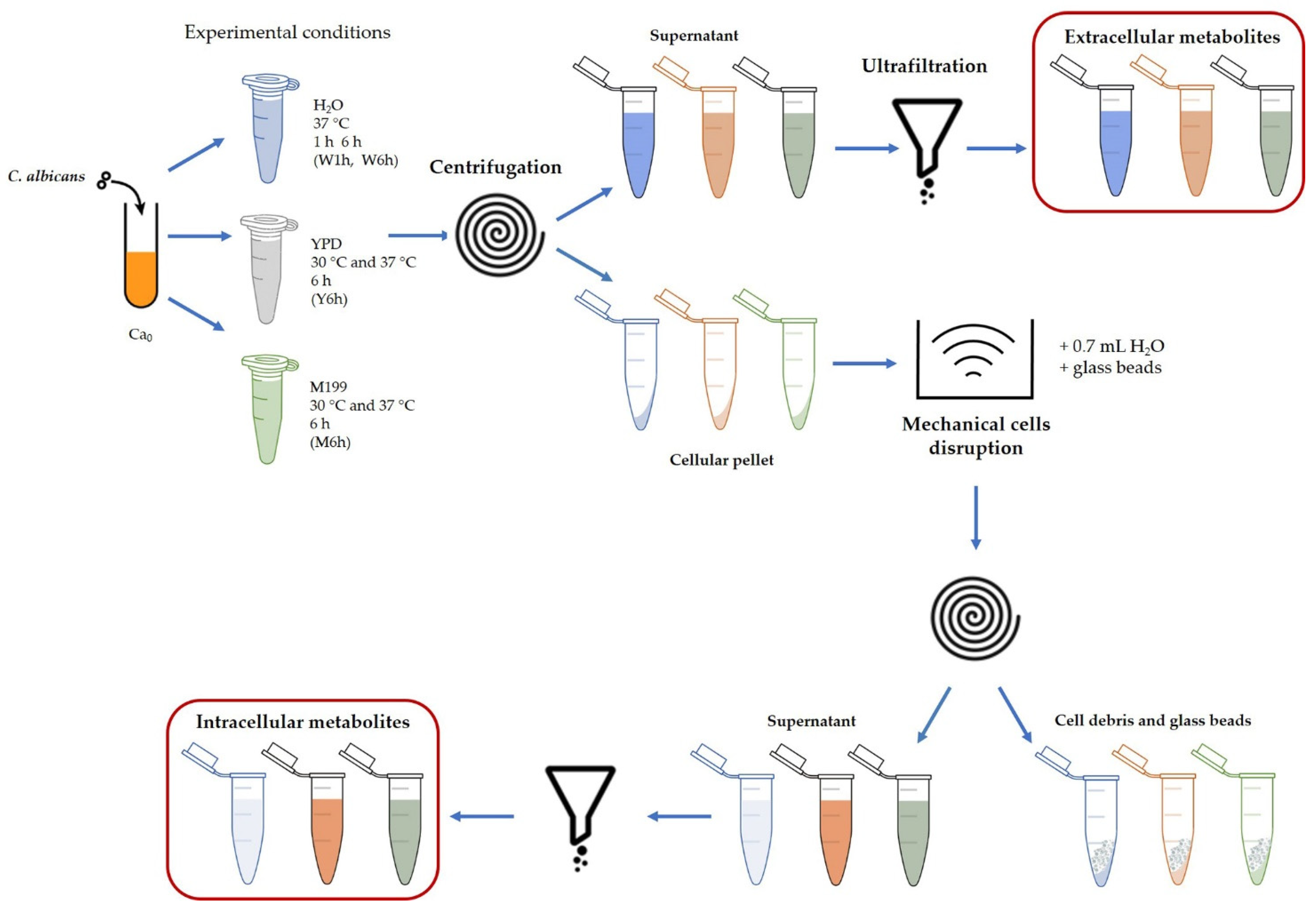
2.2. Culture Conditions and Metabolite Collection
2.2.1. Culture Conditions
2.2.2. Metabolite Collection
2.3. Sample Preparation for 1H-NMR Measurements
2.4. 1H-NMR Experiments and Analysis
2.5. Metabolite Concentrations and Statistical Analysis
3. Results
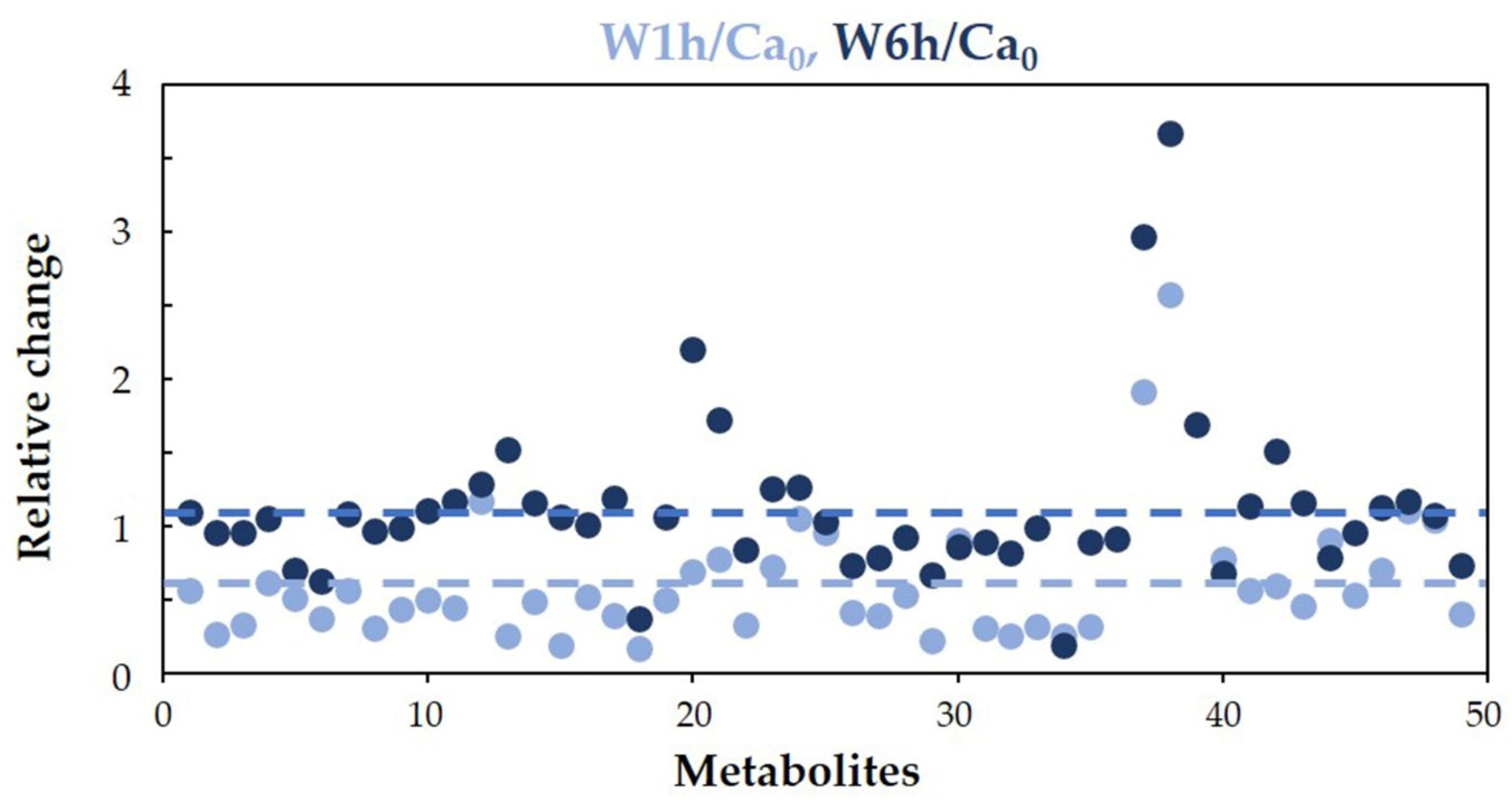
3.1. Overview of C. albicans Metabolome in Different Culture Conditions
3.2. The Metabolome of C. albicans in Water, YPD, and M199
3.2.1. Intracellular Metabolites of C. albicans Incubated in Water at 37 °C
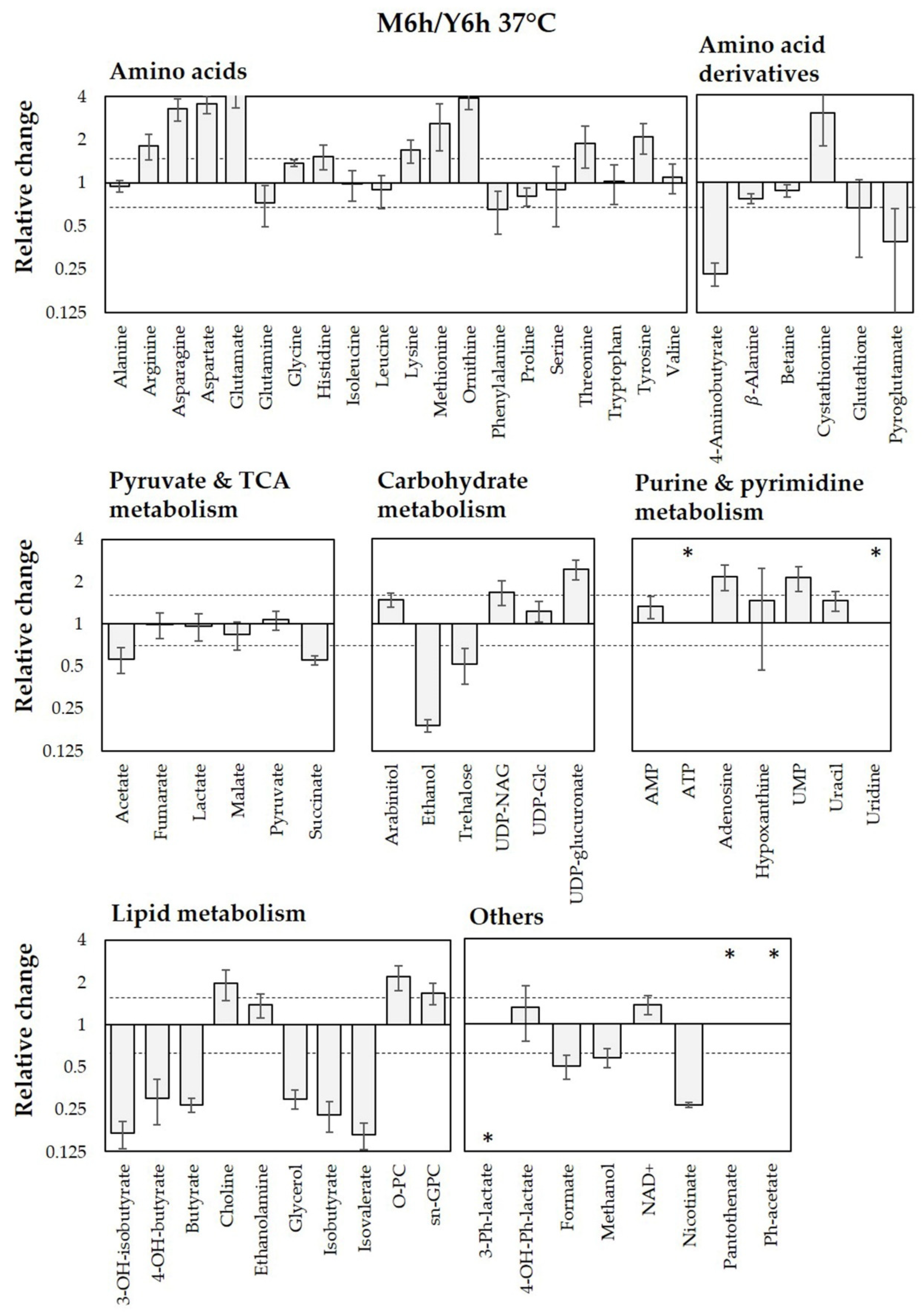
3.2.2. Intracellular Metabolites of C. albicans Incubated in YPD and M199
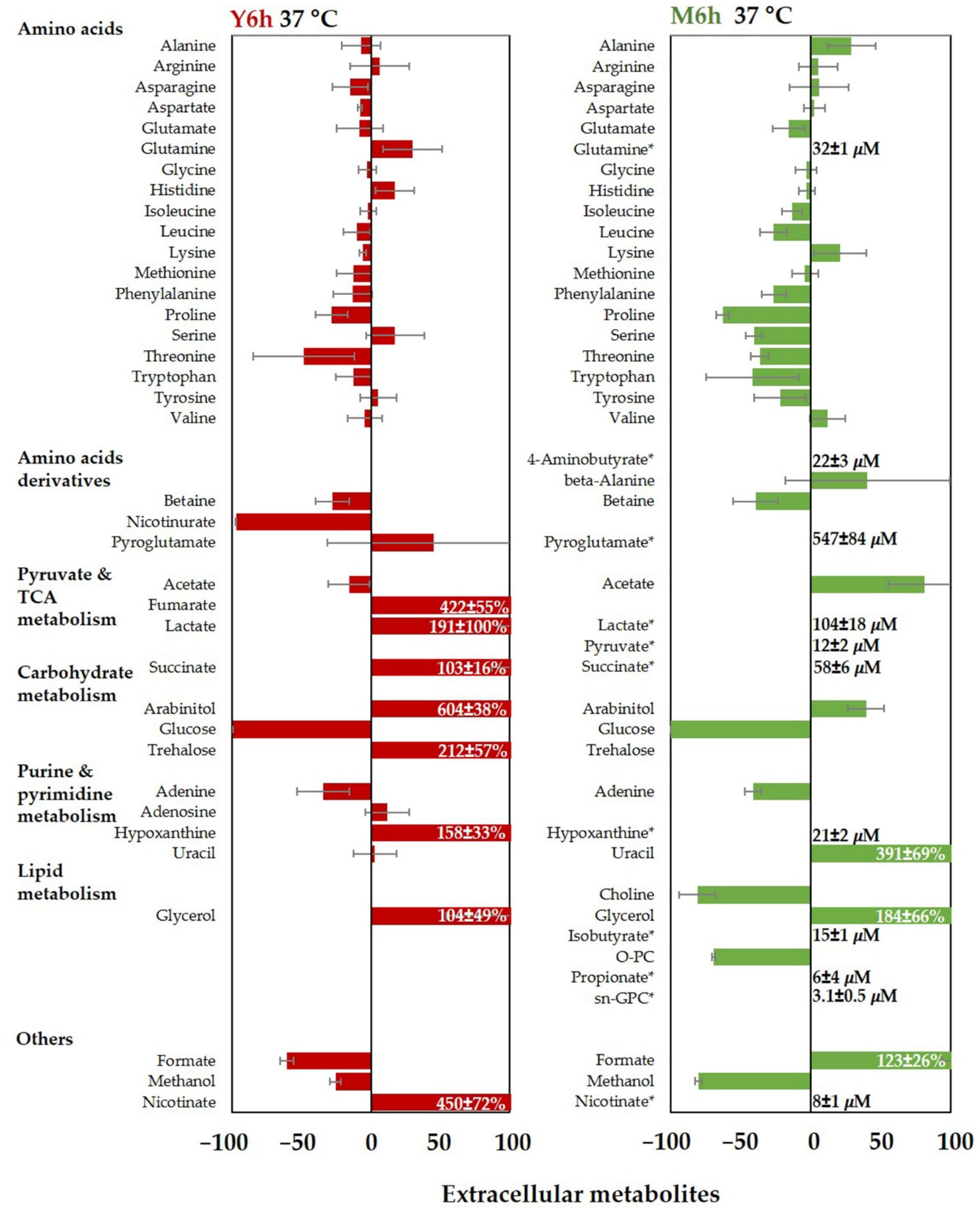
3.2.3. Extracellular Metabolites of C. albicans in Water, YPD, and M199 at 37 °C
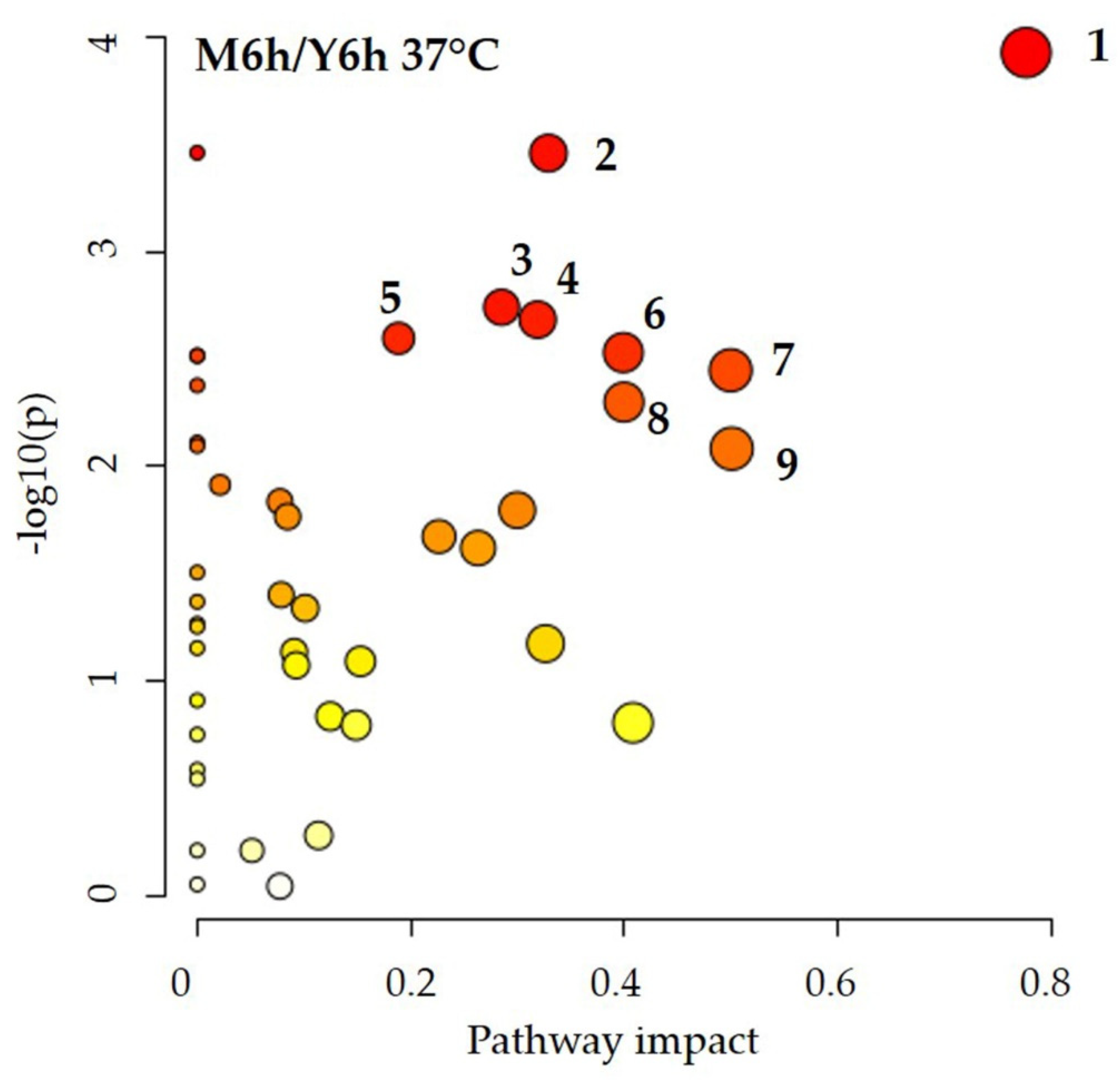
3.3. Candida albicans Energy Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhattacharya, S.; Sae-Tia, S.; Fries, B.C. Candidiasis and Mechanisms of Antifungal Resistance. Antibiotics 2020, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, C.; Solis, N.V.; Filler, S.G.; Liu, H. Synergistic regulation of hyphal elongation by hypoxia, CO(2), and nutrient conditions controls the virulence of Candida albicans. Cell Host Microbe 2013, 14, 499–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandra, S.; Linde, J.; Brock, M.; Guthke, R.; Hube, B.; Brunke, S. Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS ONE 2014, 9, e92734. [Google Scholar] [CrossRef] [Green Version]
- Alves, R.; Barata-Antunes, C.; Casal, M.; Brown, A.J.P.; Van Dijck, P.; Paiva, S. Adapting to survive: How Candida overcomes host-imposed constraints during human colonization. PLoS Pathog. 2020, 16, e1008478. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.J.; Brown, G.D.; Netea, M.G.; Gow, N.A. Metabolism impacts upon Candida immunogenicity and pathogenicity at multiple levels. Trends Microbiol. 