Comparative Genomics of Mortierellaceae Provides Insights into Lipid Metabolism: Two Novel Types of Fatty Acid Synthase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Fermentation
2.2. Biomass, Fatty Acid Measurement, and Profiling
2.3. Genome Sequencing and Assembly
2.4. Gene Prediction and Functional Annotation
2.5. Phylogenomic and Phylogenetic Analyses
2.6. Comparative Genomic Analyses
3. Results
3.1. Fatty Acid Profiles of Mortierella alpina and M. schmuckeri
3.2. Genomic Features of Mortierella alpina and M. schmuckeri
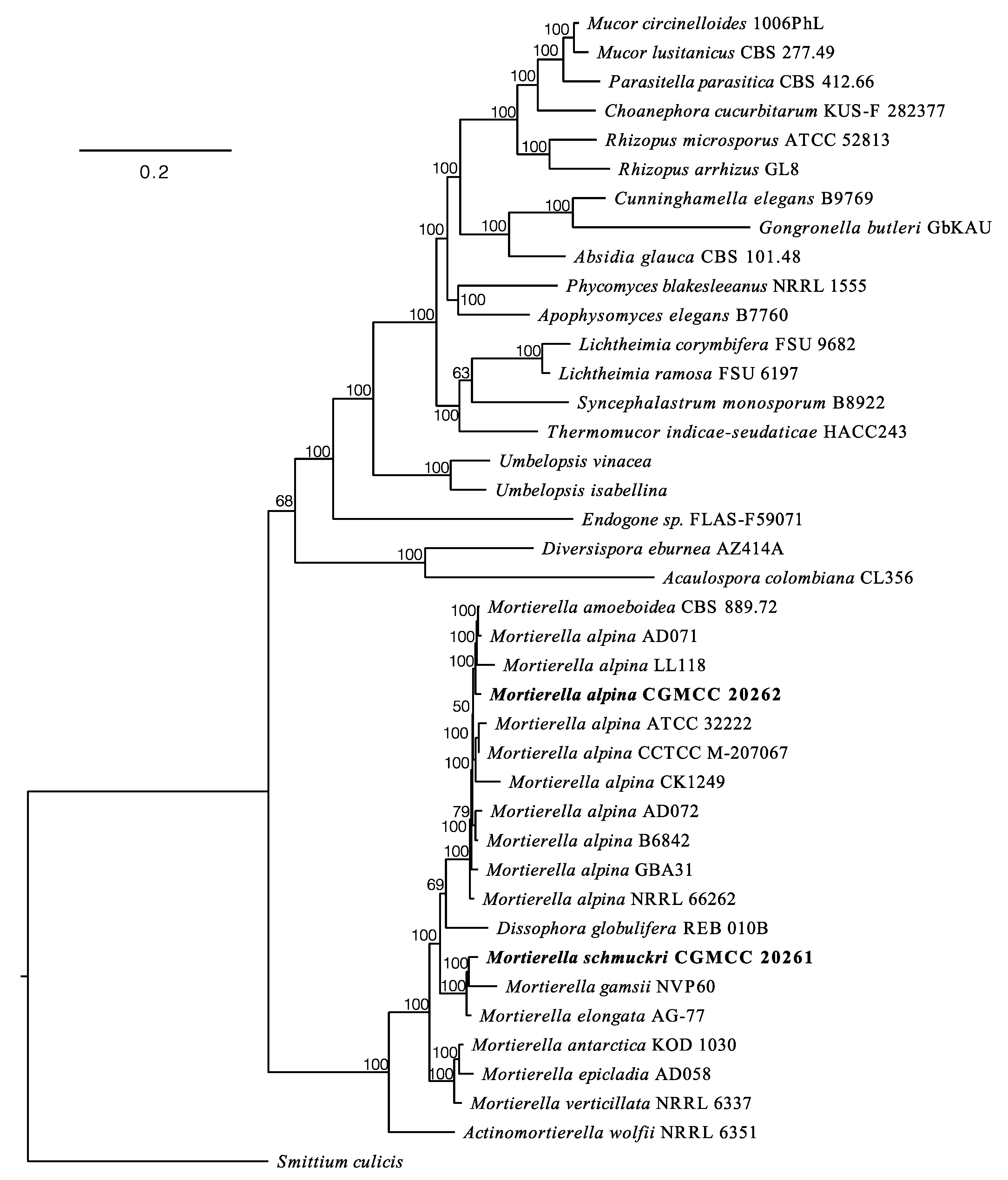
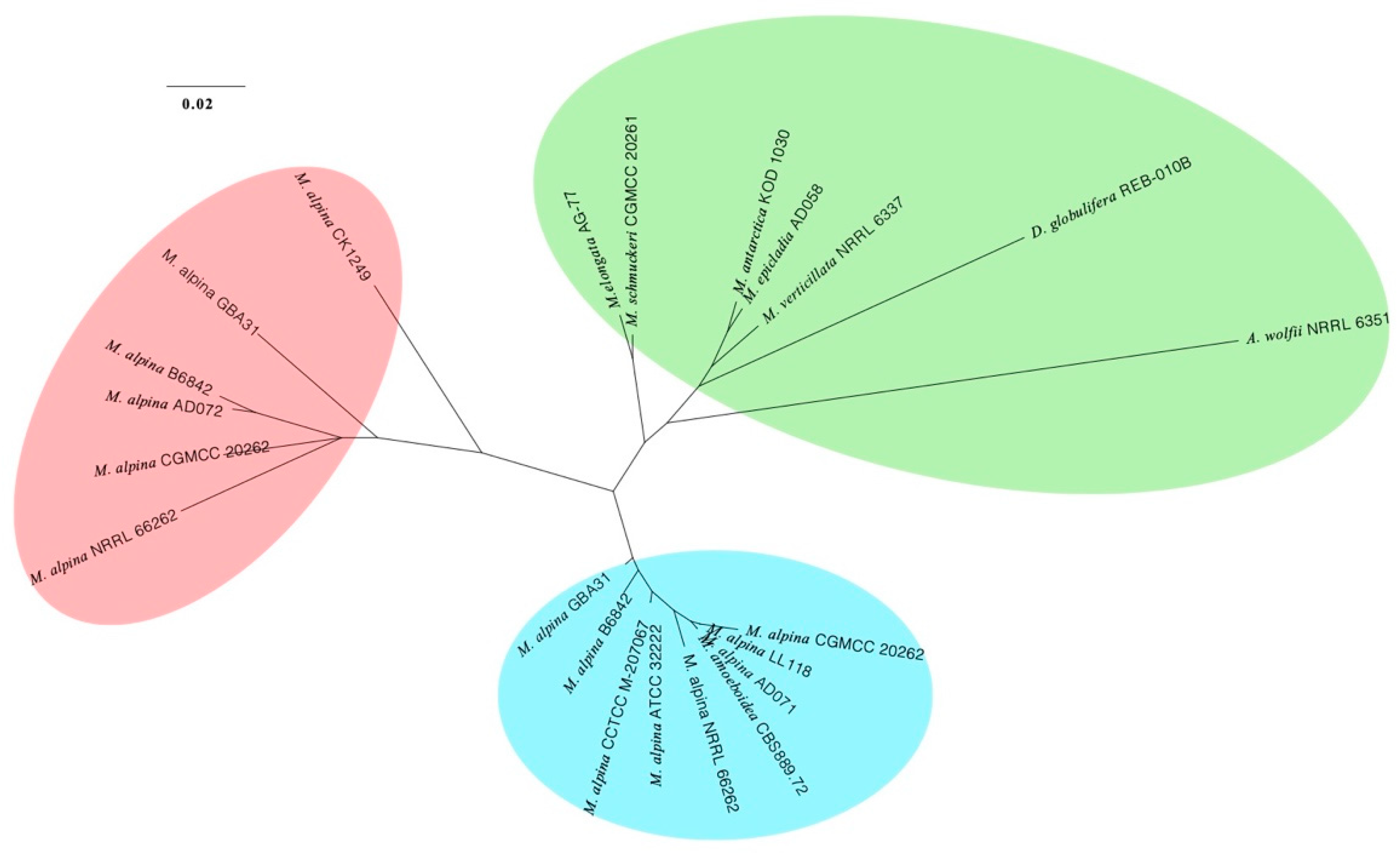
3.3. Phylogenomic Placements of Mortierella alpina and M. schmuckeri
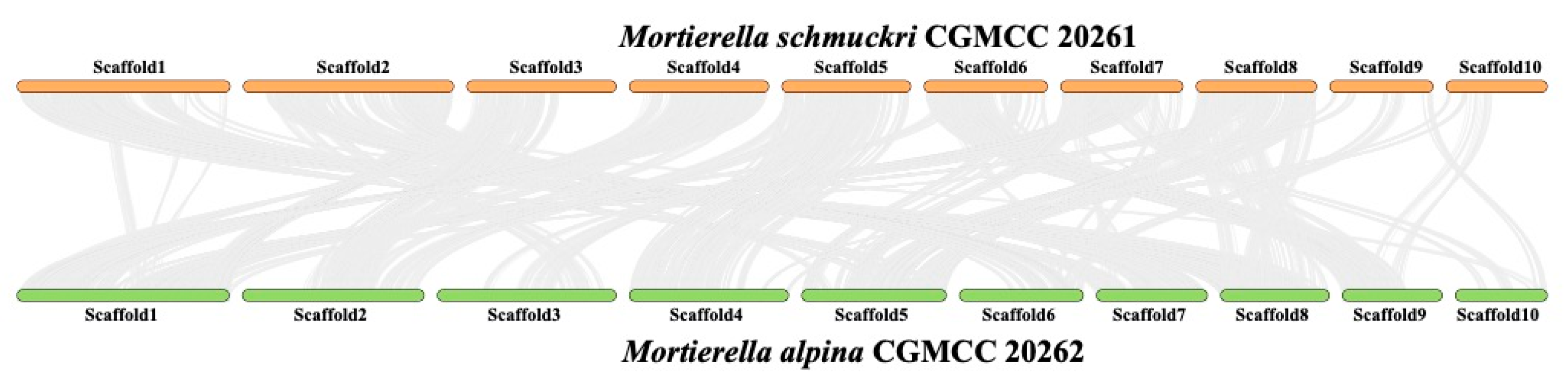
3.4. Synteny between Mortierella alpina and M. schmuckeri
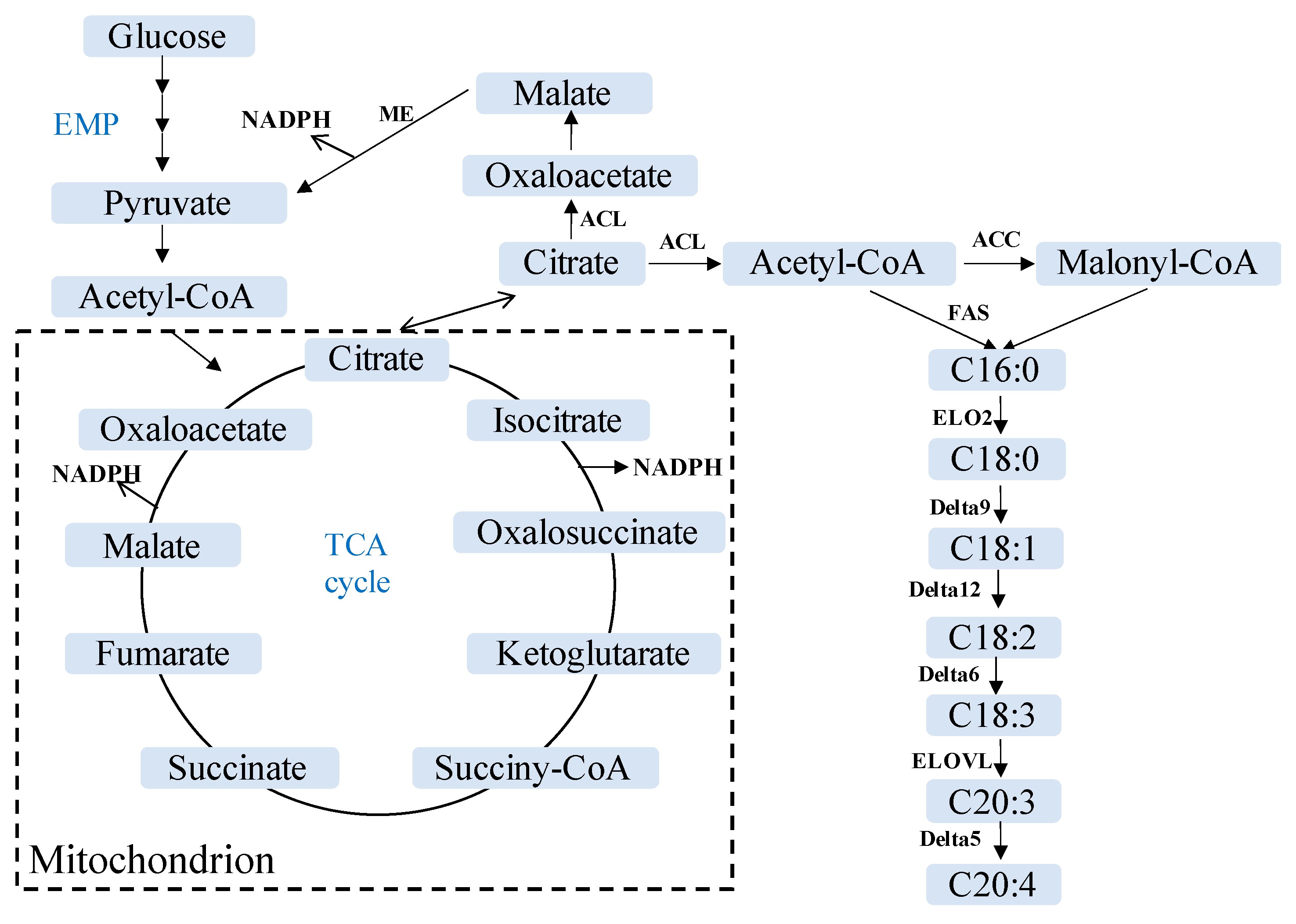
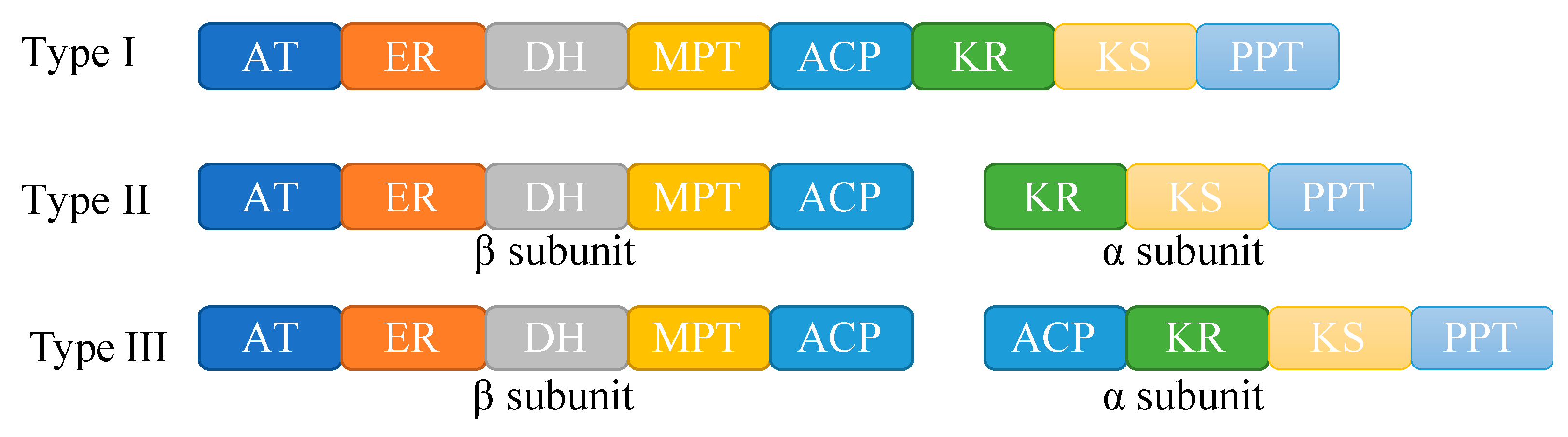
3.5. Lipid Metabolism in Mortierellaceae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, H.; Lv, M.L.; Liu, Z.; Zhang, M.Z.; Wang, Y.N.; Ju, X.; Song, Z.; Ren, L.Y.; Jia, B.S.; Qiao, M.; et al. High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnergy Res. 2021, 14, 1196–1206. [Google Scholar] [CrossRef]
