Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. pombe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fission Yeast Strains and Plasmids
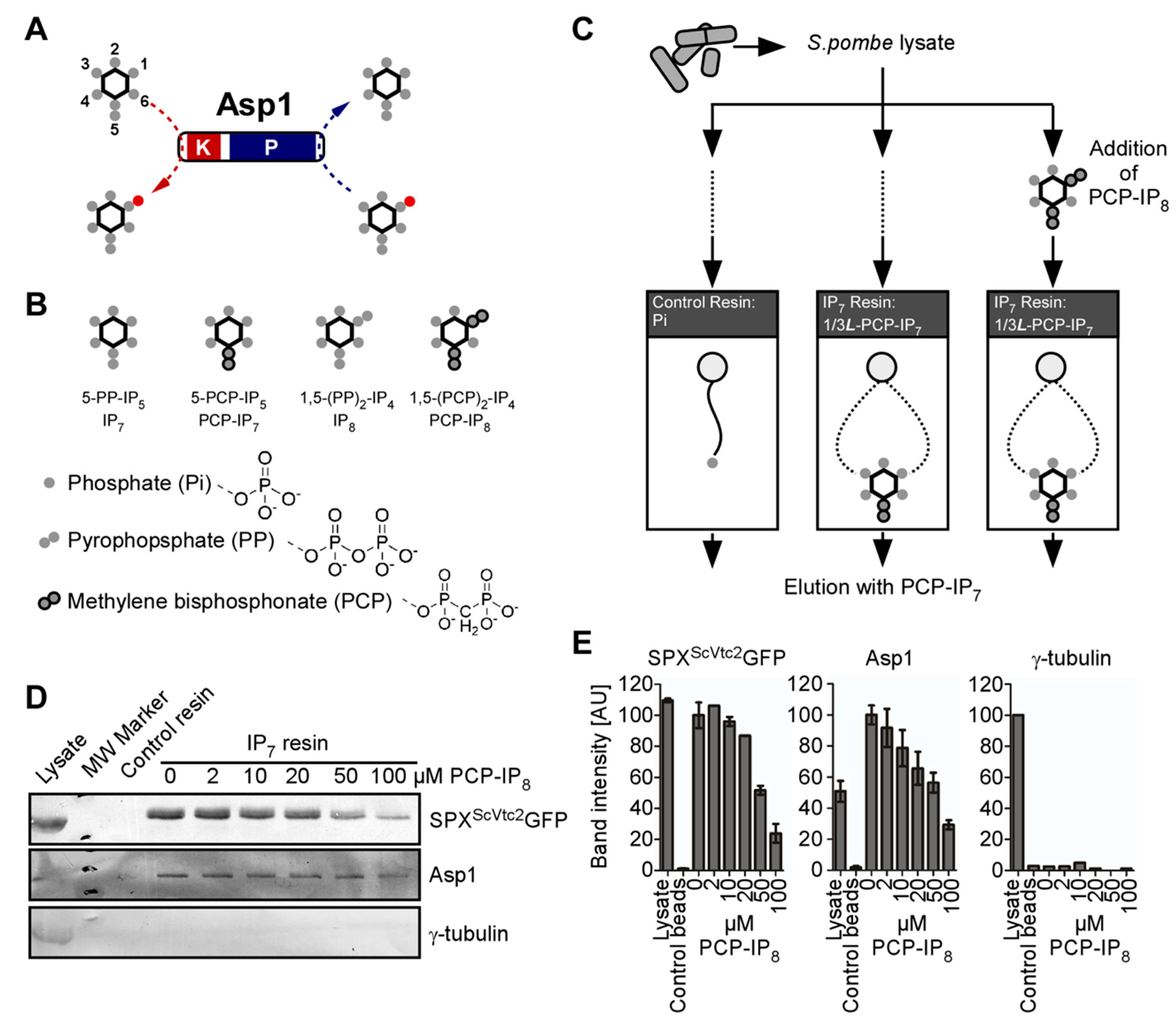
2.2. Affinity-Enrichment of IPP Binding Proteins
2.3. Microscopy
2.4. Protein Extraction, IP, and Western Blot Analysis of Mal2-GFP
2.5. qChIP
2.6. [3H]Inositol Labeling and HPLC Analysis
2.7. Quantification of IP8/IP6 Ratios
3. Results
3.1. IP8 Is Targeted by Endogenous Asp1 and Its Level Is Cell-Cycle Controlled
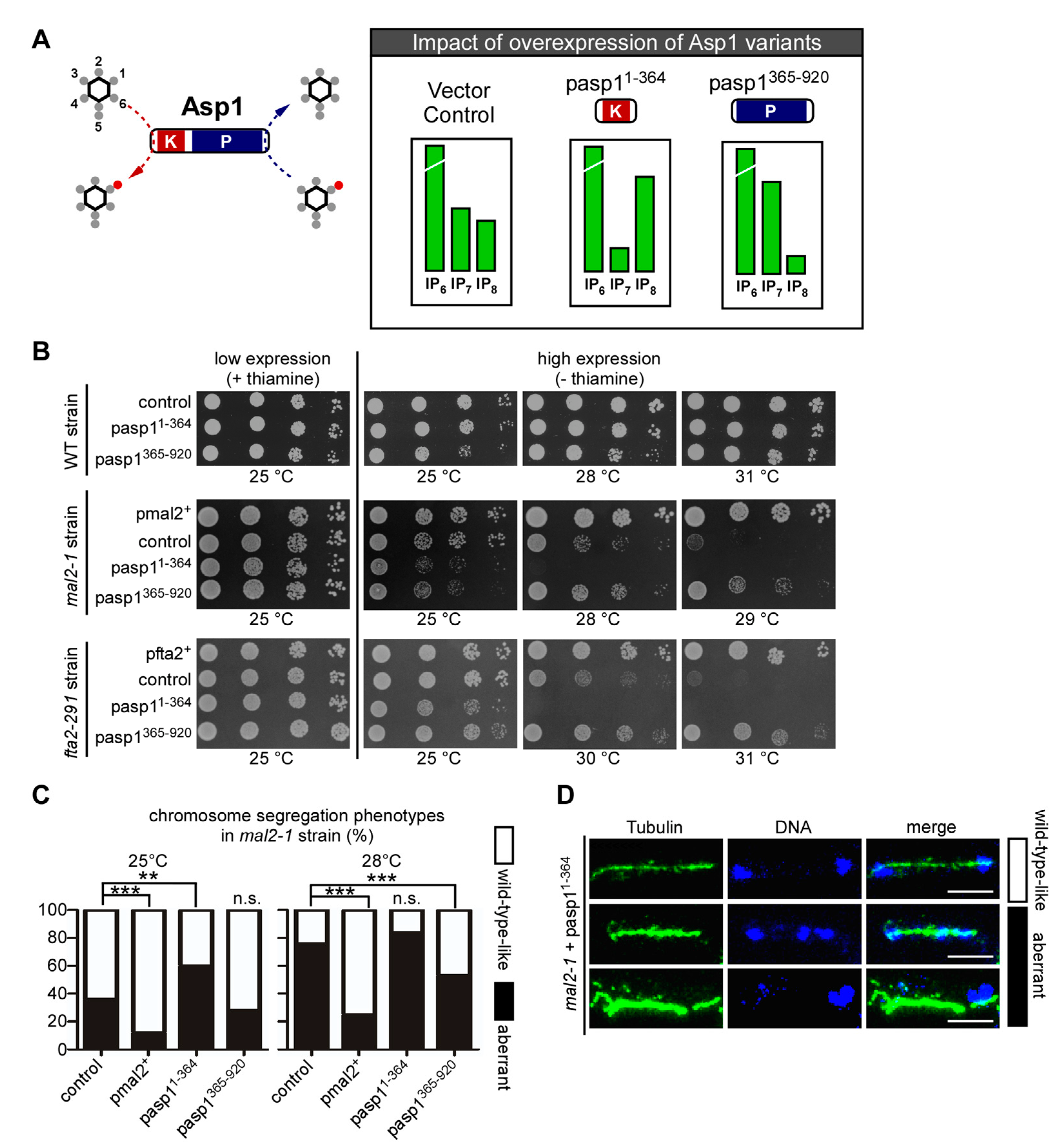
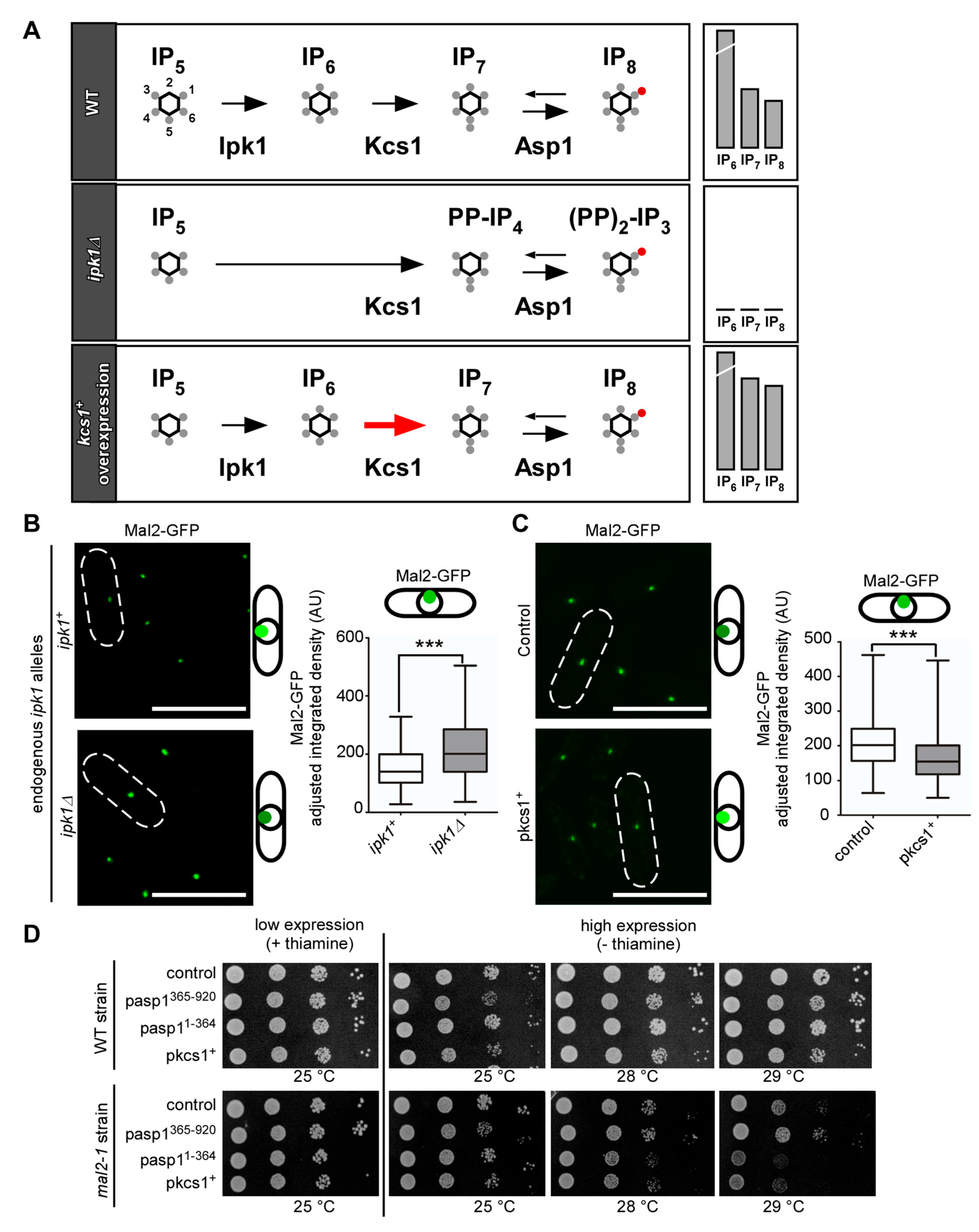
3.2. Alteration of Intracellular IP8 Levels Can Rescue Non-Growth of S. pombe CCAN Kinetochore Mutant Strains
3.3. Kinetochore-Targeting of the Mutant Mal2-1 Protein Is Subject to IP8 Levels
3.4. Wild-Type Mal2 Kinetochore-Targeting Is Modulated by Intracellular IP8 Levels
3.5. IP8, and Not IP7, Controls Kinetochore Targeting of S. pombe CCAN Components
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menniti, F.S.; Miller, R.N.; Putney, J.W., Jr.; Shears, S.B. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol. Chem. 1993, 268, 3850–3856. [Google Scholar] [CrossRef]
- Azevedo, C.; Burton, A.; Ruiz-Mateos, E.; Marsh, M.; Saiardi, A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. USA 2009, 106, 21161–21166. [Google Scholar] [CrossRef] [PubMed]
- Laha, D.; Johnen, P.; Azevedo, C.; Dynowski, M.; Weiss, M.; Capolicchio, S.; Mao, H.; Iven, T.; Steenbergen, M.; Freyer, M.; et al. VIH2 Regulates the Synthesis of Inositol Pyrophosphate InsP8 and Jasmonate-Dependent Defenses in Arabidopsis. Plant Cell 2015, 27, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Lev, S.; Li, C.; Desmarini, D.; Saiardi, A.; Fewings, N.L.; Schibeci, S.D.; Sharma, R.; Sorrell, T.C.; Djordjevic, J.T. Fungal Inositol Pyrophosphate IP7 Is Crucial for Metabolic Adaptation to the Host Environment and Pathogenicity. MBio 2015, 6, e00531-15. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Xu, J.; Cheng, W.; Rojas, T.; Chin, A.C.; Snowman, A.M.; Harraz, M.M.; Snyder, S.H. Neuronal migration is mediated by inositol hexakisphosphate kinase 1 via alpha-actinin and focal adhesion kinase. Proc. Natl. Acad. Sci. USA 2017, 114, 2036–2041. [Google Scholar] [CrossRef] [PubMed]
- Pöhlmann, J.; Fleig, U. Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell Biol. 2010, 30, 4535–4547. [Google Scholar] [CrossRef]
- Pohlmann, J.; Risse, C.; Seidel, C.; Pohlmann, T.; Jakopec, V.; Walla, E.; Ramrath, P.; Takeshita, N.; Baumann, S.; Feldbrugge, M.; et al. The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 2014, 10, e1004586. [Google Scholar] [CrossRef]
- Pulloor, N.K.; Nair, S.; McCaffrey, K.; Kostic, A.D.; Bist, P.; Weaver, J.D.; Riley, A.M.; Tyagi, R.; Uchil, P.D.; York, J.D.; et al. Human genome-wide RNAi screen identifies an essential role for inositol pyrophosphates in Type-I interferon response. PLoS Pathog. 2014, 10, e1003981. [Google Scholar] [CrossRef]
- Steidle, E.A.; Morrissette, V.A.; Fujimaki, K.; Chong, L.; Resnick, A.C.; Capaldi, A.P.; Rolfes, R.J. The InsP7 phosphatase Siw14 regulates inositol pyrophosphate levels to control localization of the general stress response transcription factor Msn2. J. Biol. Chem. 2020, 295, 2043–2056. [Google Scholar] [CrossRef]