2014, 22, 614–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudbery, P.; Gow, N.; Berman, J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004, 12, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Sudbery, P.E. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 2011, 9, 737–748. [Google Scholar] [CrossRef]
- Kornitzer, D. Regulation of Candida albicans Hyphal Morphogenesis by Endogenous Signals. J. Fungi 2019, 5, 21. [Google Scholar] [CrossRef] [Green Version]
- Rooney, P.J.; Klein, B.S. Linking fungal morphogenesis with virulence. Cell. Microbiol. 2002, 4, 127–137. [Google Scholar] [CrossRef]
- Bendel, C.M.; Hess, D.J.; Garni, R.M.; Henry-Stanley, M.; Wells, C.L. Comparative virulence of Candida albicans yeast and filamentous forms in orally and intravenously inoculated mice. Crit. Care Med. 2003, 31, 501–507. [Google Scholar] [CrossRef]
- Whiteway, M.; Oberholzer, U. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 2004, 7, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Kumamoto, C.A.; Vinces, M.D. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 2005, 7, 1546–1554. [Google Scholar] [CrossRef] [PubMed]
- Pukkila-Worley, R.; Peleg, A.Y.; Tampakakis, E.; Mylonakis, E. Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot. Cell 2009, 8, 1750–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desai, J.V. Candida albicans Hyphae: From Growth Initiation to Invasion. J. Fungi 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Rubin-Bejerano, I.; Fraser, I.; Grisafi, P.; Fink, G.R. Phagocytosis by neutrophils induces an amino acid deprivation response in Saccharomyces cerevisiae and Candida albicans. Proc. Natl. Acad. Sci. USA 2003, 100, 11007–11012. [Google Scholar] [CrossRef] [Green Version]
