Eoscyphella luciurceolata gen. and sp. nov. (Agaricomycetes) Shed Light on Cyphellopsidaceae with a New Lineage of Bioluminescent Fungi
Abstract
1. Introduction
2. Materials and Methods
2.1. Collecting Area
2.1.1. Brazilian Site of the New Luminescent Taxon
2.1.2. French Site of Maireina monacha
2.2. Morphological Analyses
2.3. Molecular Methods
2.4. Phylogenetic Analyses
3. Results
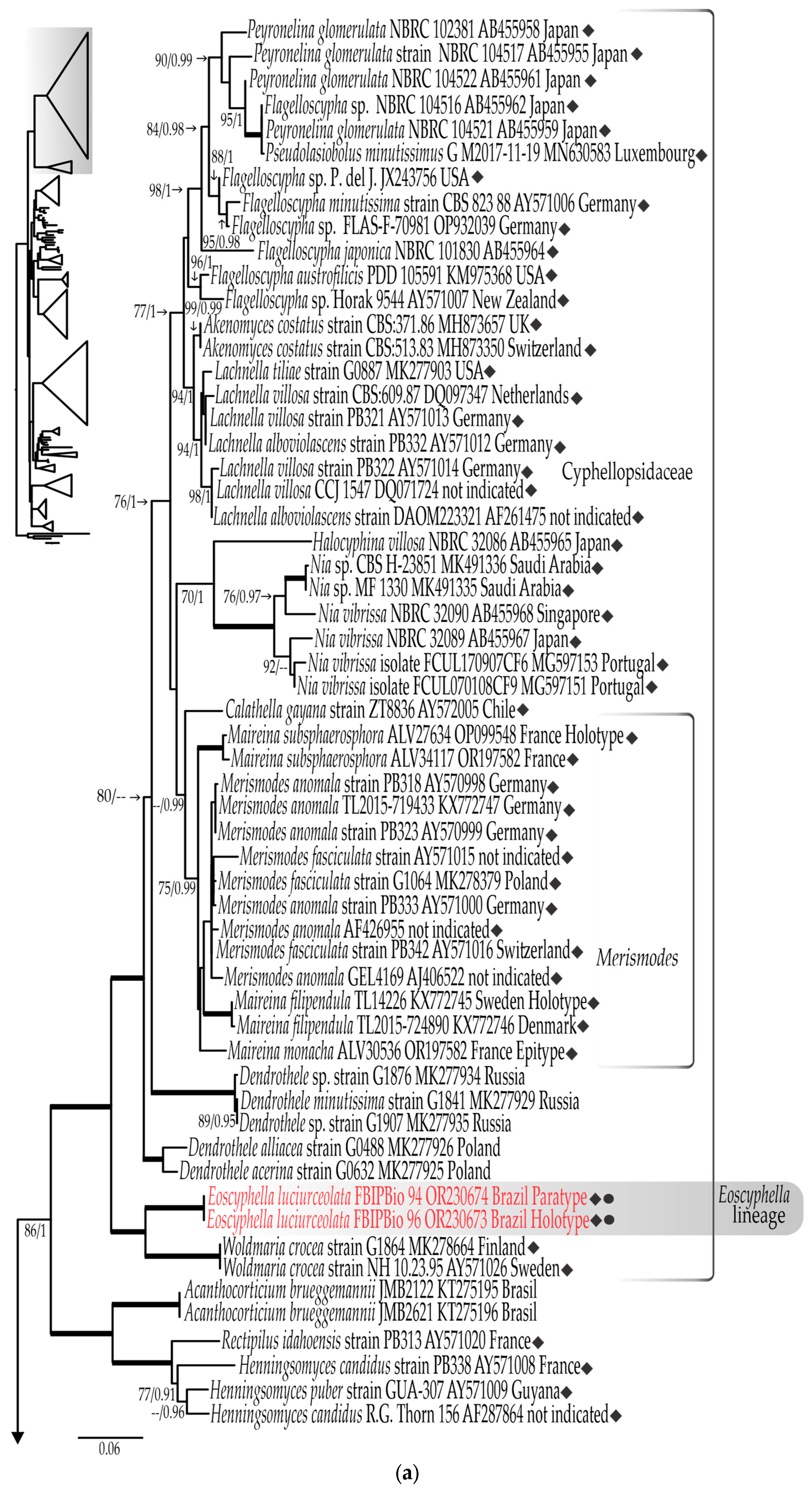
3.1. Phylogenetic Results
3.1.1. LSU rDNA Dataset
3.1.2. ITS rDNA Dataset
3.1.3. Combined LSU rDNA + ITS rDNA Dataset
3.2. Taxonomic Part
- Merismodes Earle, Bulletin of the New York Botanical Garden 5: 406 (1909) emend. Silva-Filho & Menolli
- Merismodes monacha (Speg.) Silva-Filho, Mombert & Menolli comb. nov.
