Organization and Unconventional Integration of the Mating-Type Loci in Morchella Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Species and Strains
2.2. PCR Amplification and Sequencing of The Mating Idiomorphs
2.3. Transcriptional Analyses of The Mating Genes
2.4. Comparison of The MAT Loci
3. Results
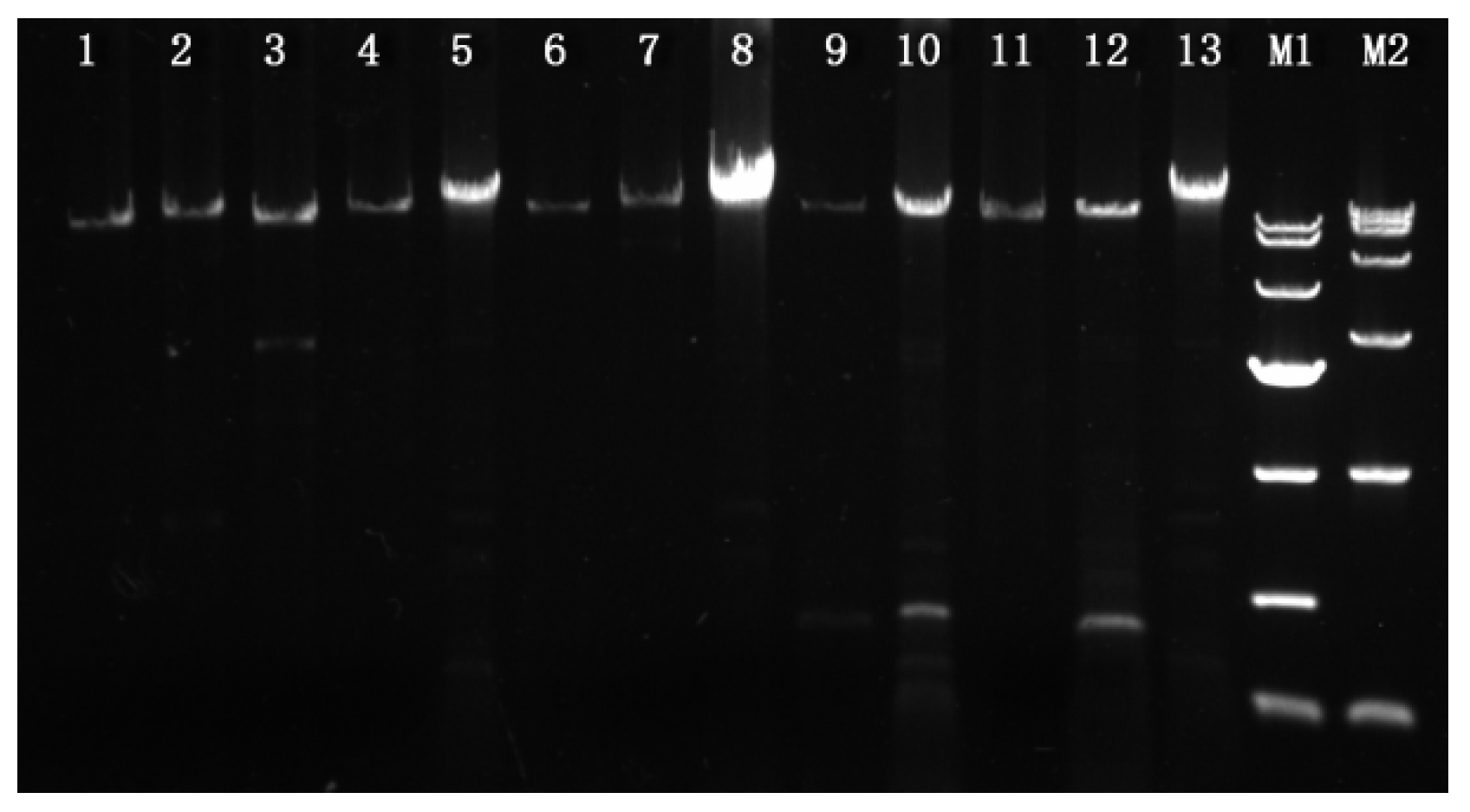
3.1. PCR Amplifications of The Mating Idiomorphs
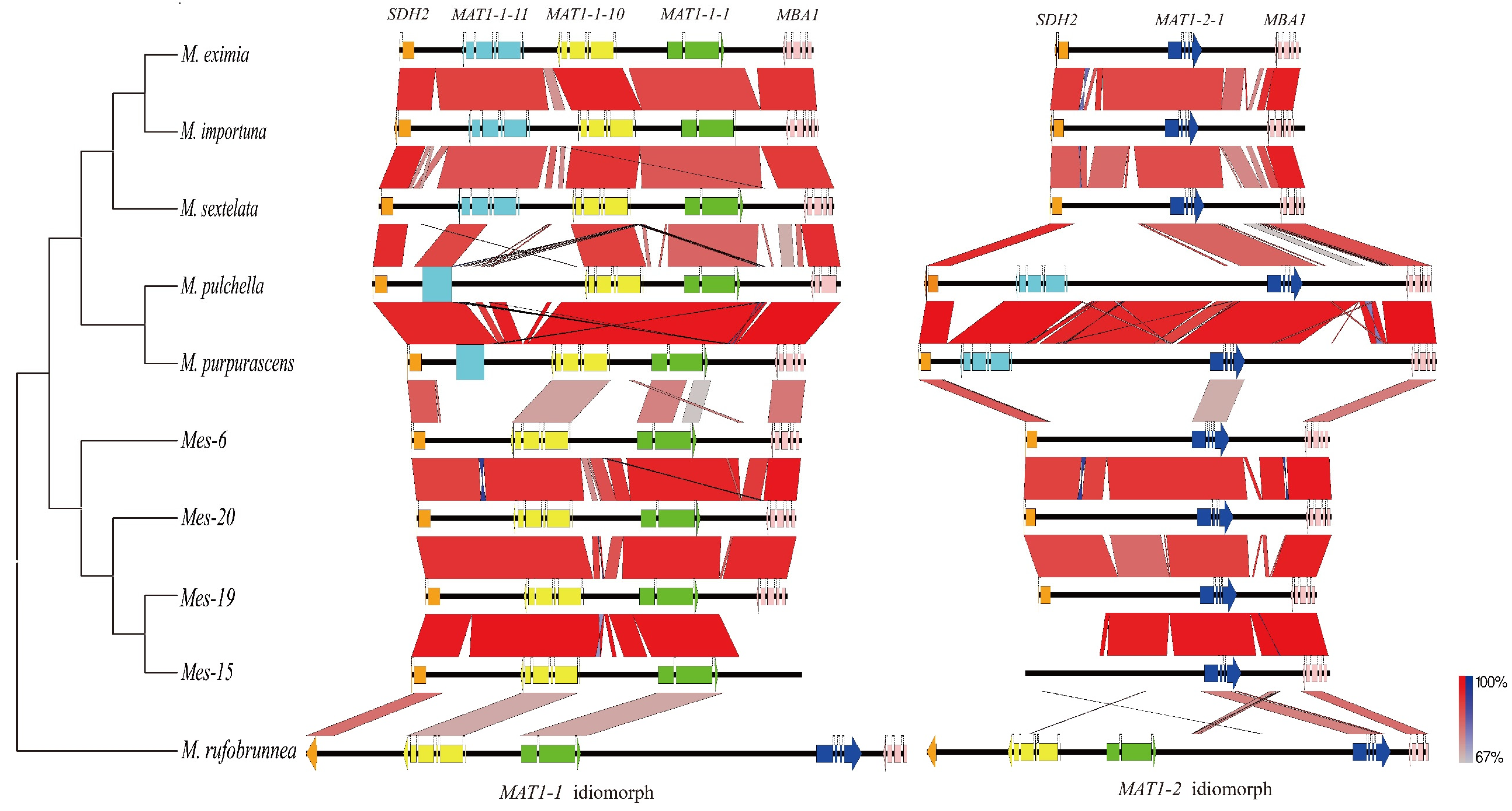
3.2. Organization of The Mating Idiomorphs
3.3. Unconventional Integration of The MAT Loci in M. purpurascena and M. pulchella
3.4. Transcription Analyses of The Mating Genes
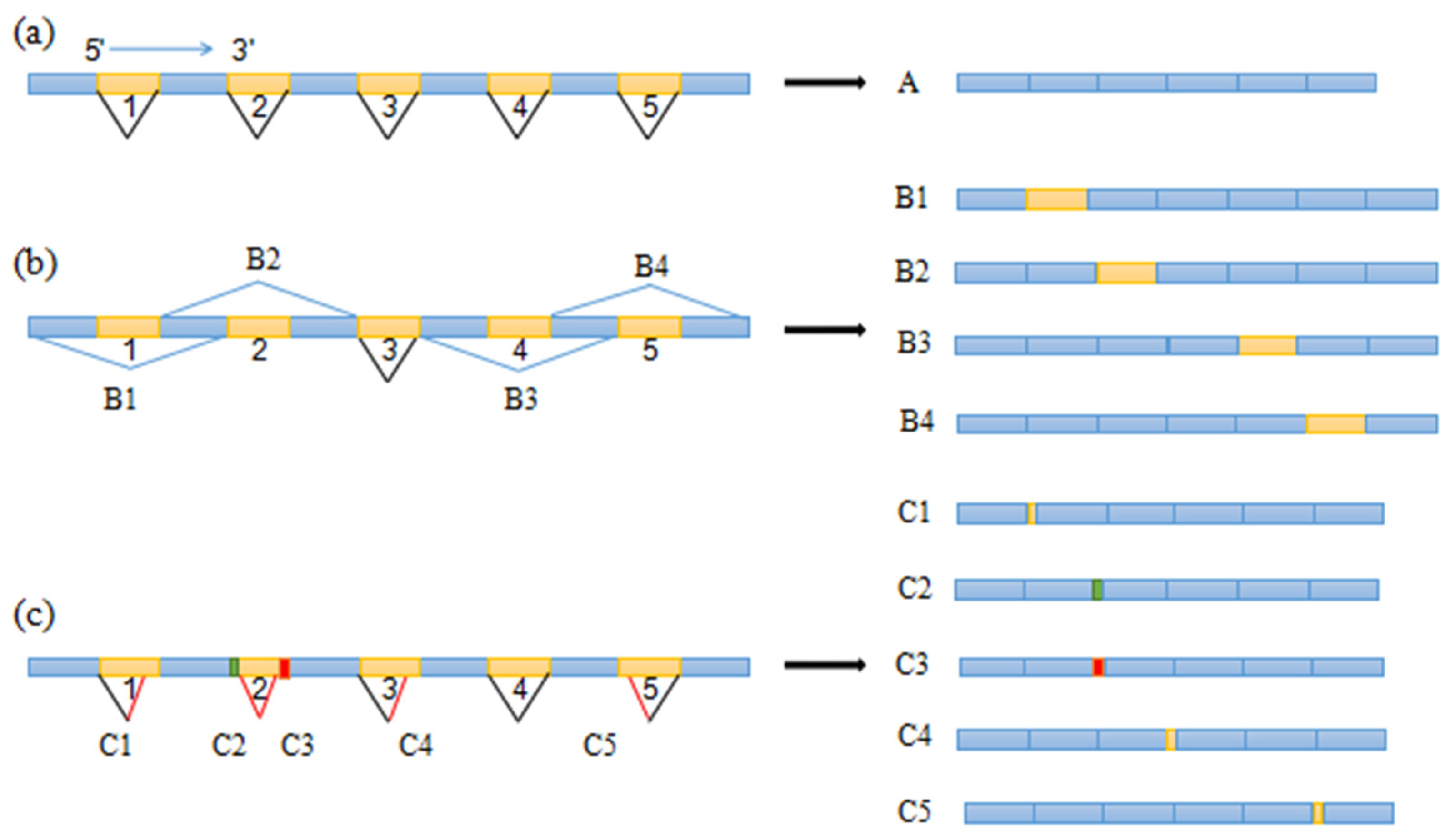
3.5. Alternative Splicing of The MAT1-1-10 Gene
4. Discussion
4.1. Mating Systems in Morchella
4.2. Unconventional Integration of the MAT Locus in Some Morel Species
4.3. Alternative Splicing of The MAT1-1-10 Gene
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chitrampalam, P.; Pryor, B.M. Characterization of mating type (MAT) alleles differentiated by a natural inversion in Sclerotinia minor. Plant Pathol. 2015, 64, 911–920. [Google Scholar] [CrossRef]
- Debuchy, R.; Turgeon, B.G. Mating-type structure, evolution, and function in Euascomycetes. In The Mycota; Springer: Berlin/Heidelberg, Germany, 2006; Volume 1, pp. 293–323. [Google Scholar]
