Genetic Diversity Analysis based on the Virulence, Physiology and Regional Variability in Different Isolates of Powdery Mildew in Pea
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Isolates and DNA Extraction
2.2. Molecular Characterization of E. pisi Using ITS Region-Specific Primers
2.3. Genetic Discrimination by RAPD Marker
2.4. Band Scoring and Data Analysis
2.5. The Efficacy of RAPD Primers for Evaluating the Diversity of E. pisi
3. Results and Discussion
3.1. Disease Symptoms
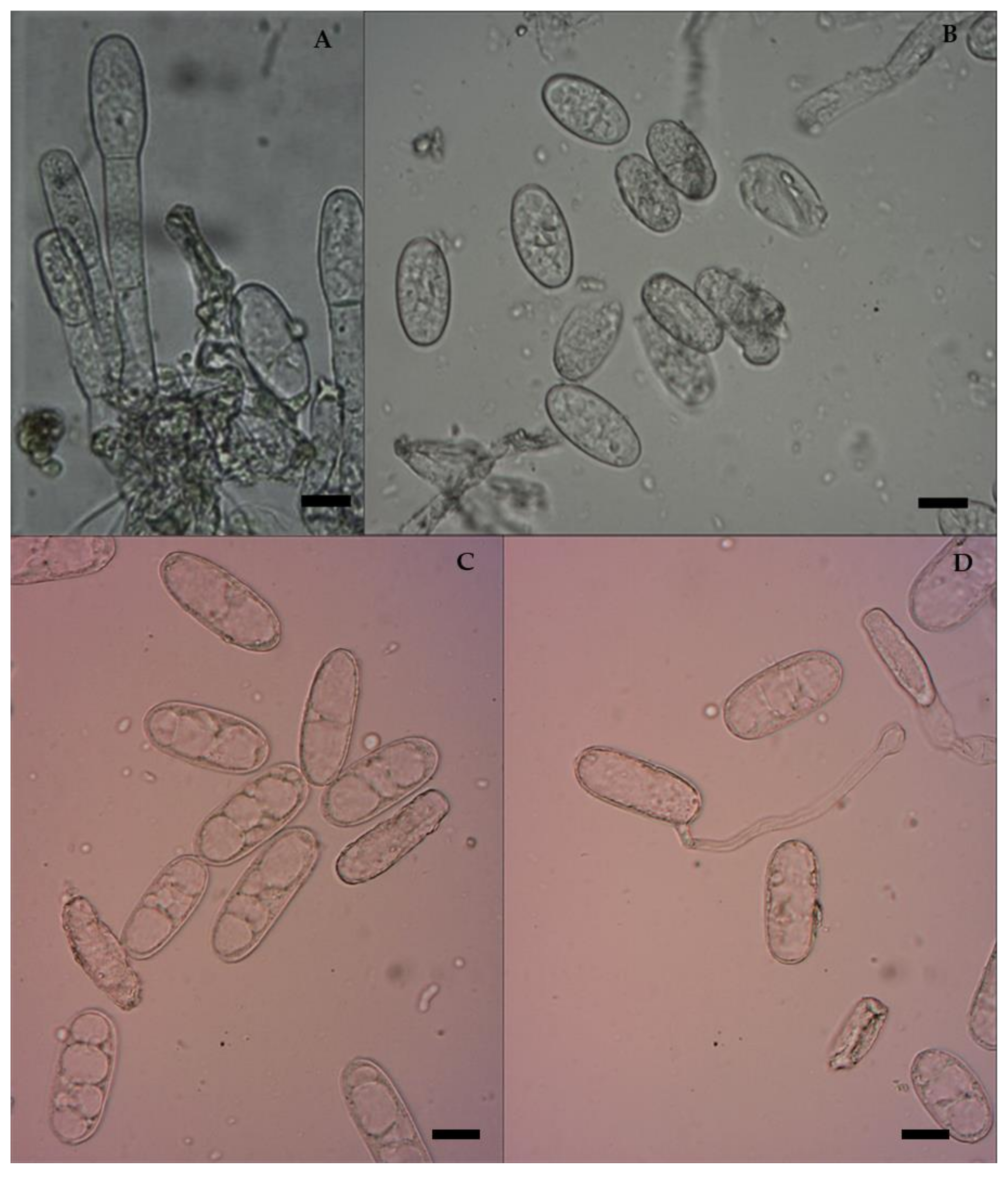
3.2. Morphological Characterization of E. pisi
3.3. Molecular Characterization of Isolates of E. pisi
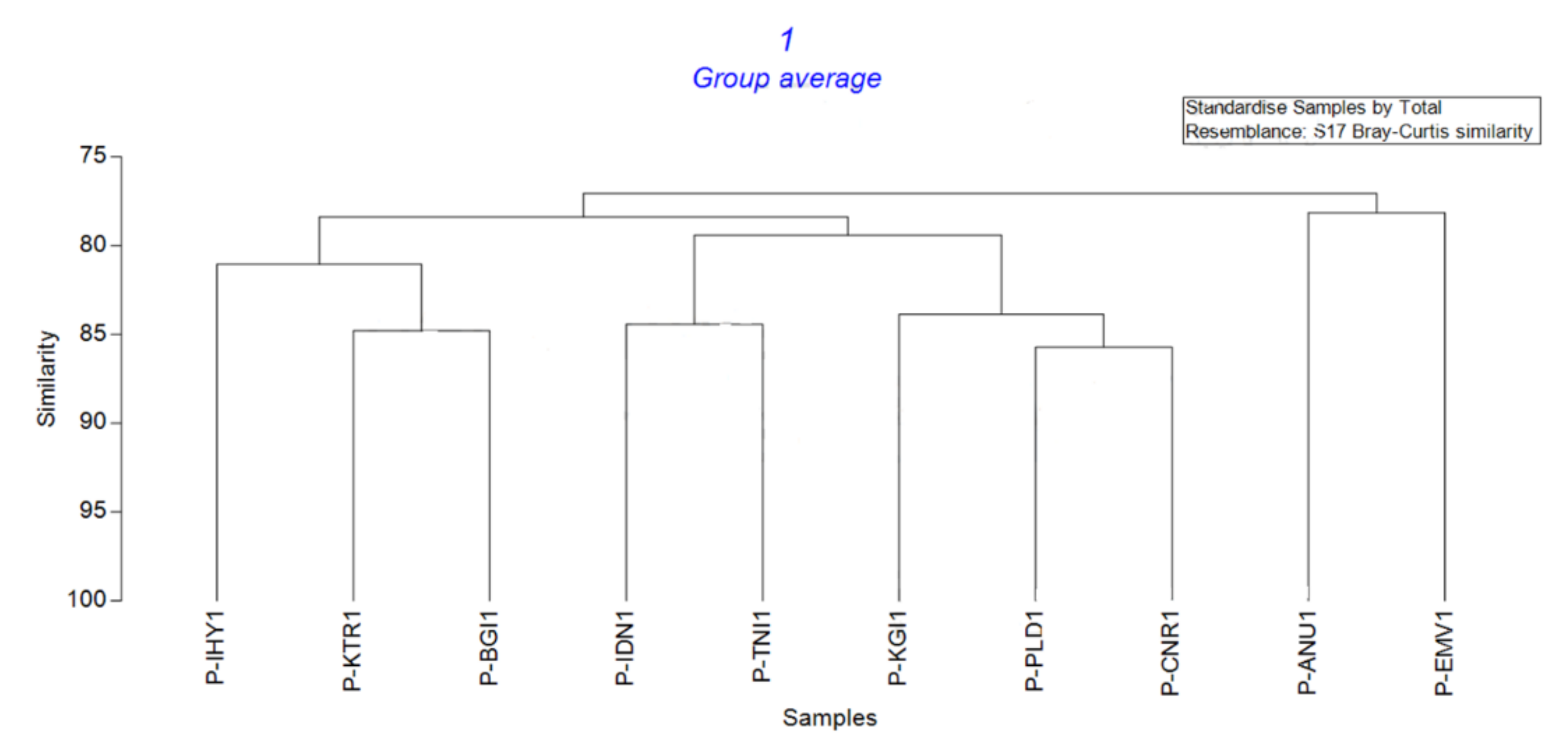
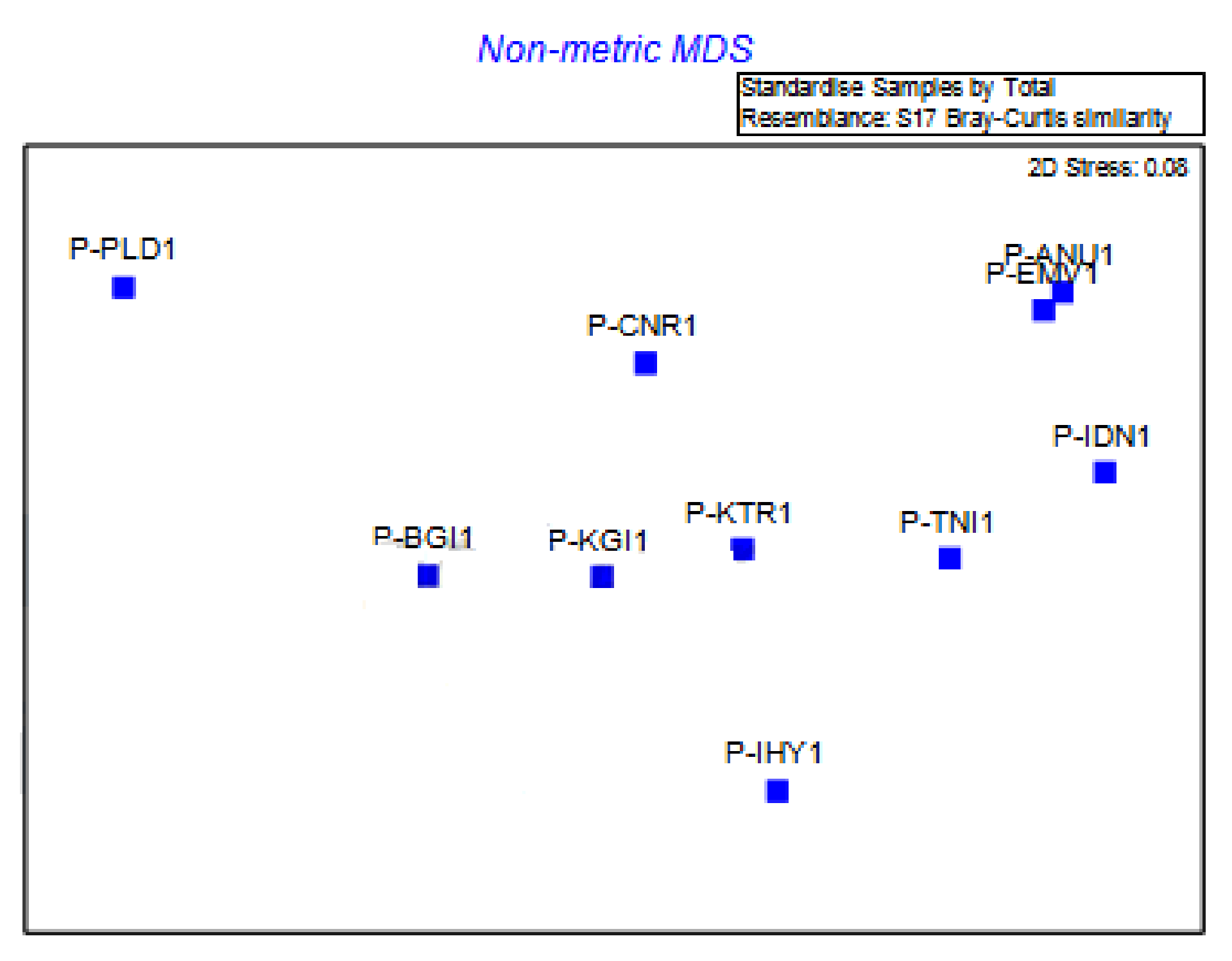
3.4. The Genetic Diversity of E. pisi in the Nilgiris
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Upadhyaya, H.D.; Dwivedi, S.L.; Ambrose, M.; Ellis, N.; Berger, J.; Smýkal, P.; Debouck, D.; Duc, G.; Dumet, D.; Flavell, A.; et al. Legume genetic resources: Management, diversity assessment, and utilization in crop improvement. Euphytica 2011, 180, 27–47. [Google Scholar] [CrossRef]
- Smýkal, P.; Aubert, G.; Burstin, J.; Coyne, C.J.; Ellis, N.T.H.; Flavell, A.J.; Ford, R.; Hýbl, M.; Macas, J.; Neumann, P.; et al. Pea (Pisum sativum L.) in the genomic era. Agronomy 2012, 2, 74–115. [Google Scholar] [CrossRef]
- Villegas-Fernández, Á.M.; Amarna, A.A.; Moral, J.; Rubiales, D. Crop diversification to control powdery mildew in pea. Agronomy 2021, 11, 690. [Google Scholar] [CrossRef]
