A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Sequence Analysis of Aokap4
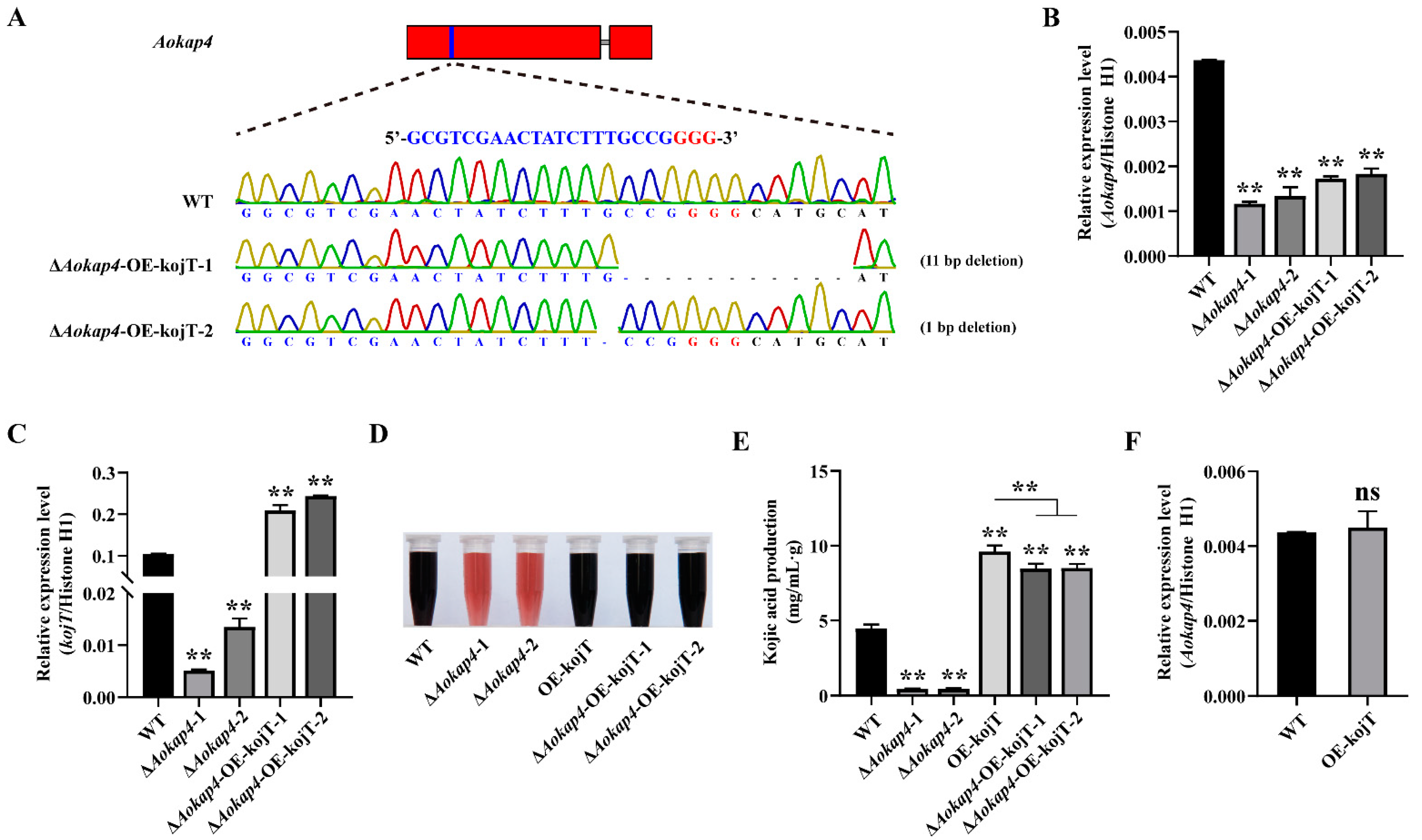
2.3. Mutagenesis of Aokap4 in A. oryzae
2.4. Phenotypic Characterization
2.5. Analysis of the Yields of Kojic Acid
2.6. Gene Expression Analysis Using RT-qPCR
2.7. Construction of Double-Deletion Mutant of Aokap4 and kojT
2.8. Overexpression of kojT in the Aokap4 Deletion Strain
2.9. Heterologous Expression of AoKap4 in Yeast
2.10. Statistical Analysis
3. Results
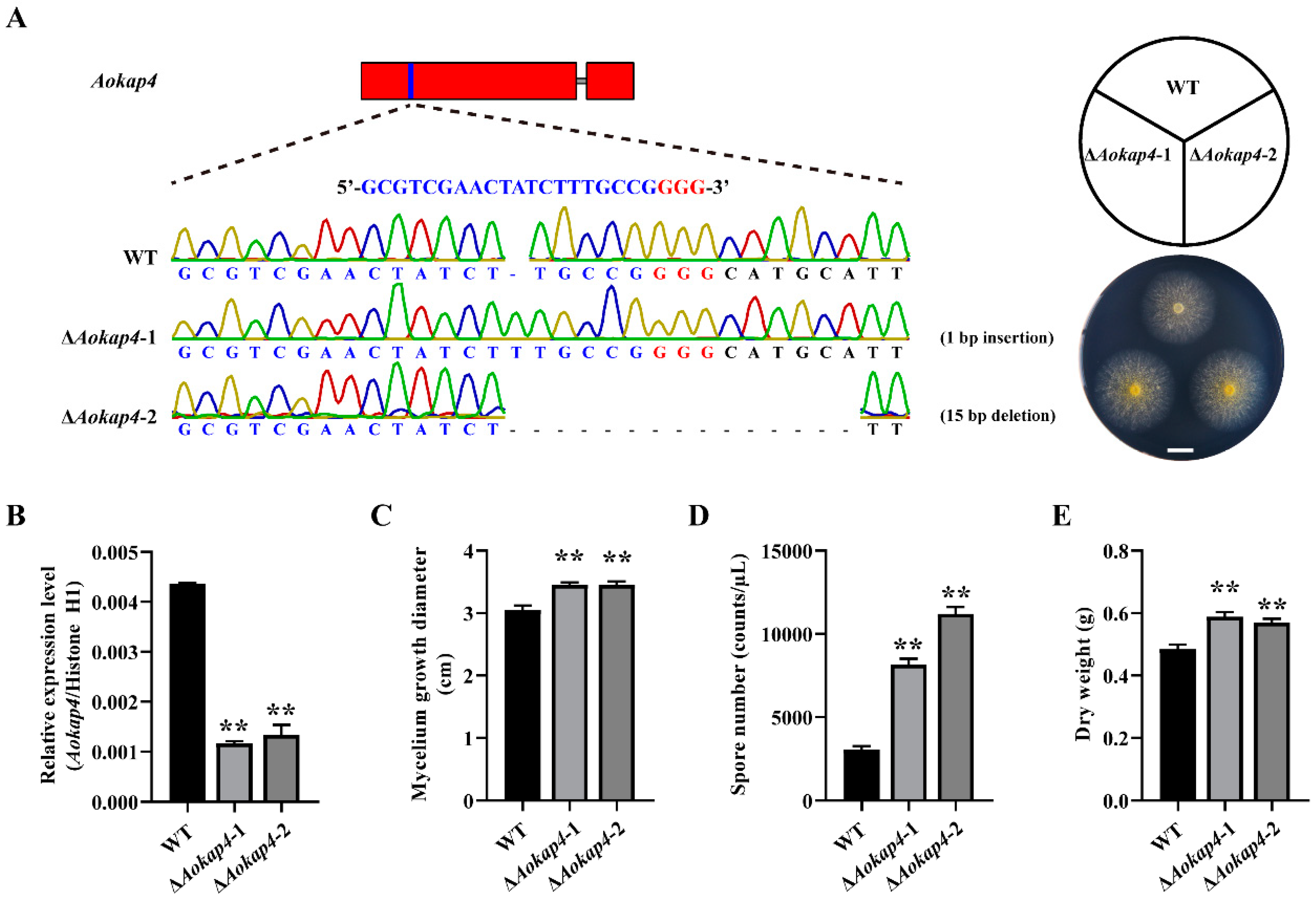
3.1. Identification and Characterization of Aokap4 Gene
3.2. Aokap4 Affected the Growth of A. oryzae
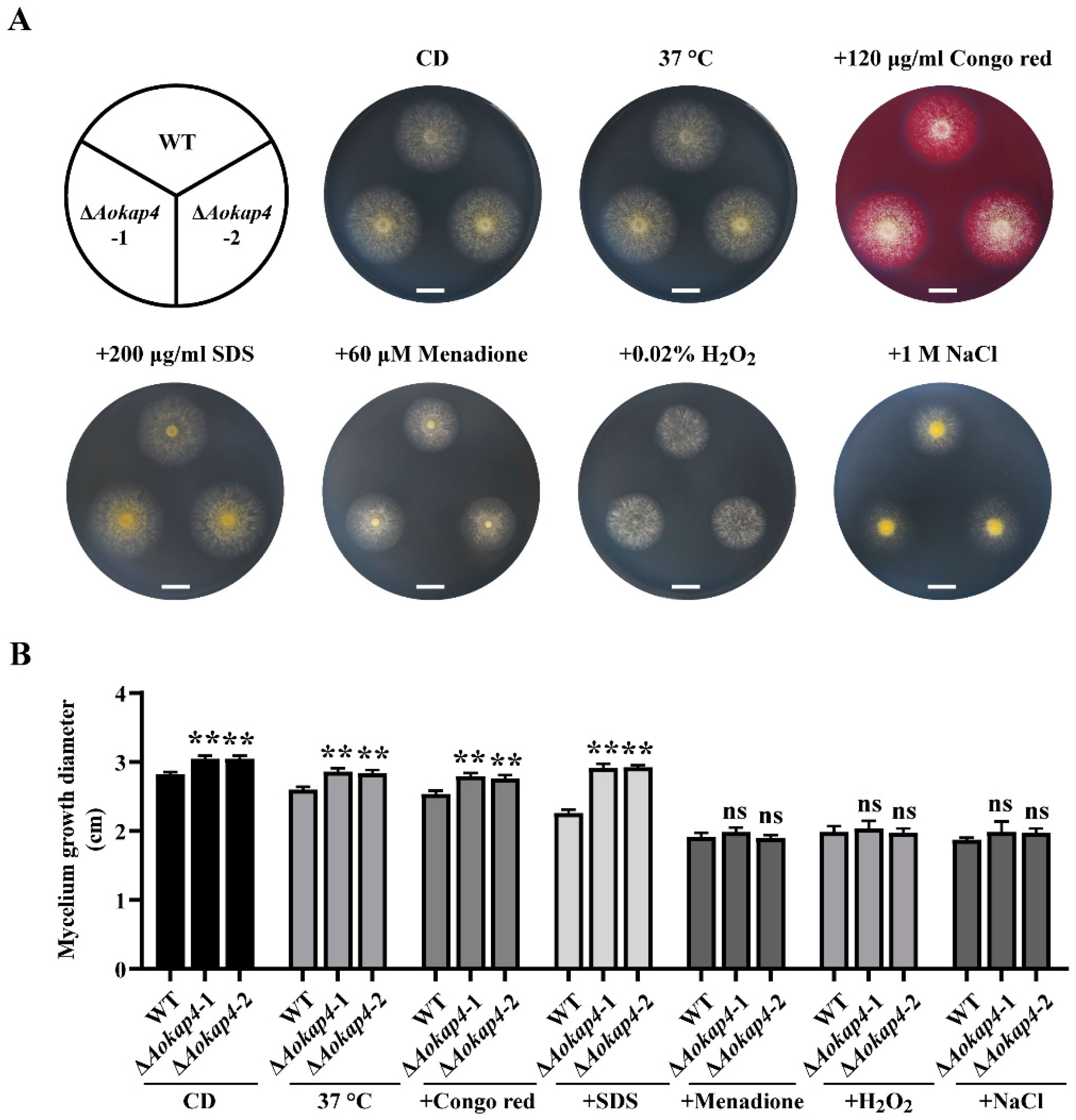
3.3. Disruption of Aokap4 Impaired the Sensitivity to Stress
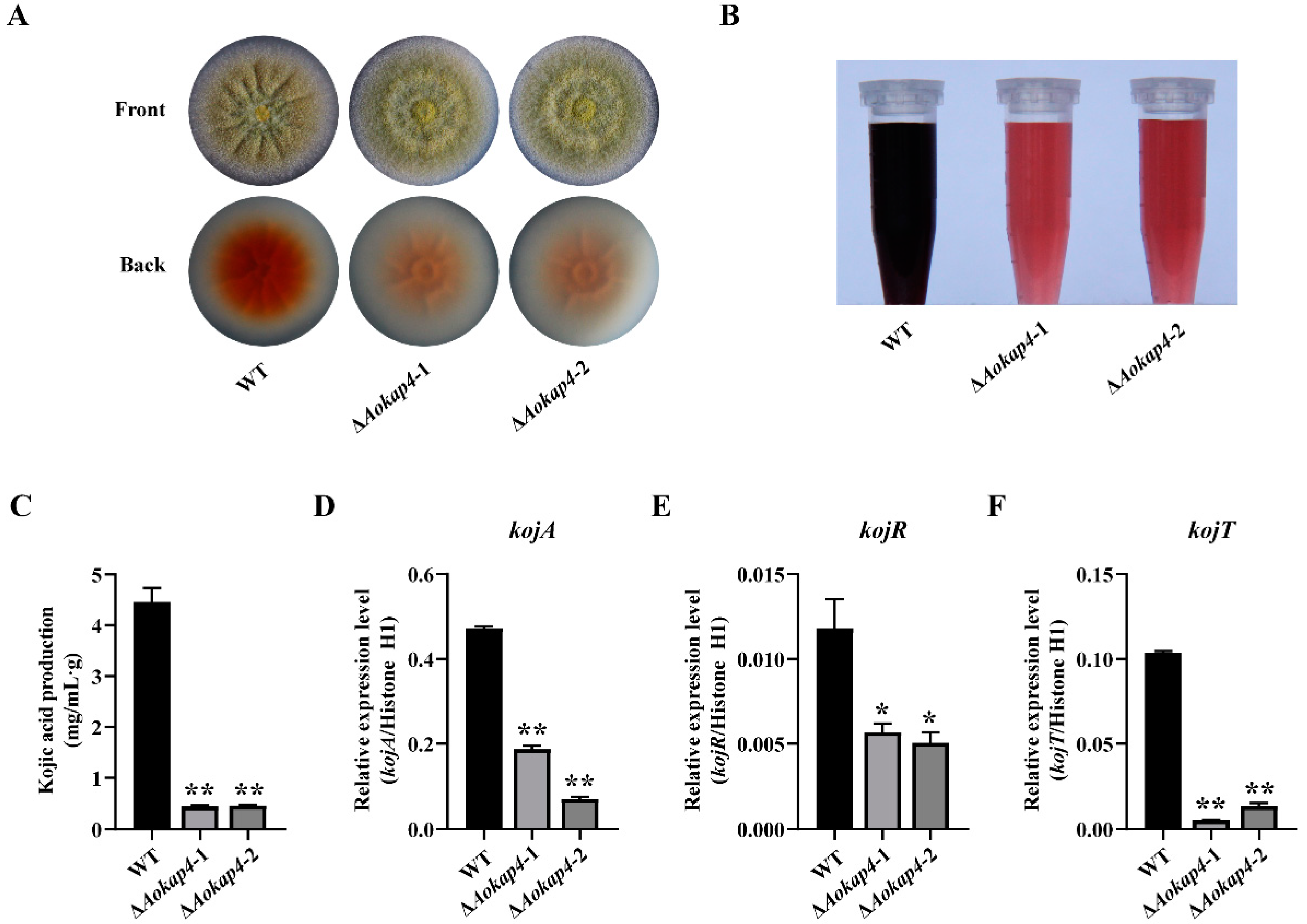
3.4. Disruption of Aokap4 Repressed Kojic Acid Production in A. oryzae
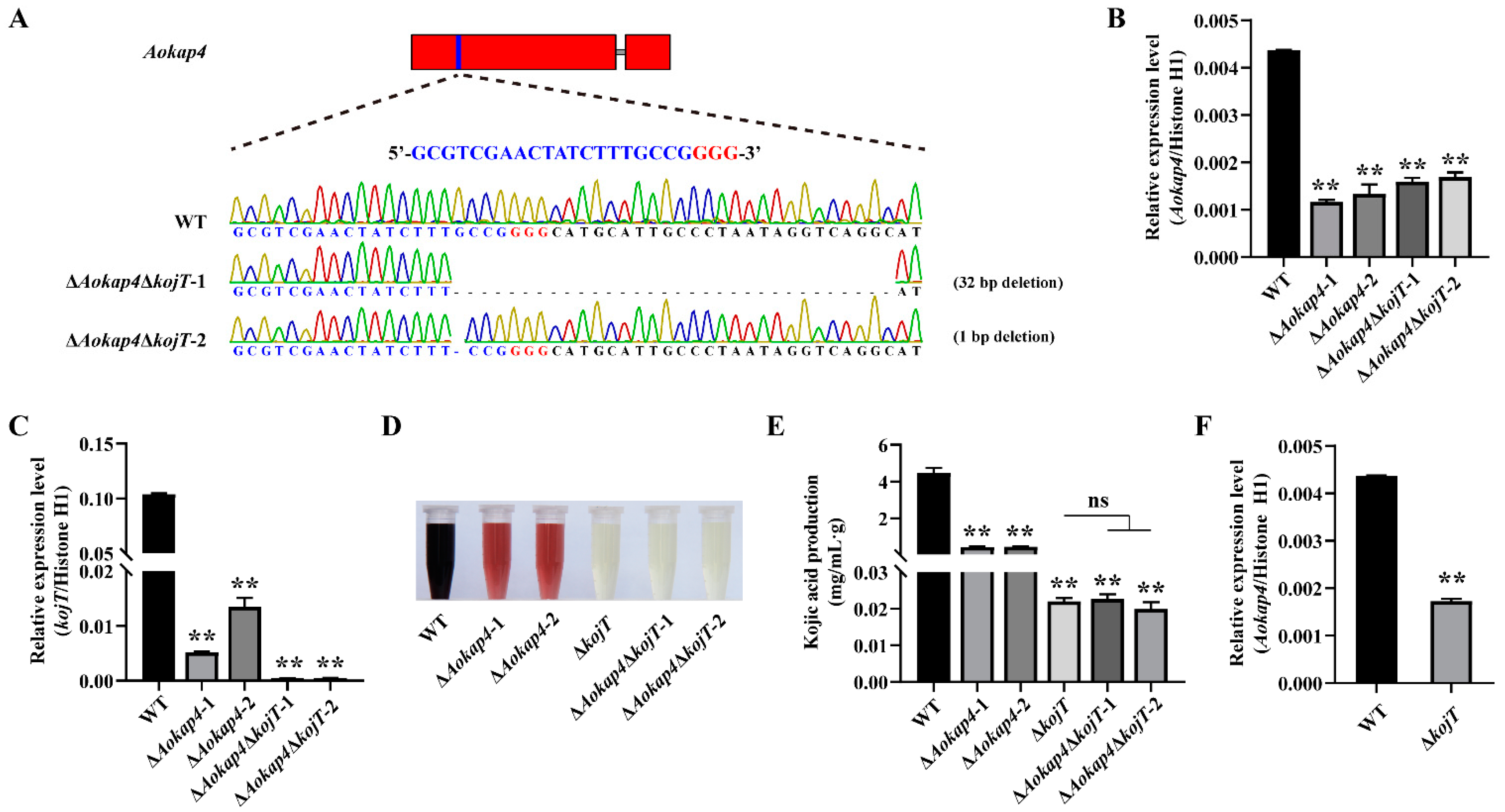
3.5. Knockout of kojT in the Aokap4-Disrupted Strain Impaired Kojic Acid Production
3.6. Overexpression of kojT in Aokap4-Deletion Mutant Could Prevent the Reduced Production of Kojic Acid
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bentley, R. From miso, sake and shoyu to cosmetics: A century of science for kojic acid. Nat. Prod. Rep. 2006, 23, 1046–1062. [Google Scholar] [CrossRef] [PubMed]