- MycoBank: MB 849402
- Merismodes filipendula (Læssøe) Silva-Filho & Menolli comb. nov.
- Merismodes subsphaerospora (Mombert) Silva-Filho, Mombert & Menolli comb. nov.
- Eoscyphella Silva-Filho, Stevani & Menolli gen. nov.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hibbett, D.S.; Bauer, R.; Binder, M.; Giachini, A.J.; Hosaka, K.; Justo, A.; Larsson, E.; Larsson, K.H.; Lawrey, J.D.; Miettinenm, O.; et al. (Eds.) Systematics and Evolution. In The Mycota; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7A. [Google Scholar]
- Kaskova, Z.M.; Dorr, F.A.; Petushkov, V.N.; Purtov, K.V.; Tsarkova, A.S.; Rodionova, N.S.; Mineev, K.S.; Guglya, E.B.; Kotlobay, A.; Baleeva, N.S.; et al. Mechanism and color modulation of fungal bioluminescence. Sci. Adv. 2017, 3, e1602847. [Google Scholar] [CrossRef]
- Kotlobay, K.; Sarkisyan, K.S.; Mokrushina, Y.A.; Marcet-Houben, M.; Serebrovskaya, E.O.; Markina, N.M.; Somermeyer, L.G.; Gorokhovatsky, A.Y.; Vvedensky, A.; Purtov, K.V.; et al. A genetically encodable bioluminescent system from fungi. Proc. Natl. Acad. Sci. USA 2018, 115, 12728–12732. [Google Scholar] [CrossRef] [PubMed]
- Desjardin, D.E.; Oliveira, A.G.; Stevani, C.V. Fungi bioluminescence revisited. Photochem. Photobiol. Sci. 2008, 7, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Heller, J.F. Ueber das Leuchten im Pflanzen und Tierreiche. Pathol. Chem. Mikr 1853, 6, 44–54, 81–90, 121–137, 161–166, 201–216, 241–251. [Google Scholar]
- Buller, A.H.R. The bioluminescence of Panus stipticus. Res. Fungi 1924, 3, 357–431. [Google Scholar]
- Bothe, F. Über das Leuchten verwesender Blätter und seine Erreger. Planta 1931, 14, 752–765. [Google Scholar] [CrossRef]
- Buller, A.H.R. Omphalia flavida, a gemmiferous and luminous leaf-spot fungus. Res. Fungi 1934, 6, 397–443. [Google Scholar]
- Wassink, W.C. Luminescence in fungi. In Bioluminescence in Action; Academic Press: New York, NY, USA, 1978. [Google Scholar]
- Corner, E.J.H. Descriptions of two luminous tropical agarics (Dictyopanus and Mycena). Mycologia 1950, 42, 423–443. [Google Scholar] [CrossRef]
- Kobayasi, Y. Contributions to the luminous fungi from Japan. J. Hattori Bot. Lab. 1951, 5, 1–6. [Google Scholar]
- Josserand, M. Sur la luminescence de “Mycena rorida” en Europe occidentale. Bull. Mens. Soc. Linn. Lyon. 1953, 22, 99–102. [Google Scholar] [CrossRef]
- Corner, E.J.H. Further descriptions of luminous agarics. Trans. Br. Myco Soc. 1954, 37, 256–271. [Google Scholar] [CrossRef]
- Haneda, Y. Luminous organisms of Japan and the Far East. In The Luminescence of Biological Systems; American Association for the Advancement of Science: Washington, DC, USA, 1955. [Google Scholar]
- Bigelow, H.E.; Miller, O.K., Jr.; Thiers, H.D. A new species of Omphalotus. Mycotaxon 1976, 3, 363–372. [Google Scholar]
- Horak, E. Mycena rorida (Fr.) Quel. and related species from the Southern Hemisphere. Ber. Schweiz. Bot. Ges. 1978, 88, 20–29. [Google Scholar]
- Zang, M. Some new species of higher fungi from Xizang (Tibet) of China. Acta Bot. Yunnan 1979, 1, 101–105. [Google Scholar]
- Corner, E.J.H. The agaric genera Lentinus, Panus and Pleurotus. Nova Hedwig. Beih. 1981, 69, 1–169. [Google Scholar]
