The Influence of Oral Terbinafine on Gut Fungal Microbiome Composition and Microbial Translocation in People Living with HIV Treated for Onychomycosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Inclusion and Exclusion Criteria
2.3. Plasma Biomarkers of Gut Damage, Microbial Translocation, and Inflammation
2.4. Total Amount of Bacteria and Fungi in Feces
2.5. Fecal Bacterial and Fungal Composition
2.6. Statistics Analyses
3. Results
3.1. Clinical Characteristics of Our Cohort
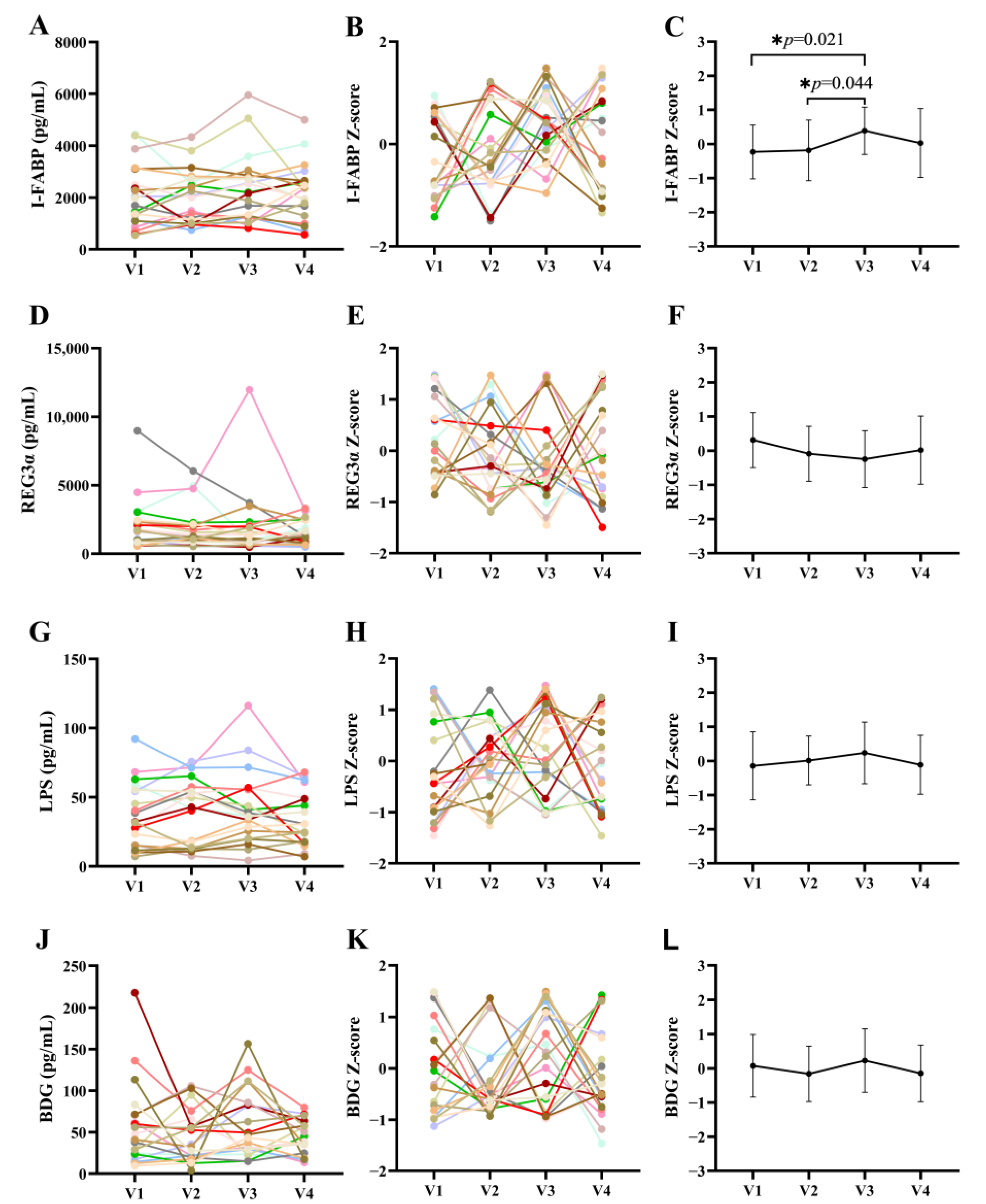
3.2. Effects of Terbinafine Treatment on Intestinal Barrier Markers in PLWH
3.3. Effects of Terbinafine Treatment on Inflammation in PLWH
3.4. Terbinafine Treatment Induces Minor Changes in Bacterial Abundance while Significantly Changing Fungal Abundance in the Gut
3.5. Terbinafine Treatment Induces Minor Changes in Bacterial and Fungal Composition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brenchley, J.M.; Price, D.A.; Schacker, T.W.; Asher, T.E.; Silvestri, G.; Rao, S.; Kazzaz, Z.; Bornstein, E.; Lambotte, O.; Altmann, D.; et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 2006, 12, 1365–1371. [Google Scholar] [CrossRef]
- Zevin, A.S.; McKinnon, L.; Burgener, A.; Klatt, N.R. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr. Opin. HIV AIDS 2016, 11, 182–190. [Google Scholar] [CrossRef]
- Yan, J.; Ouyang, J.; Isnard, S.; Zhou, X.; Harypursat, V.; Routy, J.P.; Chen, Y. Alcohol Use and Abuse Conspires with HIV Infection to Aggravate Intestinal Dysbiosis and Increase Microbial Translocation in People Living With HIV: A Review. Front. Immunol. 2021, 12, 741658. [Google Scholar] [CrossRef]
- Marchetti, G.; Tincati, C.; Silvestri, G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin. Microbiol. Rev. 2013, 26, 2–18. [Google Scholar] [CrossRef]
- Isnard, S.; Royston, L.; Scott, S.C.; Mabanga, T.; Lin, J.; Fombuena, B.; Bu, S.; Berini, C.A.; Goldberg, M.S.; Finkelman, M.; et al. Translocation of bacterial LPS is associated with self-reported cognitive abilities in men living with HIV receiving antiretroviral therapy. AIDS Res. Ther. 2023, 20, 30. [Google Scholar] [CrossRef]
- Ramendra, R.; Mancini, M.; Ayala, J.M.; Tung, L.T.; Isnard, S.; Lin, J.; Routy, J.P.; Nijnik, A.; Langlais, D. Glutathione Metabolism Is a Regulator of the Acute Inflammatory Response of Monocytes to (1→3)-β-D-Glucan. Front. Immunol. 2021, 12, 694152. [Google Scholar] [CrossRef]