- Saeedi, M.; Eslamifar, M.; Khezri, K. Kojic acid applications in cosmetic and pharmaceutical preparations. Biomed. Pharmacother. 2019, 110, 582–593. [Google Scholar] [CrossRef]
- Suryadi, H.; Irianti, M.I.; Septiarini, T.H. Methods of Random Mutagenesis of Aspergillus Strain for Increasing Kojic Acid Production. Curr. Pharm. Biotechnol. 2022, 23, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Kobayashi, A.; Ohashi, S.; Sano, M. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci. Biotechnol. Biochem. 2011, 75, 1832–1834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sano, M. Aspergillus oryzae nrtA affects kojic acid production. Biosci. Biotechnol. Biochem. 2016, 80, 1776–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawauchi, M.; Nishiura, M.; Iwashita, K. Fungus-specific sirtuin HstD coordinates secondary metabolism and development through control of LaeA. Eukaryot. Cell. 2013, 12, 1087–1096. [Google Scholar] [CrossRef] [Green Version]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal. Genet. Biol. 2010, 47, 953–961. [Google Scholar] [CrossRef]
- Marui, J.; Yamane, N.; Ohashi-Kunihiro, S.; Ando, T.; Terabayashi, Y.; Sano, M.; Ohashi, S.; Ohshima, E.; Tachibana, K.; Higa, Y.; et al. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)2Cys6 transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 2011, 112, 40–43. [Google Scholar] [CrossRef]