- Treu, R.; Agerer, A. Culture characteristics of some Mycena species. Mycotaxon 1990, 38, 279–309. [Google Scholar]
- Maas Geesteranus, R.A. Verhandelingen. In Mycenas of the Northern Hemisphere; Afdeling natuurkunde: Amsterdam, The Netherlands, 1992. [Google Scholar]
- Li, J.; Hu, X.A. New species of Lampteromyces from Hunan. Acta Sci. Nat. Univ. Norm. Hunan 1993, 16, 188–189. [Google Scholar]
- Desjardin, D.E.; Capelari, M.; Stevani, C.V. A new bioluminescent agaric from Sao Paulo, Brazil. Fung. Div. 2005, 18, 9–14. [Google Scholar]
- Desjardin, D.E.; Capelari, M.; Stevani, C.V. Bioluminescent Mycena species from São Paulo, Brazil. Mycologia 2007, 99, 317–331. [Google Scholar] [CrossRef]
- Desjardin, D.E.; Perry, B.A.; Lodge, D.J.; Stevani, C.V.; Nagasawa, E. Luminescent Mycena: New and noteworthy species. Mycologia 2010, 102, 459–477. [Google Scholar] [CrossRef]
- Desjardin, D.E.; Perry, B.A.; Stevani, C.V. New luminescent mycenoid fungi (Basidiomycota, Agaricales) from São Paulo state, Brazil. Mycologia 2016, 108, 1165–1174. [Google Scholar]
- Aravindakshan, D.M.; Kumar, T.K.A.; Manimohan, P. A new bioluminescent species of Mycena sect. Exornatae from Kerala State, India. Mycosphere 2012, 3, 556–561. [Google Scholar] [CrossRef]
- Shih, Y.S.; Chen, C.Y.; Lin, W.W.; Kao, H.W. Mycena kentingensis, a new species of luminous mushroom in Taiwan, with reference to its culture method. Mycol. Prog. 2013, 13, 429–435. [Google Scholar] [CrossRef]
- Chew, A.L.C.; Desjardin, D.E.; Tan, Y.S.; Musa, M.Y.; Sabaratnam, V. Bioluminescent fungi from Peninsular Malaysia-a taxonomic and phylogenetic overview. Fung. Div. 2015, 70, 149–187. [Google Scholar] [CrossRef]
- Cortés-Pérez, A.; Desjardin, D.E.; Perry, B.A.; Ramírez-Cruz, V.; Ramírez-Guillén, F.; Villalobos-Arámbula, A.R.; Rockefeller, A. New species and records of bioluminescent Mycena from Mexico. Mycologia 2019, 111, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Huei-Mien, K.; Isheng, J.T. Understanding and using fungal bioluminescence—Recent progress and future perspectives. Curr. Opin. Green Sustain. Chem. 2022, 33, 100570. [Google Scholar]
- Matheny, P.B.; Curtis, J.M.; Hofstetter, V.; Aime, M.C.; Moncalvo, J.M.; Ge, Z.W.; Yang, Z.L.; Slot, J.C.; Ammirati, J.F.; Baroni, T.J.; et al. Major clades of Agaricales: A multilocus phylogenetic overview. Mycologia 2006, 98, 982–995. [Google Scholar] [CrossRef]
- Mihail, J.D.; Bruhn, J.N. Dynamics of bioluminescence by Armillaria gallica, A. mellea and A. tabescens. Mycologia 2007, 99, 341–350. [Google Scholar] [CrossRef]
- Kirchmair, M.; Morandell, S.; Stolz, D.; Poder, R.; Sturmbauer, C. Phylogeny of the genus Omphalotus based on nuclear ribosomal DNA sequences. Mycologia 2004, 96, 1253–1260. [Google Scholar] [CrossRef]
- Vanden Hoek, T.L.; Erickson, T.; Hryhorczuk, D.; Narasimhan, K. Jack o’lantern mushroom poisoning. Ann. Emerg. Med. 1991, 20, 559–561. [Google Scholar] [CrossRef]
- Oliveira, A.G.; Desjardin, D.E.; Perry, B.A.; Stevani, C.V. Evidence that a single bioluminescent system is shared by all known bioluminescent fungal lineages. Photochem. Photobiol. Sci. 2012, 11, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Agerer, R. Cyphelloide Pilze aus Teneriffa. Nova Hedwig. 1978, 30, 295–342. [Google Scholar] [CrossRef]