- Oliver, J.C.; Ferreira, C.B.R.J.; Silva, N.C.; Dias, A.L.T. Candida spp. and phagocytosis: Multiple evasion mechanisms. Antonie Van Leeuwenhoek 2019, 112, 1409–1423. [Google Scholar] [CrossRef]
- Dabrowa, N.; Taxer, S.S.; Howard, D.H. Germination of Candida albicans induced by proline. Infect. Immun. 1976, 13, 830–835. [Google Scholar] [CrossRef] [Green Version]
- Holmes, A.R.; Shepherd, M.G. Proline-induced Germ-tube Formation in Candida albicans: Role of Proline Uptake and Nitrogen Metabolism. J. Gen. Microbiol. 1987, 133, 3219–3228. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, S.G.; Shafiq, A.; Hakim, S.T.; Anjum, Y.; Kazm, S.U. Effect of Growth Media, pH and Temperature on Yeast to Hyphal Transition in Candida albicans. Open J. Med. Microbiol. 2013, 3, 185–192. [Google Scholar] [CrossRef] [Green Version]
- Burgain, A.; Tebbji, F.; Khemiri, I.; Sellam, A. Metabolic Reprogramming in the Opportunistic Yeast Candida albicans in Response to Hypoxia. mSphere 2020, 5, e00913–e00919. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, R.S.; Ryan, O.; Boone, C.; Cowen, L.E. Regulatory circuitry governing morphogenesis in Saccharomyces cerevisiae and Candida albicans. Cell Cycle 2012, 11, 4294–4295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, T.-L.; Cannon, R.D.; Gallo, S.M.; Villas-Bôas, S.G. A metabolomic study of the effect of Candida albicans glutamate dehydrogenase deletion on growth and morphogenesis. NPJ Biofilms Microbiomes 2019, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, J.; Rosiana, S.; Razzaq, I.; Shapiro, R.S. Linking Cellular Morphogenesis with Antifungal Treatment and Susceptibility in Candida Pathogens. J. Fungi 2019, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Zhou, X.; Ren, B.; Cheng, L. The regulation of hyphae growth in Candida albicans. Virulence 2020, 11, 337–348. [Google Scholar] [CrossRef] [Green Version]