- Arakawa, G.Y.; Kudo, H.; Yanase, A.; Eguchi, Y.; Kodama, H.; Ogawa, M.; Koyama, Y.; Shindo, H.; Hosaka, M.; Tokuoka, M. A unique Zn(II)2-Cys6-type protein, KpeA, is involved in secondary metabolism and conidiation in Aspergillus oryzae. Fungal. Genet. Biol. 2019, 127, 35–44. [Google Scholar] [CrossRef]
- Kudo, H.; Arakawa, G.Y.; Shirai, S.; Ogawa, M.; Shindo, H.; Hosaka, M.; Tokuoka, M. New role of a histone chaperone, HirA: Involvement in kojic acid production associated with culture conditions in Aspergillus oryzae. J. Biosci. Bioeng. 2021, 133, 235–242. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhang, H.X.; Chen, Z.M.; Fan, J.X.; Chen, T.M.; Zeng, B.; Zhang, Z. Identification and characterization of a novel gene Aokap1 involved in growth and kojic acid synthesis in Aspergillus oryzae. Arch. Microbiol. 2021, 204, 67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fan, J.X.; Long, C.N.; He, B.; Hu, Z.H.; Jiang, C.M.; Li, Y.K.; Ma, L.; Wen, J.S.; Zou, X.J.; et al. Identification and characterization of the ZRT, IRT-like protein (ZIP) family genes reveal their involvement in growth and kojic acid production in Aspergillus oryzae. J. Ind. Microbiol. Biotechnol. 2019, 46, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Chen, Z.; Fan, J.; Chen, T.; Xiao, Y.; Jie, J.; Zeng, B.; Zhang, Z. Overexpression of a novel gene Aokap2 affects the growth and kojic acid production in Aspergillus oryzae. Mol. Biol. Rep. 2022, 49, 2745–2754. [Google Scholar] [CrossRef] [PubMed]