- Benjamin, B.; Garg, A.; Jork, N.; Jessen, H.J.; Schwer, B.; Shuman, S. Activities and Structure-Function Analysis of Fission Yeast Inositol Pyrophosphate (IPP) Kinase-Pyrophosphatase Asp1 and Its Impact on Regulation of pho1 Gene Expression. mBio 2022, 13, e0103422. [Google Scholar] [CrossRef]
- Guan, Z.; Zhang, Q.; Zhang, Z.; Zuo, J.; Chen, J.; Liu, R.; Savarin, J.; Broger, L.; Cheng, P.; Wang, Q.; et al. Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2—PHR2 complex. Nat. Commun. 2022, 13, 1581. [Google Scholar] [CrossRef] [PubMed]
- Moritoh, Y.; Abe, S.I.; Akiyama, H.; Kobayashi, A.; Koyama, R.; Hara, R.; Kasai, S.; Watanabe, M. The enzymatic activity of inositol hexakisphosphate kinase controls circulating phosphate in mammals. Nat. Commun. 2021, 12, 4847. [Google Scholar] [CrossRef]
- Pascual-Ortiz, M.; Walla, E.; Fleig, U.; Saiardi, A. The PPIP5K Family Member Asp1 Controls Inorganic Polyphosphate Metabolism in S. pombe. J. Fungi. 2021, 7, 626. [Google Scholar] [CrossRef] [PubMed]
- Ried, M.K.; Wild, R.; Zhu, J.; Pipercevic, J.; Sturm, K.; Broger, L.; Harmel, R.K.; Abriata, L.A.; Hothorn, L.A.; Fiedler, D.; et al. Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 2021, 12, 384. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.S.; Jessen, H.J.; Saiardi, A. The inositol hexakisphosphate kinases IP6K1 and -2 regulate human cellular phosphate homeostasis, including XPR1-mediated phosphate export. J. Biol. Chem. 2019, 294, 11597–11608. [Google Scholar] [CrossRef] [PubMed]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; et al. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Land, E.S.; Cridland, C.A.; Craige, B.; Dye, A.; Hildreth, S.B.; Helm, R.F.; Gillaspy, G.E.; Perera, I.Y. A Role for Inositol Pyrophosphates in the Metabolic Adaptations to Low Phosphate in Arabidopsis. Metabolites 2021, 11, 601. [Google Scholar] [CrossRef]
- Lee, Y.S.; Mulugu, S.; York, J.D.; O’Shea, E.K. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 2007, 316, 109–112. [Google Scholar] [CrossRef]
- Sanchez, A.M.; Garg, A.; Shuman, S.; Schwer, B. Inositol pyrophosphates impact phosphate homeostasis via modulation of RNA 3’ processing and transcription termination. Nucleic Acids Res. 2019, 47, 8452–8469. [Google Scholar] [CrossRef]
- Desmarini, D.; Lev, S.; Furkert, D.; Crossett, B.; Saiardi, A.; Kaufman-Francis, K.; Li, C.; Sorrell, T.C.; Wilkinson-White, L.; Matthews, J.; et al. IP7-SPX Domain Interaction Controls Fungal Virulence by Stabilizing Phosphate Signaling Machinery. mBio 2020, 11, e01920-20. [Google Scholar] [CrossRef]
- Pesesse, X.; Choi, K.; Zhang, T.; Shears, S.B. Signaling by higher inositol polyphosphates. Synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J. Biol. Chem. 2004, 279, 43378–43381. [Google Scholar] [CrossRef] [PubMed]
- Nagata, E.; Luo, H.R.; Saiardi, A.; Bae, B.I.; Suzuki, N.; Snyder, S.H. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J. Biol. Chem. 2005, 280, 1634–1640. [Google Scholar] [CrossRef]
- Morrissette, V.A.; Rolfes, R.J. The intersection between stress responses and inositol pyrophosphates in Saccharomyces cerevisiae. Curr. Genet. 2020, 66, 901–910. [Google Scholar] [CrossRef]