- Villa, S.; Hamideh, M.; Weinstock, A.; Qasim, M.N.; Hazbun, T.R.; Sellam, A.; Hernday, A.D.; Thangamani, S. Transcriptional control of hyphal morphogenesis in Candida albicans. FEMS Yeast Res. 2020, 20, foaa005. [Google Scholar] [CrossRef]
- Hollywood, K.; Brison, D.R.; Goodacre, R. Metabolomics: Current technologies and future trends. Proteomics 2006, 6, 4716–4723. [Google Scholar] [CrossRef]
- Grivet, J.P. NMR and microorganisms. Curr. Issues Mol. Biol. 2001, 3, 7–14. [Google Scholar] [CrossRef] [Green Version]
- Airoldi, C.; Tripodi, F.; Guzzi, C.; Nicastro, R.; Coccetti, P. NMR analysis of budding yeast metabolomics: A rapid method for sample preparation. Mol. BioSyst. 2015, 11, 379–383. [Google Scholar] [CrossRef]
- Patejko, M.; Jacyna, J.; Markuszewski, M.J. Sample preparation procedures utilized in microbial metabolomics: An overview. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017, 1043, 150–157. [Google Scholar] [CrossRef]
- Coen, M.; Bodkin, J.; Power, D.; Bubb, W.A.; Himmelreich, U.; Kuchel, P.W.; Sorrell, T.C. Antifungal effects on metabolite profiles of medically important yeast species measured by nuclear magnetic resonance spectroscopy. Antimicrob. Agents Chemother. 2006, 50, 4018–4026. [Google Scholar] [CrossRef] [Green Version]
- Oberhardt, M.A.; Palsson, B.Ø.; Papin, J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 2009, 5, 320. [Google Scholar] [CrossRef] [PubMed]
- Pawar, B.; Kanyalkar, M.; Srivastava, S. Search for novel antifungal agents by monitoring fungal metabolites in presence of synthetically designed fluconazole derivatives using NMR spectroscopy. Biochim. Biophys. Acta (BBA)-Biomembr. 2010, 1798, 2067–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.N.; Kim, J.; Kim, J.; Jung, W.H.; Lee, C.H. Influence of Iron Regulation on the Metabolome of Cryptococcus neoformans. PLoS ONE 2012, 7, e41654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caudy, A.A.; Mülleder, M.; Ralser, M. Metabolomics in Yeast. Cold Spring Harb. Protoc. 