- Agerer, R. Typusstudien an cyphelloiden Pilzen IV. Lachnella Fr. s.l. Mitt. Bot. Staatss München 1983, 19, 163–334. [Google Scholar]
- Donk, M.A. The generic names proposed for Hymenomycetes—I. “Cyphellaceae”. Reinwardtia 1951, 1, 199–220. [Google Scholar]
- Donk, M.A. Notes on “Cyphellaceae.” I. Persoonia 1959, 1, 25–110. [Google Scholar]
- Donk, M.A. A reassessment of the Cyphellaceae. Acta Bot. Neerl. 1966, 15, 95–101. [Google Scholar] [CrossRef]
- Burnett, G.T. Outlines of Botany: Including a General History of the Vegetable Kingdom, in Which Plants Are Arranged According to the System of Natural Affinities; Nabu Press: Charleston, WV, USA, 1835. [Google Scholar]
- Murrill, W.A. Notes and brief articles. A new family of Hymenomycetes. Mycologia 1916, 8, 52–56. [Google Scholar] [CrossRef]
- Cooke, W.B. The cyphellaceous fungi. A study in the Porotheleaceae. Beih. Sydowia 1961, 4, 1–144. [Google Scholar]
- Moncalvo, J.-M.; Vilgalys, R.; Redhead, S.A.; Johnson, J.E.; James, T.Y.; Aime, M.C.; Hofstetter, V.; Verduin, S.J.W.; Larsson, E.; Baroni, T.J.; et al. One hundred and seventeen clades of euagarics. Mol. Phylogenet Evol. 2002, 23, 357–400. [Google Scholar] [CrossRef]
- Bodensteiner, P.; Binder, M.; Moncalvo, J.M.; Agerer, R.; Hibbett, D.S. Phylogenetic relationships of cyphelloid homobasidiomycetes. Mol. Phylogenet Evol. 2004, 33, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Hibbett, D.S.; Larsson, K.H.; Larsson, E.; Langer, E.; Langer, G. The phylogenetic distribution of resupinate forms across the major clades of mushroom-forming fungi (Homobasidiomycetes). Syst. Biodivers. 2005, 3, 113–157. [Google Scholar] [CrossRef]
- Thorn, R.G.; Moncalvo, J.M.; Redhead, S.A.; Lodge, D.J.; Martín, M.P. A new poroid species of Resupinatus from Puerto Rico, with a reassessment of the cyphelloid genus Stigmatolemma. Mycologia 2005, 97, 1140–1151. [Google Scholar] [CrossRef]
- Baltazar, J.M.; Gorjón, S.P.; Pildain, M.B.; Rajchenberg, M.; da Silveira, M.B. Acanthocorticium brueggemanii, a new corticioid genus and species related to cyphelloid fungi in the euagarics clade (Agaricales, Basidiomycota). Botany 2015, 93, 453–463. [Google Scholar] [CrossRef]
- Lucas, A.; Dentinger, B.T.M. Rectipilus afibulatus a new cyphelloid mushroom (Agaricales) from Great Britain. Kew Bull. 2015, 70, 1–6. [Google Scholar] [CrossRef]
- Moreno, G.; Prieto, M.; Esteve-Raventós, F.; Olariaga, I. Phylogenetic assessment of Chromocyphellaceae (Agaricineae, Basidiomycota) and a new lamellate species of Chromocyphella. Mycologia 2017, 109, 578–587. [Google Scholar]
- Singer, R. The Agaricales in Modern Taxonomy, 4th ed.; Koeltz Scientific Books: Königstein, Germany, 1986. [Google Scholar]
- Handa, T.; Harada, Y. Flagelloscypha japonica: A new species of minute basidiomycete (Niaceae) from Japan. Mycoscience 2005, 46, 265–267. [Google Scholar] [CrossRef]
- Bodensteiner, P. Maireina afibulata and M. attenuatipilis, new members of the cyphelloid genus Maireina (Basidiomycota, Agaricomycetes). Mycol. Prog. 2007, 6, 221–228. [Google Scholar] [CrossRef]
- Læssøe, T.; Davey, M.L.; Petersen, J.H. A new species of Maireina on Filipendula ulmaria. Karstenia 2016, 56, 39–46. [Google Scholar] [CrossRef]