- Ghosh, S.; Shukla, D.; Suman, K.; Lakshmi, B.J.; Manorama, R.; Kumar, S.; Bhandari, R. Inositol hexakisphosphate kinase 1 maintains hemostasis in mice by regulating platelet polyphosphate levels. Blood 2013, 122, 1478–1486. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Saiardi, A. Eukaryotic Phosphate Homeostasis: The Inositol Pyrophosphate Perspective. Trends Biochem. Sci. 2017, 42, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Chong, L.S.; Perlman, D.H.; Resnick, A.C.; Fiedler, D. Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Natl. Acad. Sci. USA 2016, 113, E6757–E6765. [Google Scholar] [CrossRef]
- Saiardi, A.; Bhandari, R.; Resnick, A.C.; Snowman, A.M.; Snyder, S.H. Phosphorylation of proteins by inositol pyrophosphates. Science 2004, 306, 2101–2105. [Google Scholar] [CrossRef]
- Bhandari, R.; Saiardi, A.; Ahmadibeni, Y.; Snowman, A.M.; Resnick, A.C.; Kristiansen, T.Z.; Molina, H.; Pandey, A.; Werner, J.K., Jr.; Juluri, K.R.; et al. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. USA 2007, 104, 15305–15310. [Google Scholar] [CrossRef]
- Fridy, P.C.; Otto, J.C.; Dollins, D.E.; York, J.D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 2007, 282, 30754–30762. [Google Scholar] [CrossRef]
- Mulugu, S.; Bai, W.; Fridy, P.C.; Bastidas, R.J.; Otto, J.C.; Dollins, D.E.; Haystead, T.A.; Ribeiro, A.A.; York, J.D. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 2007, 316, 106–109. [Google Scholar] [CrossRef]
- Choi, J.H.; Williams, J.; Cho, J.; Falck, J.R.; Shears, S.B. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J. Biol. Chem. 2007, 282, 30763–30775. [Google Scholar] [CrossRef] [PubMed]
- Saiardi, A.; Erdjument-Bromage, H.; Snowman, A.M.; Tempst, P.; Snyder, S.H. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 1999, 9, 1323–1326. [Google Scholar] [CrossRef]
- Saiardi, A.; Nagata, E.; Luo, H.R.; Snowman, A.M.; Snyder, S.H. Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 2001, 276, 39179–39185. [Google Scholar] [CrossRef]
- Draskovic, P.; Saiardi, A.; Bhandari, R.; Burton, A.; Ilc, G.; Kovacevic, M.; Snyder, S.H.; Podobnik, M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem. Biol. 2008, 15, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nair, V.S.; Holland, A.A.; Capolicchio, S.; Jessen, H.J.; Johnson, M.K.; Shears, S.B. Asp1 from Schizosaccharomyces pombe binds a [2Fe-2S]2+ cluster which inhibits inositol pyrophosphate 1-phosphatase activity. Biochemistry 2015, 54, 6462–6474. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Ortiz, M.; Saiardi, A.; Walla, E.; Jakopec, V.; Kunzel, N.A.; Span, I.; Vangala, A.; Fleig, U. Asp1 Bifunctional Activity Modulates Spindle Function via Controlling Cellular Inositol Pyrophosphate Levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 2018, 38, e00047-18. [Google Scholar] [CrossRef]
- Dollins, D.E.; Bai, W.; Fridy, P.C.; Otto, J.C.; Neubauer, J.L.; Gattis, S.G.; Mehta, K.P.M.; York, J.D. Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 9356–9364. [Google Scholar] [CrossRef]
- Feoktistova, A.; McCollum, D.; Ohi, R.; Gould, K.L. Identification and characterization of Schizosaccharomyces pombe asp1+, a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 1999, 152, 895–908. [Google Scholar] [CrossRef]
- Topolski, B.; Jakopec, V.; Kunzel, N.A.; Fleig, U. Inositol Pyrophosphate Kinase Asp1 Modulates Chromosome Segregation Fidelity and Spindle Function in Schizosaccharomyces pombe. Mol. Cell. Biol. 2016, 36, 3128–3140. [Google Scholar] [CrossRef]