- Reis, C.E.R.; Valle, G.F.; Bento, H.B.; Carvalho, A.K.; Alves, T.M.; de Castro, H.F. Sugarcane by-products within the biodiesel production chain: Vinasse and molasses as feedstock for oleaginous fungi and conversion to ethyl esters. Fuel 2020, 277, 118064. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Aggelis, G. Sources of microbial oils with emphasis to Mortierella (Umbelopsis) isabellina fungus. World J. Microb. Biot. 2019, 35, 63. [Google Scholar] [CrossRef] [PubMed]
- Kothri, M.; Mavrommati, M.; Elazzazy, A.M.; Baeshen, M.N.; Moussa, T.A.; Aggelis, G. Microbial sources of polyunsaturated fatty acids (PUFAs) and the prospect of organic residues and wastes as growth media for PUFA-producing microorganisms. FEMS Microbiol. Lett. 2020, 367, fnaa028. [Google Scholar] [CrossRef] [PubMed]
- Blazeck, J.; Hill, A.; Liu, L.; Knight, R.; Miller, J.; Pan, A.; Otoupal, P.; Alper, H.S. Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nat. Commun. 2014, 5, 3131. [Google Scholar] [CrossRef] [Green Version]
- Thammarongtham, C.; Nookaew, I.; Vorapreeda, T.; Srisuk, T.; Land, M.L.; Jeennor, S.; Laoteng, K. Genome characterization of oleaginous Aspergillus oryzae BCC7051: A potential fungal-based platform for lipid production. Curr. Microbiol. 2018, 75, 57–70. [Google Scholar] [CrossRef]
- Hashem, A.H.; Suleiman, W.B.; Abu-Elrish, G.M.; Ei-Sheikh, H.H. Consolidated bioprocessing of sugarcane bagasse to microbial oil by newly isolated oleaginous fungus: Mortierella wolfii. Arab. J. Sci. Eng. 2021, 46, 199–211. [Google Scholar] [CrossRef]
- Hashem, A.H.; Abu-Elreesh, G.; El-Sheikh, H.H.; Suleiman, W.B. Isolation, identification, and statistical optimization of a psychrotolerant Mucor racemosus for sustainable lipid production. Biomass Conv. Biorefinery 2022, 1–12. [Google Scholar] [CrossRef]
- Hashem, A.H.; Hasanin, M.S.; Khalil, A.M.A.; Suleiman, W.B. Eco-green conversion of watermelon peels to single cell oils using a unique oleaginous fungus: Lichtheimia corymbifera AH13. Waste Biomass Valor 2022, 11, 5721–5732. [Google Scholar] [CrossRef]