- Law, C.J.; Maloney, P.C.; Wang, D.N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008, 62, 289–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, N. Structural Biology of the Major Facilitator Superfamily Transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Pao, S.S.; Paulsen, I.T.; Saier, M.H., Jr. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998, 62, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Pilsyk, S.; Mieczkowski, A.; Golan, M.P.; Wawrzyniak, A.; Kruszewska, J.S. Internalization of the Aspergillus nidulans AstA transporter into mitochondria depends on growth conditions and affects ATP levels and sulfite oxidase activity. Int. J. Mol. Sci. 2020, 21, 7727. [Google Scholar] [CrossRef]
- Gaur, M.; Puri, N.; Manoharlal, R.; Rai, V.; Mukhopadhayay, G.; Choudhury, D.; Prasad, R. MFS transportome of the human pathogenic yeast Candida albicans. BMC Genom. 2008, 9, 579. [Google Scholar] [CrossRef] [Green Version]
- Paulsen, I.T.; Sliwinski, M.K.; Nelissen, B.; Goffeau, A.; Saier, M.H., Jr. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998, 430, 116–125. [Google Scholar] [CrossRef] [Green Version]
- Nelissen, B.; Mordant, P.; Jonniaux, J.L.; De Wachter, R.; Goffeau, A. Phylogenetic classification of the major superfamily of membrane transport facilitators, as deduced from yeast genome sequencing. FEBS Lett. 1995, 377, 232–236. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, C.; Flores, C. Mutations in GAL2 or GAL4 alleviate catabolite repression produced by galactose in Saccharomyces cerevisiae. Enzyme. Microb. Technol. 2000, 26, 748–755. [Google Scholar] [CrossRef]
- Ozcan, S.; Johnston, M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 1995, 15, 1564–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruckeberg, A.L.; Bisson, L.F. The HXT2 gene of Saccharomyces cerevisiae is required for high-affinity glucose transport. Mol. Cell. Biol. 1990, 10, 5903–5913. [Google Scholar] [PubMed] [Green Version]
- Ferreira, C.; van Voorst, F.; Martins, A.; Neves, L.; Oliveira, R.; Kielland-Brandt, M.C.; Lucas, C.; Brandt, A. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005, 16, 2068–2076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Douglas, N.W.; Heine, M.J.; Williams, G.M.; Winther-Larsen, H.C.; Meaden, P.G. The STL1 gene of Saccharomyces cerevisiae is predicted to encode a sugar transporter-like protein. Gene 1994, 146, 215–219. [Google Scholar] [CrossRef]
- Fan, J.X.; Zhang, Z.; Long, C.N.; He, B.; Hu, Z.H.; Jiang, C.M.; Zeng, B. Identification and functional characterization of glycerol dehydrogenase reveal the role in kojic acid synthesis in Aspergillus MM. World. J. Microbiol. Biotechnol. 2020, 36, 136. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhang, H.X.; Fan, J.X.; Chen, Z.M.; Chen, T.M.; Zeng, B.; Zhang, Z. A highly efficient identification of mutants generated by CRISPR/Cas9 using the nonfunctional DsRed assisted selection in Aspergillus oryzae. World J. Microbiol. Biotechnol. 2021, 37, 132. [Google Scholar] [CrossRef]
- Bentley, R. Preparation and Analysis of Kojic Acid. Methods Enzymol. 1957, 3, 238–241. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Maruyama, J.; Kitamoto, K. Targeted gene disruption in Koji mold Aspergillus oryzae. Methods Mol. Biol. 2011, 765, 447–456. [Google Scholar]
- Zhang, Z.; Cheng, Z.J.; Gan, L.; Zhang, H.; Wu, F.Q.; Lin, Q.B.; Wang, J.L.; Wang, J.; Guo, X.P.; Zhang, X.; et al. OsHSD1, a hydroxysteroid dehydrogenase, is involved in cuticle formation and lipid homeostasis in rice. Plant Sci. 2016, 249, 35–45. [Google Scholar] [PubMed]
- Krijgsheld, P.; Bleichrodt, R.; van Veluw, G.J.; Wang, F.; Muller, W.H.; Dijksterhuis, J.; Wosten, H.A. Development in Aspergillus. Stud. Mycol. 2013, 74, 1–29. [Google Scholar] [PubMed]
- Ojeda-Lopez, M.; Chen, W.; Eagle, C.E.; Gutierrez, G.; Jia, W.L.; Swilaiman, S.S.; Huang, Z.; Park, H.S.; Yu, J.H.; Canovas, D.; et al. Evolution of asexual and sexual reproduction in the aspergilli. Stud. Mycol. 2018, 91, 37–59. [Google Scholar] [CrossRef]