- Schwer, B.; Garg, A.; Sanchez, A.M.; Bernstein, M.A.; Benjamin, B.; Shuman, S. Cleavage-Polyadenylation Factor Cft1 and SPX Domain Proteins are Agents of Inositol Pyrophosphate Toxicosis in Fission Yeast. mBio 2022, 13, e0347621. [Google Scholar] [CrossRef]
- Garg, A.; Shuman, S.; Schwer, B. A genetic screen for suppressors of hyper-repression of the fission yeast PHO regulon by Pol2 CTD mutation T4A implicates inositol 1-pyrophosphates as agonists of precocious lncRNA transcription termination. Nucleic Acids Res. 2020, 48, 10739–10752. [Google Scholar] [CrossRef] [PubMed]
- Sacristan, C.; Kops, G.J. Joined at the hip: Kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015, 25, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, C.E.; Yang, Y.; Halliday, G.M. Region- and Cell-specific Aneuploidy in Brain Aging and Neurodegeneration. Neuroscience 2018, 374, 326–334. [Google Scholar] [CrossRef]
- Coffman, V.C.; Wu, P.; Parthun, M.R.; Wu, J.Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol. 2011, 195, 563–572. [Google Scholar] [CrossRef] [PubMed]
- McKinley, K.L.; Cheeseman, I.M. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 16–29. [Google Scholar] [CrossRef]
- Musacchio, A.; Desai, A. A Molecular View of Kinetochore Assembly and Function. Biology 2017, 6, 5. [Google Scholar] [CrossRef]
- Chen, J.; Liao, A.; Powers, E.N.; Liao, H.; Kohlstaedt, L.A.; Evans, R.; Holly, R.M.; Kim, J.K.; Jovanovic, M.; Unal, E. Aurora B-dependent Ndc80 degradation regulates kinetochore composition in meiosis. Genes Dev. 2020, 34, 209–225. [Google Scholar] [CrossRef]
- Jimenez-Martin, A.; Pineda-Santaella, A.; Pinto-Cruz, J.; Leon-Perinan, D.; Garcia-Sanchez, S.; Delgado-Gestoso, D.; Marin-Toral, L.; Fernandez-Alvarez, A. The Rabl chromosome configuration masks a kinetochore reassembly mechanism in yeast mitosis. Mol. Biol. Cell 2022, 33, br8. [Google Scholar] [CrossRef]
- Fleig, U.; Sen-Gupta, M.; Hegemann, J.H. Fission yeast mal2+ is required for chromosome segregation. Mol. Cell Biol. 1996, 16, 6169–6177. [Google Scholar] [CrossRef] [Green Version]
- Kagawa, N.; Hori, T.; Hoki, Y.; Hosoya, O.; Tsutsui, K.; Saga, Y.; Sado, T.; Fukagawa, T. The CENP-O complex requirement varies among different cell types. Chromosome Res. 2014, 22, 293–303. [Google Scholar] [CrossRef]
- Poddar, A.; Roy, N.; Sinha, P. MCM21 and MCM22, two novel genes of the yeast Saccharomyces cerevisiae are required for chromosome transmission. Mol. Microbiol. 1999, 31, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Biggins, S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 2013, 194, 817–846. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Kukreja, A.A. How Kinetochore Architecture Shapes the Mechanisms of Its Function. Curr. Biol. 2017, 27, R816–R824. [Google Scholar] [CrossRef]
- Monda, J.K.; Cheeseman, I.M. The kinetochore-microtubule interface at a glance. J. Cell Sci. 2018, 131, jcs214577. [Google Scholar] [CrossRef]
- Sridhar, S.; Fukagawa, T. Kinetochore Architecture Employs Diverse Linker Strategies Across Evolution. Front. Cell Dev. Biol. 2022, 10, 862637. [Google Scholar] [CrossRef]