- Isnard, S.; Lin, J.; Bu, S.; Fombuena, B.; Royston, L.; Routy, J.P. Gut Leakage of Fungal-Related Products: Turning Up the Heat for HIV Infection. Front. Immunol. 2021, 12, 656414. [Google Scholar] [CrossRef]
- Ramendra, R.; Isnard, S.; Mehraj, V.; Chen, J.; Zhang, Y.; Finkelman, M.; Routy, J.P. Circulating LPS and (1→3)-β-D-Glucan: A Folie a Deux Contributing to HIV-Associated Immune Activation. Front. Immunol. 2019, 10, 465. [Google Scholar] [CrossRef]
- Isnard, S.; Fombuena, B.; Sadouni, M.; Lin, J.; Richard, C.; Routy, B.; Ouyang, J.; Ramendra, R.; Peng, X.; Zhang, Y.; et al. Circulating β-d-Glucan as a Marker of Subclinical Coronary Plaque in Antiretroviral Therapy-Treated People with Human Immunodeficiency Virus. Open Forum Infect. Dis. 2021, 8, ofab109. [Google Scholar] [CrossRef]
- Mehraj, V.; Ramendra, R.; Isnard, S.; Dupuy, F.P.; Ponte, R.; Chen, J.; Kema, I.; Jenabian, M.A.; Costinuik, C.T.; Lebouche, B.; et al. Circulating (1→3)-β-d-glucan Is Associated with Immune Activation During Human Immunodeficiency Virus Infection. Clin. Infect. Dis. 2020, 70, 232–241. [Google Scholar] [CrossRef]
- Mims, T.S.; Abdallah, Q.A.; Stewart, J.D.; Watts, S.P.; White, C.T.; Rousselle, T.V.; Gosain, A.; Bajwa, A.; Han, J.C.; Willis, K.A.; et al. The gut mycobiome of healthy mice is shaped by the environment and correlates with metabolic outcomes in response to diet. Commun. Biol. 2021, 4, 281. [Google Scholar] [CrossRef] [PubMed]
- Li, X.V.; Leonardi, I.; Iliev, I.D. Gut Mycobiota in Immunity and Inflammatory Disease. Immunity 2019, 50, 1365–1379. [Google Scholar] [CrossRef] [PubMed]
- Mogilnicka, I.; Ufnal, M. Gut Mycobiota and Fungal Metabolites in Human Homeostasis. Curr. Drug Targets 2019, 20, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.C.; Lam, J.M.; Leong, K.F.; Hon, K.L.; Barankin, B.; Leung, A.A.M.; Wong, A.H.C. Onychomycosis: An Updated Review. Recent. Pat. Inflamm. Allergy Drug Discov. 2020, 14, 32–45. [Google Scholar]
- Gupta, A.K.; Taborda, P.; Taborda, V.; Gilmour, J.; Rachlis, A.; Salit, I.; Gupta, M.A.; MacDonald, P.; Cooper, E.A.; Summerbell, R.C. Epidemiology and prevalence of onychomycosis in HIV-positive individuals. Int. J. Dermatol. 2000, 39, 746–753. [Google Scholar] [CrossRef]
- Ameen, M.; Lear, J.T.; Madan, V.; Mohd Mustapa, M.F.; Richardson, M. British Association of Dermatologists’ guidelines for the management of onychomycosis 2014. Br. J. Dermatol. 2014, 171, 937–958. [Google Scholar] [CrossRef]
- Haugh, M.; Helou, S.; Boissel, J.P.; Cribier, B.J. Terbinafine in fungal infections of the nails: A meta-analysis of randomized clinical trials. Br. J. Dermatol. 2002, 147, 118–121. [Google Scholar] [CrossRef]