2017, 2017, pdb.top083576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Liao, Z.; Yang, Y.; Lv, L.; Cao, Y.; Zhu, Z. Metabolomic profiling for the identification of potential biomarkers involved in a laboratory azole resistance in Candida albicans. PLoS ONE 2018, 13, e0192328. [Google Scholar] [CrossRef]
- Müller, C.; Neugebauer, T.; Zill, P.; Lass-Flörl, C.; Bracher, F.; Binder, U. Sterol Composition of Clinically Relevant Mucorales and Changes Resulting from Posaconazole Treatment. Molecules 2018, 23, 1218. [Google Scholar] [CrossRef] [Green Version]
- Castillo, S.; Patil, K.R.; Jouhten, P. Yeast Genome-Scale Metabolic Models for Simulating Genotype-Phenotype Relations. Prog. Mol. Subcell. Biol. 2019, 58, 111–133. [Google Scholar] [CrossRef]
- Chaleckis, R.; Ohashi, K.; Meister, I.; Naz, S.; Wheelock, C.E. Metabolomic Analysis of Yeast and Human Cells: Latest Advances and Challenges. Methods Mol. Biol. 2019, 2049, 233–245. [Google Scholar] [CrossRef]
- Semreen, M.H.; Soliman, S.S.M.; Saeed, B.Q.; Alqarihi, A.; Uppuluri, P.; Ibrahim, A.S. Metabolic Profiling of Candida auris, a Newly-Emerging Multi-Drug Resistant Candida Species, by GC-MS. Molecules 2019, 24, 399. [Google Scholar] [CrossRef] [Green Version]
- Sailwal, M.; Das, A.J.; Gazara, R.K.; Dasgupta, D.; Bhaskar, T.; Hazra, S.; Ghosh, D. Connecting the dots: Advances in modern metabolomics and its application in yeast system. Biotechnol. Adv. 2020, 44, 107616. [Google Scholar] [CrossRef]
- McClelland, E.E.; Ramagopal, U.A.; Rivera, J.; Cox, J.; Nakouzi, A.; Prabu, M.M.; Almo, S.C.; Casadevall, A. A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals. PLoS Pathog. 2016, 12, e1005849. [Google Scholar] [CrossRef]