- Metzenberg, R.L.; Glass, N.L. Mating type and mating strategies in Neurospora. BioEssays 1990, 12, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, B.G.; Yoder, O.C. Proposed nimenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 2000, 31, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Debuchy, R.; Berteauxlecellier, V.; Silar, P. Mating systems and sexual morphogenesis in Ascomycetes. In Cellular and Molecular Biology of Filamentous Fungi; Borkovich, K.A., Ebbole, E., Eds.; ASM Press: Washington, DC, USA, 2010; pp. 201–535. [Google Scholar]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; De Beer, Z.W.; Wingfield, B.D. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol. Rev. 2017, 31, 199–211. [Google Scholar] [CrossRef] [Green Version]
- Chai, H.M.; Chen, W.M.; Zhang, X.L.; Su, K.M.; Zhao, Y.C. Structural variation and phylogenetic analysis of the mating-type locus in the genus Morchella. Mycologia 2019, 111, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Aylward, J.; Havenga, M.; Dreyer, L.L.; Roets, F.; Wingfield, B.D.; Wingfield, M.J. Genomic characterization of mating type loci and mating type distribution in two apparently asexual plantation tree pathogens. Plant Pathol. 2020, 69, 28–37. [Google Scholar] [CrossRef]
- Wilson, A.M.; Wilken, P.M.; van der Nest, M.A.; Steenkamp, E.T.; Wingfield, M.J.; Wingfield, B.D. Homothallism: An umbrella term for describing diverse sexual behaviours. IMA Fungus 2015, 6, 207–214. [Google Scholar] [CrossRef]
- Kuo, M.; Dewsbury, D.R.; O’Donnell, K.; Carter, M.C.; Rehner, S.A.; Moore, J.D.; Moncalvo, J.M.; Canfield, S.A.; Stephenson, S.L.; Methven, A.S.; et al. Taxonomic revision of true morels (Morchella) in Canada and the United States. Mycologia 2012, 104, 1159–1177. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.; Rooney, A.P.; Mills, G.L.; Kuo, M.; Weber, N.S.; Rehner, S.A. Phylogeny and historical biogeography of true morels (Morchella) reveals an early Cretaceous origin and high continental endemism and provincialism in the Holarctic. Fungal Genet. Biol. 2011, 48, 252–265. [Google Scholar] [CrossRef]
- Du, X.H.; Zhao, Q.; O’Donnell, K.; Rooney, A.P.; Yang, Z.L. Multigene molecular phylogenetics reveals true morels (Morchella) are especially species-rich in China. Fungal Genet. Biol. 2012, 49, 455–469. [Google Scholar] [CrossRef]