- Eskat, A.; Deng, W.; Hofmeister, A.; Rudolphi, S.; Emmerth, S.; Hellwig, D.; Ulbricht, T.; Doring, V.; Bancroft, J.M.; McAinsh, A.D.; et al. Step-wise assembly, maturation and dynamic behavior of the human CENP-P/O/R/Q/U kinetochore sub-complex. PLoS ONE 2012, 7, e44717. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar]
- Moreno, M.B.; Duran, A.; Ribas, J.C. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 2000, 16, 861–872. [Google Scholar] [CrossRef]
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hostachy, S.; Utesch, T.; Franke, K.; Dornan, G.L.; Furkert, D.; Turkaydin, B.; Haucke, V.; Sun, H.; Fiedler, D. Dissecting the activation of insulin degrading enzyme by inositol pyrophosphates and their bisphosphonate analogs. Chem. Sci. 2021, 12, 10696–10702. [Google Scholar] [CrossRef] [PubMed]
- Furkert, D.; Nadler-Holly, M.; Fiedler, D. Affinity enrichment and identification of inositol poly- and pyrophosphate interactomes. STAR Protoc. 2021, 2, 100277. [Google Scholar] [CrossRef] [PubMed]
- Furkert, D.; Hostachy, S.; Nadler-Holly, M.; Fiedler, D. Triplexed Affinity Reagents to Sample the Mammalian Inositol Pyrophosphate Interactome. Cell Chem. Biol. 2020, 27, 1097–1108.e4. [Google Scholar] [CrossRef]
- Hagan, I.M.; Hyams, J.S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1988, 89, 343–357. [Google Scholar] [CrossRef]
- Woods, A.; Sherwin, T.; Sasse, R.; MacRae, T.H.; Baines, A.J.; Gull, K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 1989, 93, 491–500. [Google Scholar] [CrossRef]
- Tran, P.T.; Paoletti, A.; Chang, F. Imaging green fluorescent protein fusions in living fission yeast cells. Methods 2004, 33, 220–225. [Google Scholar] [CrossRef]
- Gould, K.L.; Moreno, S.; Owen, D.J.; Sazer, S.; Nurse, P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 1991, 10, 3297–3309. [Google Scholar] [CrossRef]
- Ekwall, K.; Partridge, J.F. Fission yeast chromosome analysis. Fluorescence in situ hybridisation (FISH) and chromatin immunoprecipitation (CHIP). In Chromosome Structural Analysis: A Practical Approach; Oxford University Press: Oxford, UK, 1999; pp. 38–57. [Google Scholar]
- Jin, Q.W.; Pidoux, A.L.; Decker, C.; Allshire, R.C.; Fleig, U. The mal2p protein is an essential component of the fission yeast centromere. Mol. Cell Biol. 2002, 22, 7168–7183. [Google Scholar] [CrossRef]
- Pidoux, A.L.; Richardson, W.; Allshire, R.C. Sim4: A novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 2003, 161, 295–307. [Google Scholar] [CrossRef]
- Choi, E.S.; Stralfors, A.; Catania, S.; Castillo, A.G.; Svensson, J.P.; Pidoux, A.L.; Ekwall, K.; Allshire, R.C. Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-ACnp1 in fission yeast. PLoS Genet. 2012, 8, e1002985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, L.; Toda, N.R.; Rappsilber, J.; Allshire, R.C. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open Biol. 2014, 4, 140043. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, C.; Saiardi, A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006, 1, 2416–2422. [Google Scholar] [CrossRef]
- Wang, H.; Falck, J.R.; Hall, T.M.; Shears, S.B. Structural basis for an inositol pyrophosphate kinase surmounting phosphate crowding. Nat. Chem. Biol. 2011, 8, 111–116. [Google Scholar] [CrossRef]