- Etxebeste, O.; Espeso, E.A. Aspergillus nidulans in the post-genomic era: A top-model filamentous fungus for the study of signaling and homeostasis mechanisms. Int. Microbiol. 2020, 23, 5–22. [Google Scholar]
- Chang, P.K.; Scharfenstein, L.L.; Luo, M.; Mahoney, N.; Molyneux, R.J.; Yu, J.; Brown, R.L.; Campbell, B.C. Loss of msnA, a putative stress regulatory gene, in Aspergillus parasiticus and Aspergillus flavus increased production of conidia, aflatoxins and kojic acid. Toxins 2011, 3, 82–104. [Google Scholar] [CrossRef] [Green Version]
- Tulha, J.; Lima, A.; Lucas, C.; Ferreira, C. Saccharomyces cerevisiae glycerol/H+ symporter Stl1p is essential for cold/near-freeze and freeze stress adaptation. A simple recipe with high biotechnological potential is given. Microb. Cell Fact. 2010, 9, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duskova, M.; Borovikova, D.; Herynkova, P.; Rapoport, A.; Sychrova, H. The role of glycerol transporters in yeast cells in various physiological and stress conditions. FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar]
- Zemancikova, J.; Papouskova, K.; Perez-Torrado, R.; Querol, A.; Sychrova, H. Stl1 transporter mediating the uptake of glycerol is not a weak point of Saccharomyces kudriavzevii’s low osmotolerance. Lett. Appl. Microbiol. 2019, 68, 81–86. [Google Scholar]