- Sulima, A.S.; Zhukov, V.A. War and Peas: Molecular Bases of Resistance to Powdery Mildew in Pea (Pisum sativum L.) and Other Legumes. Plants 2022, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Nisar, M.; Ghafoor, A.; Khan, M.R.; Qureshi, A.S. Screening of Pisum sativum L. germplasm against Erysiphe pisi Syd. Acta Biol. Crac. Ser. Bot. 2006, 48, 33–37. [Google Scholar]
- Sun, S.; Deng, D.; Duan, C.; Zong, X.; Xu, D.; He, Y.; Zhu, Z. Two novel er1 alleles conferring powdery mildew (Erysiphe pisi) resistance identified in a worldwide collection of pea (Pisum sativum L.) germplasms. Int. J. Mol. Sci. 2019, 20, 5071. [Google Scholar] [CrossRef]
- Warkentin, T.D.; Rashid, K.Y.; Xue, A.G. Fungicidal control of powdery mildew in field pea. Can. J. Plant Sci. 1996, 76, 933–935. [Google Scholar] [CrossRef]
- Baiswar, P.; Ngachan, S.V.; Verma, V.K.; Kumar, R.; Jha, A.K.; Chandra, S. Molecular evidence of Erysiphe pisi on pea and E. trifoliorum on white clover in northeast India. Australas. Plant Dis. Notes 2015, 10, 12. [Google Scholar] [CrossRef]
- Fondevilla, S.; González-Bernal, M.J.; Ben Youssef, N.O.; Rubiales, D. Development of Quantitative Real-Time PCR Assays to Quantify Erysiphe pisi and Erysiphe trifolii and Its Implementation for Monitoring Their Relative Prevalence in Pea Crops in Spain and Tunisia. Agronomy 2022, 12, 334. [Google Scholar] [CrossRef]
- Teshome, E.; Tegegn, A. Comparative Study of Powdery Mildew (Erysiphe polygoni) Disease Severity and Its Effect on Yield and Yield Components of Field Pea (Pisum sativum L.) in the Southeastern Oromia, Ethiopia. J. Plant Pathol. Microbiol. 2017, 8, 2. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rubiales, D. Powdery mildew control in pea. A review. Agron. Sustain. Dev. 2012, 32, 401–409. [Google Scholar] [CrossRef]