- Mombert, A. Maireina subsphaerospora (Niaceae, Agaricomycetes), un nouveau champignon cyphelloïde découvert en France. Bull. Mycol. Bot. Dauphiné-Savoie 2022, 246, 37–42. [Google Scholar]
- Karasiński, D.; László, G.N.; Szarkándi, J.G.; Dvořák, D.; Kolařík, M.; Holec, J. Cyphelloporia bialoviesensis (Fungi, Agaricales)—A new genus and species for a giant cyphelloid fungus from Białowieża virgin forest in Poland. Phytotaxa 2023, 589, 119–136. [Google Scholar] [CrossRef]
- Bodensteiner, P.; Maireina, W.B. Cooke. Morphologisch-Anatomische Untersuchungen an Einer Gattung Cyphelloider Homobasidiomyceten. Master’s Thesis, Fakultät für Biologie der Ludwig Maximilians Universität München, München, Germany, 2006. [Google Scholar]
- Jülich, W. Higher taxa of Basidiomycetes. Biblioth Mycol. 1982, 85, 1–485. [Google Scholar]
- Binder, M.; Hibbett, D.S.; Molitoris, H.P. Phylogenetic relationships of the marine gasteromycete Nia vibrissa. Mycologia 2001, 93, 679–688. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Binder, M. Evolution of marine mushrooms. Biol. Bull. 2001, 201, 319–322. [Google Scholar] [CrossRef][Green Version]
- Knudsen, H.; Vesterholt, J. Funga Nordica, 2nd ed.; Nordsvamp: Copenhagen, Denmark, 2012. [Google Scholar]
- Kalichman, J.; Kirk, P.M.; Matheny, P.B. A compendium of generic names of agarics and Agaricales. Taxon 2020, 69, 425–447. [Google Scholar] [CrossRef]
- Peel, M.C.; Finlayson, B.L.; McMahon, T.A. Updated world map of the Köppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007, 11, 1633–1644. [Google Scholar] [CrossRef]
- Oliveira Filho, A.; Fontes, M.A. Patterns of floristic differentiation among Atlantic Forests in southeastern Brazil, and the influence of climate. Biotropica 2000, 32, 793–810. [Google Scholar] [CrossRef]
- Aidar, M.P.M.; Godoy, J.R.L.; Bergmann, J.; Joly, C.A. Atlantic Forest succession over calcareous soil, Parque Estadual Turístico do Alto Ribeira–PETAR, SP. Rev. Bras. Bot. 2001, 24, 455–469. [Google Scholar] [CrossRef]
- Van Vooren, N.; Estival, E.; Hairaud, M.; Mombert, A.; Priou, J.-P. Ascomycètes d’Auvergne. Bull. Mycol. Bot. Dauphiné-Savoie 2022, 245, 5–24. [Google Scholar]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen Eyre: London, UK, 1978. [Google Scholar]
- Thiers, B.; New York Botanical Garden’s Virtual Herbarium. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 1 May 2023).
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Vizzini, A.; Consiglio, G.; Marchetti, M.; Borovička, J.; Campo, E.; Cooper, J.; Lebeuf, R.; Ševčíková, H. New data in Porotheleaceae and Cyphellaceae: Epitypification of Prunulus scabripes Murrill, the status of Mycopan Redhead, Moncalvo & Vilgalys and a new combination in Pleurella Horak emend. Mycol. Prog. 2022, 21, 44. [Google Scholar]
- Na, Q.; Hu, Y.; Zeng, H.; Song, Z.; Ding, H.; Cheng, X.; Ge, Y. Updated taxonomy on Gerronema (Porotheleaceae, Agaricales) with three new taxa and one new record from China. Mycokeys 2022, 89, 87–120. [Google Scholar] [CrossRef] [PubMed]
- Brian, A.P. (Department of Biological Sciences, California State University East Bay, Hayward, CA, USA); The Sequence EF514207 of G. viridilucens represents M. lucentipes. Personal communication, 2023.