- Loizides, M.; Alvarado, P.; Moreau, P.A.; Assyov, B.; Halasu, V.; Stadler, M.; Rinaldi, A.; Guilhermina, M.; Georgios, I.Z.; Borovička, J.; et al. Has taxonomic vandalism gone too far? A case study, the rise of the pay-to-publish model and the pitfalls of Morchella systematics. Mycol. Prog. 2022, 21, 7–38. [Google Scholar] [CrossRef]
- Belfiori, B.; Riccioni, C.; Paolocci, F.; Rubini, A. Characterization of the reproductive mode and life cycle of the whitish truffle T. Borchii. Mycorrhiza 2016, 26, 515–527. [Google Scholar] [CrossRef]
- Lu, Y.H.; Xia, Y.L.; Luo, F.F.; Dong, C.H.; Wang, C.S. Functional convergence and divergence of mating-type genes fulfilling in Cordyceps militaris. Fungal Genet. Biol. 2016, 88, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Du, X.H.; Yang, Z.L. Mating Systems in True Morels (Morchella). Microbiol. Mol. Biol. Rev. 2021, 85, e00220-20. [Google Scholar] [CrossRef]
- Chai, H.M.; Chen, L.J.; Chen, W.M.; Zhao, Q.; Zhang, X.L.; Su, K.M.; Zhao, Y.C. Characterization of mating-type idiomorphs suggests that Morchella importuna, Mel-20 and M. sextelata are heterothallic. Mycol. Prog. 2017, 16, 743–752. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Martin, S.H.; Wingfield, B.D.; Wingfield, M.J.; Steenkamp, E.T. Structure and evolution of the Fusarium mating type locus: New insights from the Gibberellafujikuroi complex. Fungal Genet. Biol. 2011, 48, 734–740. [Google Scholar] [CrossRef] [Green Version]
- Tsui, C.K.M.; DiGuistini, S.; Wang, Y.; Feau, N.; Dhillon, B.; Bohlmann, J.; Hamelin, R.C. Unequal recombination and evolution of the mating-type (MAT) loci in the pathogenic fungus Grosmannia clavigera and relatives. Genes Genomes Genet. 2013, 3, 465–480. [Google Scholar]
- Yin, Z.Y.; Ke, X.W.; Li, Z.P.; Chen, J.L.; Gao, X.L.; Huang, L.L. Unconventional Recombination in the Mating Type Locus of Heterothallic Apple Canker Pathogen Valsa mali. Genes Genomes Genet. 2017, 7, 1259–1265. [Google Scholar] [CrossRef] [Green Version]
- Kempken, F. Alternative splicing in ascomycetes. Appl. Microbiol. Biotechnol. 2013, 97, 4235–4241. [Google Scholar] [CrossRef] [PubMed]
- Chai, H.M.; Ma, Y.H.; Liu, P.; Chen, W.M.; Tao, N.; Zhao, Y.C. The asymmetrical distribution of opposite mating type nuclei in single-ascospore isolates revealed Morchella importuna is a pseudohomothallic fungus. Mycosystema 2022. in-print. Available online: https://kns.cnki.net/kcms/detail/11.5180.Q.20220419.1112.002.html (accessed on 14 June 2022).