- Nag, U.K.; Khare, C.P. Epidemiological studies on powdery mildew of vegetable pea. Plant Arch. 2017, 17, 171–176. [Google Scholar]
- Harveson, R.M.; Pasche, J.S.; Chen, W.; Burrows, M. (Eds.) Compendium of Pea Diseases and Pests, 3rd ed.; American Phytopathological Society: Saint Paul, MI, USA, 2021. [Google Scholar] [CrossRef]
- Kovács, G.M.; Jankovics, T.; Kiss, L. Variation in the nrDNA ITS sequences of some powdery mildew species: Do routine molecular identification procedures hide valuable information? Eur. J. Plant Pathol. 2011, 131, 135–141. [Google Scholar] [CrossRef]
- Miazzi, M.; Hajjeh, H.; Faretra, F. Observations on the population biology of the grape powdery mildew fungus Uncinula necator. J. Plant Pathol. 2003, 85, 123–129. [Google Scholar]
- Kumar, A.M.; Sharma, P. A study on corroboration between DNA markers (RAPD, ISSR, ITS) and bio-control efficacy of Trichoderma species. Fungal Genom. Biol. 2012, 1, 1–6. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Bolchacova, E.; Voigt, K.; Crous, P.W.; et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Reddy, D.C.L.; Preethi, B.; Wani, M.A.; Aghora, T.S.; Aswath, C.; Mohan, N. Screening for powdery mildew (Erysiphe pisi D.C.) resistance genelinked SCAR and SSR markers in five breeding lines of Pisum sativum L. J. Hortic. Sci. Biotechnol. 2015, 90, 78–82. [Google Scholar] [CrossRef]
- Fondevilla, S.; Rubiales, D.; Moreno, M.T.; Torres, A.M. Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi DC in pea. Mol. Breed. 2008, 22, 193–200. [Google Scholar] [CrossRef]
- Najafiniya, M.; Sharma, P. Specific PCR-based marker for detection of pathogenic groups of Fusarium oxysporum f. sp. cucumerinum in India. J. Genet. Eng. Biotechnol. 2011, 9, 29–34. [Google Scholar] [CrossRef]