- Dong, J.; Ma, G.; Sui, L.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.E.; Wittwer, C.; et al. Inositol Pyrophosphate InsP8 Acts as an Intracellular Phosphate Signal in Arabidopsis. Mol. Plant 2019, 12, 1463–1473. [Google Scholar] [CrossRef]
- Rustici, G.; Mata, J.; Kivinen, K.; Lio, P.; Penkett, C.J.; Burns, G.; Hayles, J.; Brazma, A.; Nurse, P.; Bahler, J. Periodic gene expression program of the fission yeast cell cycle. Nat. Genet. 2004, 36, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.; Thuriaux, P.; Nasmyth, K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. MGG 1976, 146, 167–178. [Google Scholar] [CrossRef]
- Nasmyth, K.; Nurse, P. Cell division cycle mutants altered in DNA replication and mitosis in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. MGG 1981, 182, 119–124. [Google Scholar] [CrossRef]
- Fantes, P. Epistatic gene interactions in the control of division in fission yeast. Nature 1979, 279, 428–430. [Google Scholar] [CrossRef]
- Hirano, T.; Funahashi, S.; Uemura, T.; Yanagida, M. Isolation and characterization of Schizosaccharomyces pombe cutmutants that block nuclear division but not cytokinesis. EMBO J. 1986, 5, 2973–2979. [Google Scholar] [CrossRef]
- Hagan, I.M.; Grallert, A. Spatial control of mitotic commitment in fission yeast. Biochem. Soc. Trans. 2013, 41, 1766–1771. [Google Scholar] [CrossRef] [PubMed]
- Russell, P.; Nurse, P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 1986, 45, 145–153. [Google Scholar] [CrossRef]
- Matsuo, Y.; Maurer, S.P.; Surrey, T.; Toda, T. Purification and characterisation of the fission yeast Ndc80 complex. Protein Expr. Purif. 2017, 135, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Obuse, C.; Iwasaki, O.; Kiyomitsu, T.; Goshima, G.; Toyoda, Y.; Yanagida, M. A conserved Mis12 centromere complex is linked to heterochromatic HP1 and outer kinetochore protein Zwint-1. Nat. Cell Biol. 2004, 6, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Jakopec, V.; Topolski, B.; Fleig, U. Sos7, an essential component of the conserved Schizosaccharomyces pombe Ndc80-MIND-Spc7 complex, identifies a new family of fungal kinetochore proteins. Mol. Cell Biol. 2012, 32, 3308–3320. [Google Scholar] [CrossRef]
- Liu, X.; McLeod, I.; Anderson, S.; Yates, J.R., 3rd; He, X. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005, 24, 2919–2930. [Google Scholar] [CrossRef]
- Folco, H.D.; Campbell, C.S.; May, K.M.; Espinoza, C.A.; Oegema, K.; Hardwick, K.G.; Grewal, S.I.S.; Desai, A. The CENP-A N-tail confers epigenetic stability to centromeres via the CENP-T branch of the CCAN in fission yeast. Curr. Biol. 2015, 25, 348–356. [Google Scholar] [CrossRef]
- Funabiki, H.; Hagan, I.; Uzawa, S.; Yanagida, M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993, 121, 961–976. [Google Scholar] [CrossRef]
- Fernandez-Alvarez, A.; Cooper, J.P. The functionally elusive RabI chromosome configuration directly regulates nuclear membrane remodeling at mitotic onset. Cell Cycle 2017, 16, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Kerres, A.; Jakopec, V.; Beuter, C.; Karig, I.; Pohlmann, J.; Pidoux, A.; Allshire, R.; Fleig, U. Fta2, an essential fission yeast kinetochore component, interacts closely with the conserved Mal2 protein. Mol. Biol. Cell 2006, 17, 4167–4178. [Google Scholar] [CrossRef]