- Hashem, A.H.; Suleiman, W.B.; Abu-elreesh, G.; Shehabeldine, A.M.; Khalil, A.M.A. Sustainable lipid production from oleaginous fungus Syncephalastrum racemosum using synthetic and watermelon peel waste media. Bioresour. Technol. Rep. 2020, 12, 100569. [Google Scholar] [CrossRef]
- Gad, A.M.; Suleiman, W.B.; El-Sheikh, H.H.; Eimezayen, H.A.; Beltagy, A.M. Characterization of cellulase from Geotrichum candidum strain Gad1 approaching bioethanol production. Arab. J. Sci. Eng. 2022, 47, 6837–6850. [Google Scholar] [CrossRef]
- Wang, L.; Chen, W.; Feng, Y.; Ren, Y.; Gu, Z.; Chen, H.; Wang, H.; Thomas, M.J.; Zhang, B.; Berquin, I.M. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS ONE 2011, 6, e28319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bánki, O.; Roskov, Y.; Döring, M.; Ower, G.; Vandepitte, L.; Hobern, D.; Remsen, D.; Schalk, P.; DeWalt, R.; Keping, M.; et al. Catalogue of Life Checklist, Version 2022-03-21; Catalogue of Life. 2022. Available online: https://www.catalogueoflife.org/ (accessed on 2 July 2022).