- Smith, R.L.; Sawbridge, T.; Mann, R.; Kaur, J.; May, T.W.; Edwards, J. Rediscovering an old foe: Optimised molecular methods for DNA extraction and sequencing applications for fungarium specimens of powdery mildew (Erysiphales). PLoS ONE 2020, 15, e0232535. [Google Scholar] [CrossRef]
- Qiu, P.L.; Liu, S.Y.; Bradshaw, M.; Rooney-Latham, S.; Takamatsu, S.; Bulgakov, T.S.; Tang, S.R.; Feng, J.; Jin, D.N.; Aroge, T.; et al. Multi-locus phylogeny and taxonomy of an unresolved, heterogeneous species complex within the genus Golovinomyces (Ascomycota, Erysiphales), including G. ambrosiae, G. circumfusus and G. spadiceus. BMC Microbiol. 2020, 20, 51. [Google Scholar] [CrossRef] [PubMed]
- Devi, J.; Mishra, G.P.; Sagar, V.; Kaswan, V.; Dubey, R.K.; Singh, P.M.; Sharma, S.K.; Behera, T.K. Gene-Based Resistance to Erysiphe Species Causing Powdery Mildew Disease in Peas (Pisum sativum L.). Genes 2022, 13, 316. [Google Scholar] [CrossRef] [PubMed]
- Bahadur, A.; Singh, U.P.; Singh, D.P.; Sarma, B.K.; Singh, K.P.; Singh, A.; Aust, H.J. Control of Erysiphe pisi Causing Powdery Mildew of Pea by Cashewnut Shell Extract. Mycobiology 2008, 36, 60–65. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ghafoor, A.; McPhee, K. Marker assisted selection (MAS) for developing powdery mildew resistant pea cultivars. Euphytica 2012, 186, 593–607. [Google Scholar] [CrossRef]
- Nongmaithem, N.; Basudha, C.; Sharma, S.K. Incidence of Rust, Powdery Mildew and Wilt in Pea and Broad bean Plant of Manipur, India. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2611–2616. [Google Scholar] [CrossRef]