- He, X.S.; Zhao, M.; Wang, Y.Z.; Zhang, N.; Tan, R.H.; Wang, Y.; Zhu, W.K.; Chen, B. Study on the hyphal fusion and non-fusion phenomena between morels. Edible Med. Mushrooms 2019, 27, 244–252. [Google Scholar]
- Liu, W.; Cai, Y.L.; He, P.X.; Bian, Y.B. Cultivation of monosporic and hybrid populations and polarity analysis of Morchella importuna. J. Fungal Res. 2019, 17, 43–49. [Google Scholar]
- Tan, F.H. Expounding the development period of morel cultivation technology using exogenous nutrition bag in China. Edible Med. Mushrooms 2019, 27, 257–263. [Google Scholar]
- Nagel, J.H.; Wingfield, M.J.; Slippers, B. Evolution of the mating types and mating strategies in prominent genera in the Botryosphaeriaceae. Fungal Genet. Biol. 2018, 114, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Li, J.Q.; Wingfield, B.D.; Wingfield, M.J.; Barnes, I.; Fourie, A.; Crous, P.W.; Chen, S.F. Mating genes in Calonectria and evidence for a heterothallic ancestral state. Persoonia 2020, 45, 163–176. [Google Scholar] [CrossRef]
- Lumley, A.J.; Michalczyk, L.; Kitson, J.J.; Spurgin, L.G.; Morrison, C.A.; Godwin, J.L.; Dickinson, M.E.; Martin, O.Y.; Emerson, B.C.; Chapman, T.; et al. Sexual selection protects against extinction. Nature 2015, 522, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, F.E.; Duhamel, M.; Carpentier, F.; Hood, M.E.; Foulongne-Oriol, M.; Silar, P.; Malagnac, F.; Grognet, P.; Giraud, T. Recombination suppression and evolutionary strata around mating-type loci in fungi: Documenting patterns and understanding evolutionary and mechanistic causes. New Phytol. 2021, 229, 2470–2491. [Google Scholar] [CrossRef]
- Petters-Vandresen, D.A.; Rossi, B.J.; Groenewald, J.Z.; Crous, P.W.; Machado, M.A.; Stukenbrock, E.H.; Glienke, C. Mating-type locus rearrangements and shifts in thallism states in Citrus associated Phyllosticta species. Fungal Genet. Biol. 2020, 144, 103444. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef] [Green Version]
- Sablok, G.; Powell, B.; Braessler, J.; Yu, F.; Min, X.J. Comparative landscape of alternative splicing in fruit plants. Curr. Plant Biol. 2017, 9, 29–36. [Google Scholar] [CrossRef]
- Grutzmann, K.; Szafranski, K.; Pohl, M.; Voigt, K.; Petzold, A.; Schuster, S. Fungal alternative splicing is associated with multicellular complexity and virulence: A genome-wide multi-species study. DNA Res. 2014, 21, 27–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzafar, S.; Sharma, R.D.; Chauhan, N.; Prasad, R. Intron distribution and emerging role of alternative splicing in fungi. FEMS Microbiol. Lett. 2021, 368, fnab135. [Google Scholar] [CrossRef]
- Xie, B.B.; Li, D.; Shi, W.L.; Qin, Q.L.; Wang, X.W.; Rong, J.C.; Sun, C.Y.; Huang, F.; Zhang, X.Y.; Dong, X.W.; et al. Deep RNA sequencing reveals a high frequency of alternative splicing events in the fungus Trichoderma longibrachiatum. BMC Genom. 2015, 16, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Hilarion, S.; Paulet, D.; Lee, K.T.; Hon, C.C.; Janbon, G. Intron retention-dependent gene regulation in Cryptococcus neoformans. Sci. Rep. 2016, 6, 32252. [Google Scholar] [CrossRef]