- Eroshin, V.; Dedyukhina, E.; Chistyakova, T.; Zhelifonova, V.; Kurtzman, C.; Bothast, R. Arachidonic-acid production by species of Mortierella. World J. Microbiol. Biotechnol. 1996, 12, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Botha, A.; Paul, I.; Roux, C.; Kock, J.L.; Coetzee, D.J.; Strauss, T.; Maree, C. An isolation procedure for arachidonic acid producing Mortierella species. Antonie. Leeuw. 1999, 75, 253–256. [Google Scholar] [CrossRef]
- Kikukawa, H.; Sakuradani, E.; Ando, A.; Shimizu, S.; Ogawa, J. Arachidonic acid production by the oleaginous fungus Mortierella alpina 1S-4: A review. J. Adv. Res. 2018, 11, 15–22. [Google Scholar] [CrossRef]
- Ishikura, Y.; Ikeda, G.; Akimoto, K.; Hata, M.; Kusumoto, A.; Kidokoro, A.; Kontani, M.; Kawashima, H.; Kiso, Y.; Koga, Y. Arachidonic acid supplementation decreases P300 latency and increases P300 amplitude of event-related potentials in healthy elderly men. Neuropsychobiology 2009, 60, 73–79. [Google Scholar] [CrossRef]
- Tallima, H.; El Ridi, R. Arachidonic acid: Physiological roles and potential health benefits–a review. J. Adv. Res. 2018, 11, 33–41. [Google Scholar] [CrossRef]
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. In Advances in Applied Microbiology; Academic Press: San Diego, CA, USA, 2002; Volume 51, pp. 1–51. [Google Scholar]
- Ho, S.-Y.; Jiang, Y.; Chen, F. Polyunsaturated fatty acids (PUFAs) content of the fungus Mortierella alpina isolated from soil. J. Agric. Food Chem. 2007, 55, 3960–3966. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef] [Green Version]
- Kikukawa, H.; Sakuradani, E.; Ando, A.; Okuda, T.; Shimizu, S.; Ogawa, J. Microbial production of dihomo-γ-linolenic acid by Δ5-desaturase gene-disruptants of Mortierella alpina 1S-4. J. Biosci. Bioeng. 2016, 122, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Tang, X.; Lu, H.; Zhang, H.; Chen, Y.Q.; Chen, H.; Chen, W. Role of adenosine monophosphate deaminase during fatty acid accumulation in oleaginous fungus Mortierella alpina. J. Agric. Food Chem. 2019, 67, 9551–9559. [Google Scholar] [CrossRef] [PubMed]
- Rong, C.; Chen, H.; Tang, X.; Gu, Z.; Zhao, J.; Zhang, H.; Chen, Y.; Chen, W. Structural determinants of substrate specificity of omega-3 desaturases from Mortierella alpina and Rhizophagus irregularis by domain-swapping and molecular docking. Int. J. Mol. Sci. 2019, 20, 1603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorapreeda, T.; Thammarongtham, C.; Palasak, T.; Srisuk, T.; Jenjaroenpun, P.; Wongsurawat, T.; Nookaew, I.; Laoteng, K. Systematic genome analysis of a novel arachidonic acid-producing strain uncovered unique metabolic traits in the production of acetyl-CoA-derived products in Mortierella fungi. Gene 2020, 741, 144559. [Google Scholar] [CrossRef] [PubMed]
- Etienne, K.A.; Chibucos, M.C.; Su, Q.; Orvis, J.; Daugherty, S.; Ott, S.; Sengamalay, N.A.; Fraser, C.M.; Lockhart, S.R.; Bruno, V.M. Draft genome sequence of Mortierella alpina isolate CDC-B6842. Genome Announc. 2014, 2, e01180-13. [Google Scholar] [CrossRef] [Green Version]
- Vandepol, N.; Liber, J.; Desirò, A.; Na, H.; Kennedy, M.; Barry, K.; Grigoriev, I.V.; Miller, A.N.; O’Donnell, K.; Stajich, J.E. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020, 104, 267–289. [Google Scholar] [CrossRef]
- Yang, S.; Vinatzer, B.A. Draft Genome sequence of Mortierella alpina Strain LL118, isolated from an Aspen (Populus tremuloides) leaf litter sample. Microbiol. Resour. Announc. 2021, 10, e00864-21. [Google Scholar] [CrossRef]
- Seif, E.; Leigh, J.; Liu, Y.; Roewer, I.; Forget, L.; Lang, B.F. Comparative mitochondrial genomics in zygomycetes: Bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 2005, 33, 734–744. [Google Scholar] [CrossRef] [Green Version]