- Savelieff, M.G.; Pappalardo, L. Novel cutting-edge metabolite-based diagnostic tools for aspergillosis. PLoS Pathog. 2017, 13, e1006486. [Google Scholar] [CrossRef] [Green Version]
- Vadlapudi, V.; Borah, N.; Yellusani, K.R.; Gade, S.; Reddy, P.; Rajamanikyam, M.; Vempati, L.N.S.; Gubbala, S.P.; Chopra, P.; Upadhyayula, S.M.; et al. Aspergillus Secondary Metabolite Database, a resource to understand the Secondary metabolome of Aspergillus genus. Sci. Rep. 2017, 7, 7325. [Google Scholar] [CrossRef] [Green Version]
- Ząbek, A.; Klimek-Ochab, M.; Jawień, E.; Młynarz, P. Biodiversity in targeted metabolomics analysis of filamentous fungal pathogens by 1H NMR-based studies. World J. Microbiol. Biotechnol. 2017, 33, 132. [Google Scholar] [CrossRef] [Green Version]
- Brandt, P.; Garbe, E.; Vylkova, S. Catch the wave: Metabolomic analyses in human pathogenic fungi. PLoS Pathog. 2020, 16, e1008757. [Google Scholar] [CrossRef]
- Oliver, J.C.; Laghi, L.; Parolin, C.; Foschi, C.; Marangoni, A.; Liberatore, A.; Dias, A.L.T.; Cricca, M.; Vitali, B. Metabolic profiling of Candida clinical isolates of different species and infection sources. Sci. Rep. 2020, 10, 16716. [Google Scholar] [CrossRef]
- Han, T.-l.; Cannon, R.D.; Villas-Bôas, S.G. Metabolome analysis during the morphological transition of Candida albicans. Metabolomics 2012, 8, 1204–1217. [Google Scholar] [CrossRef]
- Han, T.-L.; Tumanov, S.; Cannon, R.D.; Villas-Boas, S.G. Metabolic Response of Candida albicans to Phenylethyl Alcohol under Hyphae-Inducing Conditions. PLoS ONE 2013, 8, e71364. [Google Scholar] [CrossRef]
- Kamthan, M.; Kamthan, A.; Ruhela, D.; Maiti, P.; Bhavesh, N.S.; Datta, A. Upregulation of galactose metabolic pathway by N-acetylglucosamine induced endogenous synthesis of galactose in Candida albicans. Fungal Genet. Biol. 2013, 54, 15–24. [Google Scholar] [CrossRef]