- Haring, M.; Offermann, S.; Danker, T.; Horst, I.; Peterhansel, C.; Stam, M. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 2007, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitzberger, F.; Harrison, S.C. RWD domain: A recurring module in kinetochore architecture shown by a Ctf19-Mcm21 complex structure. EMBO Rep. 2012, 13, 216–222. [Google Scholar] [CrossRef] [PubMed]
- Pesenti, M.E.; Prumbaum, D.; Auckland, P.; Smith, C.M.; Faesen, A.C.; Petrovic, A.; Erent, M.; Maffini, S.; Pentakota, S.; Weir, J.R.; et al. Reconstitution of a 26-Subunit Human Kinetochore Reveals Cooperative Microtubule Binding by CENP-OPQUR and NDC80. Mol. Cell 2018, 71, 923–939.e10. [Google Scholar] [CrossRef] [PubMed]
- Schmitzberger, F.; Richter, M.M.; Gordiyenko, Y.; Robinson, C.V.; Dadlez, M.; Westermann, S. Molecular basis for inner kinetochore configuration through RWD domain-peptide interactions. EMBO J. 2017, 36, 3458–3482. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, B.; Wente, S.R. Dual functions for the Schizosaccharomyces pombe inositol kinase Ipk1 in nuclear mRNA export and polarized cell growth. Eukaryot. Cell 2009, 8, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef]
- Kim, D.U.; Hayles, J.; Kim, D.; Wood, V.; Park, H.O.; Won, M.; Yoo, H.S.; Duhig, T.; Nam, M.; Palmer, G.; et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 2010, 28, 617–623. [Google Scholar] [CrossRef]
- Banfic, H.; Crljen, V.; Lukinovic-Skudar, V.; Dembitz, V.; Lalic, H.; Bedalov, A.; Visnjic, D. Inositol pyrophosphates modulate cell cycle independently of alteration in telomere length. Adv. Biol. Regul. 2016, 60, 22–28. [Google Scholar] [CrossRef]
- Banfic, H.; Bedalov, A.; York, J.D.; Visnjic, D. Inositol pyrophosphates modulate S phase progression after pheromone-induced arrest in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 1717–1725. [Google Scholar] [CrossRef]
- Safrany, S.T.; Caffrey, J.J.; Yang, X.; Bembenek, M.E.; Moyer, M.B.; Burkhart, W.A.; Shears, S.B. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998, 17, 6599–6607. [Google Scholar] [CrossRef]
- Safrany, S.T.; Ingram, S.W.; Cartwright, J.L.; Falck, J.R.; McLennan, A.G.; Barnes, L.D.; Shears, S.B. The diadenosine hexaphosphate hydrolases from Schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase. Overlapping substrate specificities in a MutT-type protein. J. Biol. Chem. 1999, 274, 21735–21740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonetti, A.; Szijgyarto, Z.; Bosch, D.; Loss, O.; Azevedo, C.; Saiardi, A. Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 2011, 286, 31966–31974. [Google Scholar] [CrossRef] [PubMed]
- Steidle, E.A.; Chong, L.S.; Wu, M.; Crooke, E.; Fiedler, D.; Resnick, A.C.; Rolfes, R.J. A Novel Inositol Pyrophosphate Phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the beta-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5). J. Biol. Chem. 2016, 291, 6772–6783. [Google Scholar] [CrossRef] [PubMed]
- Ohkura, H.; Hagan, I.M.; Glover, D.M. The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995, 9, 1059–1073. [Google Scholar] [CrossRef]
- Kim, J.; Goshima, G. Mitotic spindle formation in the absence of Polo kinase. Proc. Natl. Acad. Sci. USA 2022, 119, e2114429119. [Google Scholar] [CrossRef]