- Uehling, J.; Gryganskyi, A.; Hameed, K.; Tschaplinski, T.; Misztal, P.; Wu, S.; Desirò, A.; Vande Pol, N.; Du, Z.; Zienkiewicz, A. Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ. Microbiol. 2017, 19, 2964–2983. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, X.Y.; Huang, B. The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito (Mortierellales: Mortierellaceae). Mitochondrial DNA B 2022, 7, 374–376. [Google Scholar] [CrossRef]
- Kendrick, A.; Ratledge, C. Desaturation of polyunsaturated fatty acids in Mucor circinelloides and the involvement of a novel membrane-bound malic enzyme. Eur. J. Biochem. 1992, 209, 667–673. [Google Scholar] [CrossRef]
- Betina, V.; Koman, V. Changes in the lipid composition during the photo-induced conidiation of Trichoderma viride. Folia Microbiol. 1980, 25, 295. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.S.; Alexander, D.H.; Marks, P.; Klammer, A.A.; Drake, J.; Heiner, C.; Clum, A.; Copeland, A.; Huddleston, J.; Eichler, E.E. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanke, M.; Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 2003, 19, ii215–ii225. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Entwustke, S.; Wu, P.Z.; Yang, Z.L.; Busk, P.K.; Xu, Y.; Yin, Y.B. dbCAN2: A meta server for automated carbohydrate-activate enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [Green Version]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Ou, S.; Su, W.; Liao, Y.; Chougule, K.; Agda, J.R.; Hellinga, A.J.; Lugo, C.S.B.; Elliott, T.A.; Ware, D.; Peterson, T. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 2019, 20, 275. [Google Scholar] [CrossRef] [Green Version]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Stærfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, R.; Serra, F.; Tárraga, J.; Medina, I.; Carbonell, J.; Pulido, L.; de María, A.; Capella-Gutíerrez, S.; Huerta-Cepas, J.; Gabaldón, T. Phylemon 2.0: A suite of web-tools for molecular evolution, phylogenetics, phylogenomics and hypotheses testing. Nucleic Acids Res. 2011, 39, W470–W474. [Google Scholar] [CrossRef] [Green Version]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [Green Version]
- James, T.Y.; Pelin, A.; Bonen, L.; Ahrendt, S.; Sain, D.; Corradi, N.; Stajich, J.E. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr. Biol. 2013, 23, 1548–1553. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, S.; Burmester, A.; Wöstemeyer, J. Complete mitochondrial DNA sequence of the mucoralean fungus Absidia glauca, a model for studying host-parasite interactions. Genome Announc. 2016, 4, e00153-16. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Desirò, A.; Na, H.; Sandor, L.; Lipzen, A.; Clum, A.; Barry, K.; Grigoriev, I.V.; Martin, F.M.; Stajich, J.E. Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytol. 2019, 222, 511–525. [Google Scholar] [CrossRef] [Green Version]
- Schwartze, V.U.; Winter, S.; Shelest, E.; Marcet-Houben, M.; Horn, F.; Wehner, S.; Linde, J.; Valiante, V.; Sammeth, M.; Riege, K. Gene expansion shapes genome architecture in the human pathogen Lichtheimia corymbifera: An evolutionary genomics analysis in the ancient terrestrial Mucorales (Mucoromycotina). PLoS Genet. 2014, 10, e1004496. [Google Scholar] [CrossRef]
- Linde, J.; Schwartze, V.; Binder, U.; Lass-Flörl, C.; Voigt, K.; Horn, F. De novo whole-genome sequence and genome annotation of Lichtheimia ramosa. Genome Announc. 2014, 2, e00888-14. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Billmyre, R.B.; Li, A.; Carson, S.; Sykes, S.M.; Huh, E.Y.; Mieczkowski, P.; Ko, D.C.; Cuomo, C.A.; Heitman, J. Analysis of a food-borne fungal pathogen outbreak: Virulence and genome of a Mucor circinelloides isolate from yogurt. MBio 2014, 5, e01390-14. [Google Scholar] [CrossRef] [Green Version]