- Irimia, M.; Rukov, J.L.; Penny, D.; Roy, S.W. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 2007, 7, 188. [Google Scholar] [CrossRef] [Green Version]
- Zavolan, M.; Kondo, S.; Schonbach, C.; Adachi, J.; Hume, D.A.; RIKEN GER Group and GSL Members; Hayashizaki, Y.; Gaasterland, T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003, 13, 1290–1300. [Google Scholar] [CrossRef] [Green Version]

| Strain | Species | Code | Ascocarp | Origin | Mating-Type Idiomorph |
|---|---|---|---|---|---|
| YAASMHL1 | M. sextelata | Mel-6 | YAASMHL | Sichuan, China | MAT1-1 |
| YAASMHL47 | MAT1-2 | ||||
| YAASMQM-23 | M. eximia | Mel-7 | YAASMQM | Tibet, China | MAT1-1 |
| YAASMQM-4 | MAT1-2 | ||||
| YAASM23-11 | M. purpurascena | Mel-20 | YAASM23 | Yunnan, China | MAT1-1 |
| YAASM23-22 | MAT1-2 | ||||
| YAASM131-9 | M. pulchella | Mel-31 | YAASM131 | Yunnan, China | MAT1-1 |
| YAASM131-12 | MAT1-2 | ||||
| YAASM50-38 | Mes-6 | YAASM50 | Shanxi, China | MAT1-1 | |
| YAASM50-09 | MAT1-2 | ||||
| M115 | Mes-15 | YAASM2689 | Yunnan, China | MAT1-1, MAT1-2 | |
| YAASM43-24 | Mes-19 | YAASM43 | Sichuan, China | MAT1-1 | |
| YAASM43-08 | MAT1-2 | ||||
| YAASMVR | M. rufobrunnea | American | MAT1-1, MAT1-2 |
| Species | Code | MAT1-1 Idiomorph | MAT1-2 Idiomorph | ||||
|---|---|---|---|---|---|---|---|
| Gene Arrangements | Lengths (kb) | GenBank Accession | Gene Arrangements | Lengths (kb) | GenBank Accession | ||
| M. sextelata | Mel-6 | MAT1-1-1, MAT1-1-10, MAT1-1-11 | 11.5 | MN589921 | MAT1-2-1 | 7.1 | MN589922 |
| M. eximia | Mel-7 | MAT1-1-1, MAT1-1-10, MAT1-1-11 | 10.2 | ON622485 | MAT1-2-1 | 6.6 | ON622484 |
| M. importuna | Mel-10 | MAT1-1-1, MAT1-1-10, MAT1-1-11 | 10.5 | KY782630 | MAT1-2-1 | 6.7 | KY782629 |
| M. purpurascena | Mel-20 | MAT1-1-1, MAT1-1-10, truncated MAT1-1-11 | 9.4 | MN589929 | MAT1-2-1, MAT1-1-11 | 15.9 | MN589930 |
| M. pulchella | Mel-31 | MAT1-1-1, MAT1-1-10, truncated MAT1-1-11 | 11.8 | ON622483 | MAT1-2-1, MAT1-1-11 | 15.9 | ON622482 |
| Mes-6 | MAT1-1-1, MAT1-1-10 | 8.0 | MN589927 | MAT1-2-1 | 7.3 | MN589928 | |
| Mes-15 | MAT1-1-1, MAT1-1-10 | - | KY782631 | MAT1-2-1 | - | KY782632 | |
| Mes-19 | MAT1-1-1, MAT1-1-10 | 7.2 | MN589925 | MAT1-2-1 | 6.4 | MN589926 | |
| Mes-20 | MAT1-1-1, MAT1-1-10 | 7.8 | MN589923 | MAT1-2-1 | 7.5 | MN589924 | |
| M. rufobrunnea | MAT1-1-1, MAT1-1-10, MAT1-2-1 | 16.7 | MN589931 | ||||
| Species | Code | MAT1-1-1 | MAT1-2-1 | MAT1-1-11 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA (bp) | Intron Numbers | Amino Acids (aa) | DNA (bp) | Intron Numbers | Amino Acids (aa) | DNA (bp) | Intron Numbers | Amino Acids (aa) | ||
| M. sextelata | Mel-6 | 1730 | 2 | 537 | 1199 | 3 | 329 | 1823 | 5 | 511 |