- Abasova, L.V.; Aghayeva, D.N.; Takamatsu, S. Notes on powdery mildews of the genus Erysiphe from Azerbaijan. Curr. Res. Environ. Appl. Mycol. 2018, 8, 30–53. [Google Scholar] [CrossRef]
- Attanayake, R.N.; Glawe, D.A.; McPhee, K.E.; Dugan, F.M.; Chen, W. Erysiphe trifolii—A newly recognized powdery mildew pathogen of pea. Plant Pathol. 2010, 59, 712–720. [Google Scholar] [CrossRef]
- Ellingham, O.; David, J.; Culham, A. Enhancing identification accuracy for powdery mildews using previously underexploited DNA loci. Mycologia 2019, 111, 798–812. [Google Scholar] [CrossRef]
- Hsiao, H.; Ariyawansa, H.A.; Hsu, C.; Wang, C.; Shen, Y. New Records of Powdery Mildews from Taiwan: Erysiphe ipomoeae comb. nov., E. aff. betae on Buckwheat, and E. neolycopersici comb. nov. on Cardiospermum halicacabum. Diversity 2022, 14, 204. [Google Scholar] [CrossRef]
- Komínková, E.; Dreiseitl, A.; Maleèková, E.; Doležel, J.; Valárik, M. Genetic diversity of blumeria graminis f. sp. hordei in central Europe and its comparison with australian population. PLoS ONE 2016, 11, e0167099. [Google Scholar] [CrossRef]



| S. No. | Primers | Sequence 5′-3′ |
|---|---|---|
| 1. | OPA-01 | CAGGCCCTTC |
| 2. | OPA-03 | AGTCAGCCAC |
| 3. | OPA-05 | AGGGGTCTTG |
| 4. | OPA-07 | GAAACGGGTG |
| 5. | OPA-09 | GGGTAACGCC |
| 6. | OPA-18 | AGGTGACCGT |
| 7. | OPB-02 | TGATCCCTGG |
| 8. | OPC-08 | TGGACCGGTG |
| 9. | OPC-12 | TGTCATCCCC |
| 10. | OPE-01 | CCCAAGGTCC |
| 11. | OPF-01 | ACGGATCCTG |
| 12. | OPF-06 | GGGAATTCGG |
| 13. | OPF-10 | GGAAGCTTGG |
| 14. | OPF-12 | GGCTGCAGAA |
| 15. | OPF-14 | TGCTGCAGGT |
| 16. | OPG-05 | CTGAGACGGA |
| 17. | OPG-08 | TCACGTCCAC |
| 18. | OPG-11 | TGCCCGTCGT |
| 19. | OPG-16 | AGCGTCCTCC |