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 2016, 26, 1577–1584. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, M.H.; Kaul, D.; Muto, C.; Cheng, S.J.; Richter, R.A.; Bruno, V.M.; Liu, G.; Beyhan, S.; Sundermann, A.J.; Mounaud, S. Genetic diversity of clinical and environmental Mucorales isolates obtained from an investigation of mucormycosis cases among solid organ transplant recipients. Microb. Genom. 2020, 6, mgen000473. [Google Scholar] [CrossRef]
- Horn, F.; Üzüm, Z.; Möbius, N.; Guthke, R.; Linde, J.; Hertweck, C. Draft genome sequences of symbiotic and nonsymbiotic Rhizopus microsporus strains CBS 344.29 and ATCC 62417. Genome Announc. 2015, 3, e01370-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; White, M.M.; Kvist, S.; Moncalvo, J.-M. Genome-wide survey of gut fungi (Harpellales) reveals the first horizontally transferred ubiquitin gene from a mosquito host. Mol. Biol. Evol. 2016, 33, 2544–2554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeda, I.; Tamano, K.; Yamane, N.; Ishii, T.; Miura, A.; Umemura, M.; Terai, G.; Baker, S.E.; Koike, H.; Machida, M. Genome sequence of the Mucoromycotina fungus Umbelopsis isabellina, an effective producer of lipids. Genome Announc. 2014, 2, e00071-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Dulermo, T.; Nicaud, J.-M. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab. Eng. 2011, 13, 482–491. [Google Scholar] [CrossRef]
- Beopoulos, A.; Cescut, J.; Haddouche, R.; Uribelarrea, J.-L.; Molina-Jouve, C.; Nicaud, J.M. Yarrowia lipolytica as a model for bio-oil production. Prog. Lipid Res. 2009, 48, 375–387. [Google Scholar] [CrossRef]
- Rodríguez-Frómeta, R.A.; Gutiérrez, A.; Torres-Martínez, S.; Garre, V. Malic enzyme activity is not the only bottleneck for lipid accumulation in the oleaginous fungus Mucor circinelloides. Appl. Microbiol. Biot. 2013, 97, 3063–3072. [Google Scholar] [CrossRef]
- Jareonkitmongkol, S.; Sakuradani, E.; Shimizu, S. A novel Δ5-desaturase-defective mutant of Mortierella alpina 1S-4 and its dihomo-γ-linolenic acid productivity. Appl. Environ. Microb. 1993, 59, 4300–4304. [Google Scholar] [CrossRef] [Green Version]
- Parker-Barnes, J.M.; Das, T.; Bobik, E.; Leonard, A.E.; Thurmond, J.M.; Chaung, L.T.; Huang, Y.S.; Mukerji, P. Identification and characterization of an enzyme involved in the elongation of n-6 and n-3 polyunsaturated fatty acids. Proc. Natl. Acad. Sci. USA 2000, 97, 8284–8289. [Google Scholar] [CrossRef] [Green Version]
- Sakuradani, E.; Nojiri, M.; Suzuki, H.; Shimizu, S. Identification of a novel fatty acid elongase with a wide substrate specificity from arachidonic acid-producing fungus Mortierella alpina 1S-4. Appl. Microbiol. Biot. 2009, 84, 709–716. [Google Scholar] [CrossRef]
- Shin, G.H.; Veen, M.; Stahl, U.; Lang, C. Overexpression of genes of the fatty acid biosynthetic pathway leads to accumulation of sterols in Saccharomyces cerevisiae. Yeast 2012, 29, 371–383. [Google Scholar] [CrossRef] [PubMed]
- Tamano, K.; Bruno, K.S.; Karagiosis, S.A.; Culley, D.E.; Deng, S.; Collett, J.R.; Umemura, M.; Koike, H.; Baker, S.E.; Machida, M. Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl. Microbiol. Biot. 2013, 97, 269–281. [Google Scholar] [CrossRef] [PubMed]

| Species | Strains | BioSample | References |
|---|---|---|---|
| Absidia glauca | CBS 101.48 | SAMEA3923633 | [56] |
| Acaulospora morrowiae | CL551 | SAMEA8911292 | |
| Apophysomyces elegans | B7760 | SAMN02351510 | |
| Actinomortierella wolfii * | NRRL 6351 | SAMN05720777 | [27] |
| Choanephora cucurbitarum | KUS-F282377 | SAMN04532838 | |
| Cunninghamella elegans | B9769 | SAMN02351511 | |