- Kanamaru, K. Roles of the His-Asp phosphorelay signal transduction system in controlling cell growth and development in Aspergillus nidulans. Biosci. Biotechnol. Biochem. 2011, 75, 1–6. [Google Scholar]
- Sienko, M.; Natorff, R.; Skoneczny, M.; Kruszewska, J.; Paszewski, A.; Brzywczy, J. Regulatory mutations affecting sulfur metabolism induce environmental stress response in Aspergillus nidulans. Fungal Genet. Biol. 2014, 65, 37–47. [Google Scholar] [CrossRef]
- Saito, H.; Posas, F. Response to hyperosmotic stress. Genetics 2012, 192, 289–318. [Google Scholar] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.; Chen, Z.; Li, Y.; Zeng, B.; Zhang, Z. A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae. J. Fungi 2022, 8, 885. https://doi.org/10.3390/jof8080885
Chen T, Chen Z, Li Y, Zeng B, Zhang Z. A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae. Journal of Fungi. 2022; 8(8):885. https://doi.org/10.3390/jof8080885
Chicago/Turabian StyleChen, Tianming, Ziming Chen, Yuzhen Li, Bin Zeng, and Zhe Zhang. 2022. "A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae" Journal of Fungi 8, no. 8: 885. https://doi.org/10.3390/jof8080885
APA StyleChen, T., Chen, Z., Li, Y., Zeng, B., & Zhang, Z. (2022). A Novel Major Facilitator Superfamily Transporter Gene Aokap4 near the Kojic Acid Gene Cluster Is Involved in Growth and Kojic Acid Production in Aspergillus oryzae. Journal of Fungi, 8(8), 885. https://doi.org/10.3390/jof8080885