| 20. | OPL-05 | ACGCAGGCAC |
| S. No. | Primers | Sum of Banding Pattern | Polymorphic Banding Pattern | Monomorphic Banding Pattern | PIC Value | EMR |
|---|---|---|---|---|---|---|
| 1. | OPA-01 | 10 | 9 | 1 | 0.903 | 9.05 |
| 2. | OPA-03 | 6 | 5 | 1 | 0.822 | 5.38 |
| 3. | OPA-05 | 8 | 7 | 1 | 0.849 | 0.015 |
| 4. | OPA-07 | 7 | 6 | 1 | 0.820 | 7.80 |
| 5. | OPA-09 | 10 | 9 | 1 | 0.829 | 9.05 |
| 6. | OPA-18 | 4 | 3 | 1 | 0.749 | 3.56 |
| 7. | OPB-02 | 4 | 3 | 1 | 0.749 | 3.12 |
| 8. | OPC-08 | 10 | 9 | 1 | 0.944 | 13.94 |
| 9. | OPC-12 | 10 | 9 | 1 | 0.884 | 9.05 |
| 10. | OPE-01 | 10 | 9 | 1 | 0.800 | 8.10 |
| 11. | OPF-01 | 10 | 9 | 1 | 0.898 | 9.05 |
| 12. | OPF-06 | 10 | 9 | 1 | 0.899 | 9.05 |
| 13. | OPF-10 | 14 | 13 | 1 | 0.931 | 13.22 |
| 14. | OPF-12 | 6 | 5 | 1 | 0.830 | 9.20 |
| 15. | OPF-14 | 7 | 6 | 1 | 0.848 | 5.40 |
| 16. | OPG-05 | 7 | 6 | 1 | 0.855 | 5.93 |
| 17. | OPG-08 | 8 | 7 | 1 | 0.869 | 7.27 |
| 18. | OPG-11 | 7 | 6 | 1 | 0.821 | 5.14 |
| 19. | OPG-16 | 7 | 6 | 1 | 0.791 | 5.40 |
| 20. | OPL-05 | 7 | 6 | 1 | 0.786 | 5.14 |
| Total | 162 | 142 | 20 |
| Isolate | P-ANU1 | P-EMV1 | P-IDN1 | P-TNI1 | P-IHY1 | P-KTR1 | P-BGI1 | P-PLD1 | P-CNR1 | P-KGI1 |
|---|---|---|---|---|---|---|---|---|---|---|
| P-ANU1 | 100 | |||||||||
| P-EMV1 | 78.15 | 100 | ||||||||
| P-IDN1 | 79.83 | 80.36 | 100 | |||||||
| P-TNI1 | 77.87 | 73.77 | 84.43 | 100 | ||||||
| P-IHY1 | 78.15 | 74.58 | 76.27 | 78.69 | 100 | |||||
| P-KTR1 | 76.80 | 76.80 | 76.00 | 79.20 | 81.60 | 100 | ||||
| P-BGI1 | 78.99 | 79.13 | 80.00 | 79.51 | 80.51 | 84.80 | 100 | |||
| P-PLD1 | 77.18 | 76.37 | 76.37 | 81.48 | 77.19 | 79.52 | 79.69 | 100 | ||