| Dissophora globulifera * | REB 010B | SAMN05720531 | [27] |
| Diversispora eburnea | AZ414A | SAMEA8911293 | |
| Endogone sp. | FLAS-F59071 | SAMN09071421 | [57] |
| Gongronella butleri | GbKAU | SAMN15221701 | |
| Lichtheimia corymbifera | FSU 9682 | SAMEA2189700 | [58] |
| L. ramosa | FSU 6197 | SAMN05179542 | [59] |
| Mortierella alpina * | CGMCC 20262 | SAMN29490473 | This study |
| M. alpina * | AD071 | SAMN05720461 | [27] |
| M. alpina * | AD072 | SAMN05720462 | [27] |
| M. alpina * | ATCC 32222 | SAMN02981246 | [12] |
| M. alpina * | B6842 | SAMN02370960 | [26] |
| M. alpina * | CCTCC M-207067 | SAMN03658567 | |
| M. alpina * | CK1249 | SAMN05720518 | [27] |
| M. alpina * | GBA31 | SAMN05720773 | [27] |
| M. alpina * | LL118 | SAMN20056918 | [28] |
| M. alpina * | NRRL 66262 | SAMN10361219 | [27] |
| M. amoeboidea * | CBS 889.72 | SAMN19911466 | [31] |
| M. antarctica * | KOD1030 | SAMN05720520 | [27] |
| M. elongata * | AG-77 | SAMN02745706 | [30] |
| M. epicladia * | AD058 | SAMN05720441 | [27] |
| M. gamsii * | NVP60 | SAMN05720530 | [27] |
| M. verticillata * | NRRL 6337 | SAMN00699802 | [29] |
| M. schmuckeri * | CGMCC 20261 | SAMN29492047 | This study |
| Mucor circinelloides | 1006PhL | SAMN00103456 | [60] |
| M. lusitanicus | CBS 277.49 | SAMN00120579 | [61] |
| Parasitella parasitica | CBS 412.66 | SAMEA278055 | [56] |
| Rhizopus arrhizus | GL8 | SAMN14162349 | [62] |
| R. microsporus | ATCC 52813 | SAMN06821222 | [63] |
| Phycomyces blakesleeanus | NRRL 1555 | SAMN00189023 | [61] |
| Smittium culicis | GSMNP | SAMN04489870 | [64] |
| Syncephalastrum monosporum | B8922 | SAMN02370995 | |
| Thermomucor indicae-seudaticae | HACC 243 | SAMN03070115 | |
| Umbelopsis isabellina | WA0000067209 | SAMN16393839 | [65] |
| U. vinacea | WA0000051536 | SAMN16393840 |
| Species | M. alpina CGMCC 20262 | M. schmuckeri CGMCC 20261 | |
|---|---|---|---|
| Genome size (Mb) | 40.22 | 49.24 | |
| Scaffolds | 28 | 25 | |
| Largest scaffolds (Mb) | 4.35 | 4.78 | |
| GC (%) | 50.92 | 47.46 | |
| N50 (Mb) | 2.53 | 2.71 | |
| L50 | 6 | 8 | |
| Assembly BUSCO coverage (%) | 97.4 | 97.6 | |
| PCG models | 11,761 | 13,051 | |
| Pfam | 8507 | 8636 | |
| NT | 4026 | 4434 | |
| NR | 4904 | 6180 | |
| GO | 5411 | 5571 | |
| Eggnog | 9076 | 9436 | |
| KEGG | 4858 | 4921 | |
| CAZymes | 229 | 222 | |
| Gene clusters of secondary metabolites | |||
| Terpene | 4 | 2 | |
| Fungal-RiPP | 0 | 1 | |
| NRPS | 15 | 0 | |
| NRPS-like | 1 | 2 | |
| Siderophore | 2 | 0 | |
| Repetitive elements (% in genomes) | 7.29 | 7.45 | |
| ncRNA | |||
| rRNA | 38 | 29 | |
| tRNA | 226 | 262 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Nie, Y.; Jiang, Y.; Wang, S.; Zhang, T.-Y.; Liu, X.-Y. Comparative Genomics of Mortierellaceae Provides Insights into Lipid Metabolism: Two Novel Types of Fatty Acid Synthase. J. Fungi 2022, 8, 891. https://doi.org/10.3390/jof8090891
Zhao H, Nie Y, Jiang Y, Wang S, Zhang T-Y, Liu X-Y. Comparative Genomics of Mortierellaceae Provides Insights into Lipid Metabolism: Two Novel Types of Fatty Acid Synthase. Journal of Fungi. 2022; 8(9):891. https://doi.org/10.3390/jof8090891
Chicago/Turabian StyleZhao, Heng, Yong Nie, Yang Jiang, Shi Wang, Tian-Yu Zhang, and Xiao-Yong Liu. 2022. "Comparative Genomics of Mortierellaceae Provides Insights into Lipid Metabolism: Two Novel Types of Fatty Acid Synthase" Journal of Fungi 8, no. 9: 891. https://doi.org/10.3390/jof8090891
APA StyleZhao, H., Nie, Y., Jiang, Y., Wang, S., Zhang, T.-Y., & Liu, X.-Y. (2022). Comparative Genomics of Mortierellaceae Provides Insights into Lipid Metabolism: Two Novel Types of Fatty Acid Synthase. Journal of Fungi, 8(9), 891. https://doi.org/10.3390/jof8090891