| M. eximia | Mel-7 | 1676 | 2 | 519 | 1203 | 3 | 330 | 1825 | 5 | 511 |
| M. importuna | Mel-10 | 1694 | 2 | 525 | 1197 | 3 | 329 | 1825 | 5 | 512 |
| M. purpurascena | Mel-20 | 1651 | 2 | 511 | 1247 | 3 | 344 | 1838 | 5 | 511 |
| M. pulchella | Mel-31 | 1645 | 2 | 509 | 1247 | 3 | 344 | 1828 | 5 | 511 |
| Mes-6 | 1757 | 2 | 547 | 1328 | 3 | 385 | ||||
| Mes-15 | 1766 | 2 | 543 | 1304 | 3 | 377 | ||||
| Mes-19 | 1751 | 2 | 545 | 1301 | 3 | 376 | ||||
| Mes-20 | 1751 | 2 | 545 | 1286 | 3 | 371 | ||||
| M. rufobrunnea | 1760 | 2 | 546 | 1343 | 3 | 384 | ||||
| Species | Code | Total Numbers of Clones | Types of Splicing a | Numbers of Clones | Intron Numbers | DNA (bp) | Amino Acids |
|---|---|---|---|---|---|---|---|
| M. sextelata | Mel-6 | 21 | A | 11 | 5 | 1723 | 482 |
| B4 | 3 | 4 | 1689 | 490 | |||
| B3 + B4 | 3 | 3 | 1689 | 510 | |||
| B2 + B4 | 1 | 3 | - b | - | |||
| B2 + B3 + B4 | 3 | 2 | - | - | |||
| M. eximia | Mel-7 | 10 | B4 | 10 | 4 | 1661 | 479 |
| M. importuna | Mel-10 | 10 | B4 | 10 | 4 | 1688 | 488 |
| M. purpurascena | Mel-20 | 10 | A | 7 | 5 | 1737 | 482 |
| C3 | 3 | 5 | 1737 | 486 | |||
| M. pulchella | Mel-31 | 10 | C3 | 6 | 5 | 1737 | 478 |
| C4 | 2 | 5 | 1737 | 482 | |||
| C3 + C4 | 2 | 5 | 1737 | 486 | |||
| Mes-6 | 10 | A | 2 | 5 | 1762 | 493 | |
| C1 | 1 | 5 | - | - | |||
| C1 + B4 | 1 | 4 | - | - | |||
| B4 | 2 | 4 | 1654 | 478 | |||
| C5 | 3 | 5 | 1704 | 473 | |||
| C5-2 c | 1 | 5 | 1733 | 476 | |||
| Mes-15 | 10 | A | 8 | 5 | 1760 | 491 | |
| C5 | 1 | 5 | - | - | |||
| C1 + C5 | 1 | 5 | - | - | |||
| Mes-19 | 14 | A | 11 | 5 | 1760 | 491 | |
| B4 | 3 | 4 | 1760 | 512 | |||
| Mes-20 | 10 | A | 1 | 5 | 1757 | 491 | |
| B4 | 6 | 4 | 1648 | 476 | |||
| C5 | 1 | 5 | 1699 | 420 | |||
| B1 + C5 | 1 | 4 | 1699 | 489 | |||
| B1 + B4 + C2 | 1 | 3 | 1649 | 474 | |||
| M. rufobrunnea | Mruf | 11 | B4 | 4 | 4 | 1698 | 485 |
| C3 | 3 | 5 | 1850 | 515 | |||
| C3 + B4 | 3 | 4 | 1698 | 489 | |||
| B1 + C3 | 1 | 4 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chai, H.; Liu, P.; Ma, Y.; Chen, W.; Tao, N.; Zhao, Y. Organization and Unconventional Integration of the Mating-Type Loci in Morchella Species. J. Fungi 2022, 8, 746. https://doi.org/10.3390/jof8070746
Chai H, Liu P, Ma Y, Chen W, Tao N, Zhao Y. Organization and Unconventional Integration of the Mating-Type Loci in Morchella Species. Journal of Fungi. 2022; 8(7):746. https://doi.org/10.3390/jof8070746
Chicago/Turabian StyleChai, Hongmei, Ping Liu, Yuanhao Ma, Weimin Chen, Nan Tao, and Yongchang Zhao. 2022. "Organization and Unconventional Integration of the Mating-Type Loci in Morchella Species" Journal of Fungi 8, no. 7: 746. https://doi.org/10.3390/jof8070746
APA StyleChai, H., Liu, P., Ma, Y., Chen, W., Tao, N., & Zhao, Y. (2022). Organization and Unconventional Integration of the Mating-Type Loci in Morchella Species. Journal of Fungi, 8(7), 746. https://doi.org/10.3390/jof8070746