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molec Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molec Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Stamatakis, A. RaxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.l.; Doall, O.R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes version 3.0: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Earle, F.S. The Genera of North American Gill Fungi; Bulletin of the New York Botanical Garden: New York, NY, USA, 1909. [Google Scholar]
- Olariaga, I.; Huhtinen, S.; Læssøe, T.; Petersen, J.H.; Hansen, K. Phylogenetic origins and family classification of typhuloid fungi, with emphasis on Ceratellopsis, Macrotyphula and Typhula (Basidiomycota). Stud. Mycol. 2020, 96, 155–184. [Google Scholar] [CrossRef]
- Reid, D.A. Notes on some fungi in Michigan—I. “Cyphellaceae”. Persoonia 1964, 3, 97–154. [Google Scholar]
- Agerer, R.; Prillinger, H.-J.; Noll, H.-P. Studien zur Sippenstruktur der Gattung Cyphellopsis—I. Darstellung zweier Ausgangssippen. Z. Mykol. 1980, 46, 177–207. [Google Scholar]
- Singer, R. The Agaricales in Modern Taxonomy, 3rd ed.; J. Cramer: Lehre, Germany, 1975. [Google Scholar]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.M.; Lee, H.H.; Lin, C.I.; Liu, Y.C.; Lu, M.R.; Hsieh, J.A.; Chang, C.C.; Wu, P.H.; Lu, M.J.; Li, J.Y.; et al. Mycena genomes resolve the evolution of fungal bioluminescence. Proc. Natl. Acad. Sci. USA 2020, 117, 31267–31277. [Google Scholar] [CrossRef]
- Oliveira, J.J.S.; Vargas-Isla, R.; Cabral, T.S.; Cardoso, J.S.; Andriolli, F.S.; Rodrigues, D.P.; Ikeda, T.; Clement, C.R.; Ishikawa, N.K. The Amazonian luminescent Mycena cristinae sp. nov. from Brazil. Mycoscience 2021, 62, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Borges, M.E.A. Diversidade de fungos bioluminescentes do gênero Mycena (Basidiomycota, Mycenaceae) da Mata Atlântica catarinense, Santa Catarina, Brasil. Master’s Thesis, Universidade Federal de Santa Catarina, Florianópolis, Santa Catarina, Brasil, 2020. [Google Scholar]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva-Filho, A.G.S.; Mombert, A.; Nascimento, C.C.; Nóbrega, B.B.; Soares, D.M.M.; Martins, A.G.S.; Domingos, A.H.R.; Santos, I.; Della-Torre, O.H.P.; Perry, B.A.; et al. Eoscyphella luciurceolata gen. and sp. nov. (Agaricomycetes) Shed Light on Cyphellopsidaceae with a New Lineage of Bioluminescent Fungi. J. Fungi 2023, 9, 1004. https://doi.org/10.3390/jof9101004
Silva-Filho AGS, Mombert A, Nascimento CC, Nóbrega BB, Soares DMM, Martins AGS, Domingos AHR, Santos I, Della-Torre OHP, Perry BA, et al. Eoscyphella luciurceolata gen. and sp. nov. (Agaricomycetes) Shed Light on Cyphellopsidaceae with a New Lineage of Bioluminescent Fungi. Journal of Fungi. 2023; 9(10):1004. https://doi.org/10.3390/jof9101004
Chicago/Turabian StyleSilva-Filho, Alexandre G. S., Andgelo Mombert, Cristiano C. Nascimento, Bianca B. Nóbrega, Douglas M. M. Soares, Ana G. S. Martins, Adão H. R. Domingos, Isaias Santos, Olavo H. P. Della-Torre, Brian A. Perry, and et al. 2023. "Eoscyphella luciurceolata gen. and sp. nov. (Agaricomycetes) Shed Light on Cyphellopsidaceae with a New Lineage of Bioluminescent Fungi" Journal of Fungi 9, no. 10: 1004. https://doi.org/10.3390/jof9101004
APA StyleSilva-Filho, A. G. S., Mombert, A., Nascimento, C. C., Nóbrega, B. B., Soares, D. M. M., Martins, A. G. S., Domingos, A. H. R., Santos, I., Della-Torre, O. H. P., Perry, B. A., Desjardin, D. E., Stevani, C. V., & Menolli, N., Jr. (2023). Eoscyphella luciurceolata gen. and sp. nov. (Agaricomycetes) Shed Light on Cyphellopsidaceae with a New Lineage of Bioluminescent Fungi. Journal of Fungi, 9(10), 1004. https://doi.org/10.3390/jof9101004