- Liboro, K.; Yu, S.-R.; Lim, J.; So, Y.-S.; Bahn, Y.-S.; Eoh, H.; Park, H. Transcriptomic and Metabolomic Analysis Revealed Roles of Yck2 in Carbon Metabolism and Morphogenesis of Candida albicans. Front. Cell. Infect. Microbiol. 2021, 11, 636834. [Google Scholar] [CrossRef]
- Schaller, M.; Zakikhany, K.; Naglik, J.R.; Weindl, G.; Hube, B. Models of oral and vaginal candidiasis based on in vitro reconstituted human epithelia. Nat. Protoc. 2006, 1, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, M.D.; Beynon, R.J.; Gethings, L.A.; Claydon, A.J.; Langridge, J.I.; Vissers, J.P.C.; Brown, A.J.P.; Hammond, D.E. Specificity of the osmotic stress response in Candida albicans highlighted by quantitative proteomics. Sci. Rep. 2018, 8, 14492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulain, D. Candida albicans, plasticity and pathogenesis. Crit. Rev. Microbiol. 2015, 41, 208–217. [Google Scholar] [CrossRef]
- Chaieb, K.; Kouidhi, B.; Zmantar, T.; Mahdouani, K.; Bakhrouf, A. Starvation survival of Candida albicans in various water microcosms. J. Basic Microbiol. 2011, 51, 357–363. [Google Scholar] [CrossRef]
- Almshawit, H.; Pouniotis, D.; Macreadie, I. Cell density impacts on Candida glabrata survival in hypo-osmotic stress. FEMS Yeast Res. 2014, 14, 508–516. [Google Scholar] [CrossRef] [Green Version]
- D’Enfert, C.; Kaune, A.K.; Alaban, L.R.; Chakraborty, S.; Cole, N.; Delavy, M.; Kosmala, D.; Marsaux, B.; Fróis-Martins, R.; Morelli, M.; et al. The impact of the Fungus-Host-Microbiota interplay upon Candida albicans infections: Current knowledge and new perspectives. FEMS Microbiol. Rev. 2021, 45, fuaa060. [Google Scholar] [CrossRef]
- Kayingo, G.; Wong, B. The MAP kinase Hog1p differentially regulates stress-induced production and accumulation of glycerol and d-arabitol in Candida albicans. Microbiology 2005, 151, 2987–2999. [Google Scholar] [CrossRef] [Green Version]