- Zong, G.; Jork, N.; Hostachy, S.; Fiedler, D.; Jessen, H.J.; Shears, S.B.; Wang, H. New structural insights reveal an expanded reaction cycle for inositol pyrophosphate hydrolysis by human DIPP1. FASEB J. 2021, 35, e21275. [Google Scholar] [CrossRef]
- Marquez-Monino, M.A.; Ortega-Garcia, R.; Shipton, M.L.; Franco-Echevarria, E.; Riley, A.M.; Sanz-Aparicio, J.; Potter, B.V.L.; Gonzalez, B. Multiple substrate recognition by yeast diadenosine and diphosphoinositol polyphosphate phosphohydrolase through phosphate clamping. Sci. Adv. 2021, 7, eabf6744. [Google Scholar] [CrossRef]
- Pintard, L.; Archambault, V. A unified view of spatio-temporal control of mitotic entry: Polo kinase as the key. Open Biol. 2018, 8, 180114. [Google Scholar] [CrossRef]
- Navarro, A.P.; Cheeseman, I.M. Kinetochore assembly throughout the cell cycle. Semin. Cell Dev. Biol. 2021, 117, 62–74. [Google Scholar] [CrossRef]
- Gascoigne, K.E.; Cheeseman, I.M. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol. 2013, 201, 23–32. [Google Scholar] [CrossRef]
- Navarro, A.P.; Cheeseman, I.M. Dynamic cell cycle-dependent phosphorylation modulates CENP-L-CENP-N centromere recruitment. Mol. Biol. Cell 2022, 33, mbcE22060239. [Google Scholar] [CrossRef]
- Klemm, C.; Thorpe, P.H.; Olafsson, G. Cell-cycle phospho-regulation of the kinetochore. Curr. Genet. 2021, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Nabetani, A.; Koujin, T.; Tsutsumi, C.; Haraguchi, T.; Hiraoka, Y. A conserved protein, Nuf2, is implicated in connecting the centromere to the spindle during chromosome segregation: A link between the kinetochore function and the spindle checkpoint. Chromosoma 2001, 110, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.S.; Toda, T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr. Biol. 2011, 21, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Kerres, A.; Jakopec, V.; Fleig, U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol. Biol. Cell 2007, 18, 2441–2454. [Google Scholar] [CrossRef]
- Hayashi, T.; Fujita, Y.; Iwasaki, O.; Adachi, Y.; Takahashi, K.; Yanagida, M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 2004, 118, 715–729. [Google Scholar] [CrossRef]
- Takahashi, K.; Chen, E.S.; Yanagida, M. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 2000, 288, 2215–2219. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuenzel, N.A.; Alcázar-Román, A.R.; Saiardi, A.; Bartsch, S.M.; Daunaraviciute, S.; Fiedler, D.; Fleig, U. Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. pombe. J. Fungi 2022, 8, 933. https://doi.org/10.3390/jof8090933
Kuenzel NA, Alcázar-Román AR, Saiardi A, Bartsch SM, Daunaraviciute S, Fiedler D, Fleig U. Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. pombe. Journal of Fungi. 2022; 8(9):933. https://doi.org/10.3390/jof8090933
Chicago/Turabian StyleKuenzel, Natascha Andrea, Abel R. Alcázar-Román, Adolfo Saiardi, Simon M. Bartsch, Sarune Daunaraviciute, Dorothea Fiedler, and Ursula Fleig. 2022. "Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. pombe" Journal of Fungi 8, no. 9: 933. https://doi.org/10.3390/jof8090933
APA StyleKuenzel, N. A., Alcázar-Román, A. R., Saiardi, A., Bartsch, S. M., Daunaraviciute, S., Fiedler, D., & Fleig, U. (2022). Inositol Pyrophosphate-Controlled Kinetochore Architecture and Mitotic Entry in S. pombe. Journal of Fungi, 8(9), 933. https://doi.org/10.3390/jof8090933