| P-CNR1 | 74.62 | 73.08 | 76.15 | 83.85 | 76.92 | 83.08 | 76.92 | 85.71 | 100 | |
| P-KGI1 | 78.13 | 77.34 | 75.78 | 82.81 | 75.78 | 81.25 | 75.78 | 82.35 | 85.39 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seethapathy, P.; Sankaralingam, S.; Pandita, D.; Pandita, A.; Loganathan, K.; Wani, S.H.; El-Ansary, D.O.; Sharma, H.; Casini, R.; Mahmoud, E.A.; et al. Genetic Diversity Analysis based on the Virulence, Physiology and Regional Variability in Different Isolates of Powdery Mildew in Pea. J. Fungi 2022, 8, 798. https://doi.org/10.3390/jof8080798
Seethapathy P, Sankaralingam S, Pandita D, Pandita A, Loganathan K, Wani SH, El-Ansary DO, Sharma H, Casini R, Mahmoud EA, et al. Genetic Diversity Analysis based on the Virulence, Physiology and Regional Variability in Different Isolates of Powdery Mildew in Pea. Journal of Fungi. 2022; 8(8):798. https://doi.org/10.3390/jof8080798
Chicago/Turabian StyleSeethapathy, Parthasarathy, Subbiah Sankaralingam, Deepu Pandita, Anu Pandita, Kousalya Loganathan, Shabir Hussain Wani, Diaa O. El-Ansary, Hanoor Sharma, Ryan Casini, Eman A. Mahmoud, and et al. 2022. "Genetic Diversity Analysis based on the Virulence, Physiology and Regional Variability in Different Isolates of Powdery Mildew in Pea" Journal of Fungi 8, no. 8: 798. https://doi.org/10.3390/jof8080798
APA StyleSeethapathy, P., Sankaralingam, S., Pandita, D., Pandita, A., Loganathan, K., Wani, S. H., El-Ansary, D. O., Sharma, H., Casini, R., Mahmoud, E. A., & Elansary, H. O. (2022). Genetic Diversity Analysis based on the Virulence, Physiology and Regional Variability in Different Isolates of Powdery Mildew in Pea. Journal of Fungi, 8(8), 798. https://doi.org/10.3390/jof8080798