- Han, T.L.; Cannon, R.D.; Villas-Boas, S.G. The metabolic response of Candida albicans to farnesol under hyphae-inducing conditions. FEMS Yeast Res. 2012, 12, 879–889. [Google Scholar] [CrossRef] [Green Version]
- Garbe, E.; Vylkova, S. Role of Amino Acid Metabolism in the Virulence of Human Pathogenic Fungi. Curr. Clin. Micro. Rpt. 2019, 6, 108–119. [Google Scholar] [CrossRef] [Green Version]


| Medium | Glucose | Ethanol (mM) 6 h—30 °C | Ethanol (mM) 6 h—37 °C | ||||
|---|---|---|---|---|---|---|---|
| (mM) | Extracellular | Intracellular # | Production § | Extracellular | Intracellular # | Production § | |
| water | - | 3.6 | 1.1 | ||||
| YPD | 78.6 | 159.6 | 6.2 | 105.5% | 168.0 | 6.5 | 111.0% |
| M199 | 4.7 | 8.2 | 2.1 | 109.6% | 9.7 | 2.0 | 124.5% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallo, M.; Giovati, L.; Magliani, W.; Pertinhez, T.A.; Conti, S.; Ferrari, E.; Spisni, A.; Ciociola, T. Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions. J. Fungi 2022, 8, 723. https://doi.org/10.3390/jof8070723
Gallo M, Giovati L, Magliani W, Pertinhez TA, Conti S, Ferrari E, Spisni A, Ciociola T. Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions. Journal of Fungi. 2022; 8(7):723. https://doi.org/10.3390/jof8070723
Chicago/Turabian StyleGallo, Mariana, Laura Giovati, Walter Magliani, Thelma A. Pertinhez, Stefania Conti, Elena Ferrari, Alberto Spisni, and Tecla Ciociola. 2022. "Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions" Journal of Fungi 8, no. 7: 723. https://doi.org/10.3390/jof8070723
APA StyleGallo, M., Giovati, L., Magliani, W., Pertinhez, T. A., Conti, S., Ferrari, E., Spisni, A., & Ciociola, T. (2022). Metabolic Plasticity of Candida albicans in Response to Different Environmental Conditions. Journal of Fungi, 8(7), 723. https://doi.org/10.3390/jof8070723