- Kreijkamp-Kaspers, S.; Hawke, K.; Guo, L.; Kerin, G.; Bell-Syer, S.E.; Magin, P.; Bell-Syer, S.V.; van Driel, M.L. Oral antifungal medication for toenail onychomycosis. Cochrane Database Syst. Rev. 2017, 7, CD010031. [Google Scholar] [CrossRef]
- Darkes, M.J.; Scott, L.J.; Goa, K.L. Terbinafine: A review of its use in onychomycosis in adults. Am. J. Clin. Dermatol. 2003, 4, 39–65. [Google Scholar] [CrossRef]
- Krishnan-Natesan, S. Terbinafine: A pharmacological and clinical review. Expert. Opin. Pharmacother. 2009, 10, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Balfour, J.A.; Faulds, D. Terbinafine. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in superficial mycoses. Drugs 1992, 43, 259–284. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi Omran, S.; Rezaei Dastjerdi, M.; Zuashkiani, M.; Moqarabzadeh, V.; Taghizadeh-Armaki, M. In Vitro Antifungal Susceptibility of Candida Species Isolated from Iranian Patients with Denture Stomatitis. Biomed. Res. Int. 2018, 2018, 3086586. [Google Scholar] [CrossRef] [PubMed]
- Sancak, B.; Ayhan, M.; Karaduman, A.; Arikan, S. In vitro activity of ketoconazole, itraconazole and terbinafine against Malassezia strains isolated from neonates. Mikrobiyol. Bul. 2005, 39, 301–308. [Google Scholar]
- Leber, R.; Fuchsbichler, S.; Klobucnikova, V.; Schweighofer, N.; Pitters, E.; Wohlfarter, K.; Lederer, M.; Landl, K.; Ruckenstuhl, C.; Hapala, I.; et al. Molecular mechanism of terbinafine resistance in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2003, 47, 3890–3900. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358. [Google Scholar] [CrossRef]
- Osborne, C.S.; Leitner, I.; Favre, B.; Ryder, N.S. Amino acid substitution in Trichophyton rubrum squalene epoxidase associated with resistance to terbinafine. Antimicrob. Agents Chemother. 2005, 49, 2840–2844. [Google Scholar] [CrossRef]
- Majid, I.; Sheikh, G.; Kanth, F.; Hakak, R. Relapse after Oral Terbinafine Therapy in Dermatophytosis: A Clinical and Mycological Study. Indian J. Dermatol. 2016, 61, 529–533. [Google Scholar]
- Blanchard, G.; Amarov, B.; Fratti, M.; Salamin, K.; Bontems, O.; Chang, Y.T.; Sabou, A.M.; Kunzle, N.; Monod, M.; Guenova, E. Reliable and rapid identification of terbinafine resistance in dermatophytic nail and skin infections. J. Eur. Acad. Dermatol. Venereol. 2023. [Google Scholar] [CrossRef]
- Gianni, C. Update on antifungal therapy with terbinafine. G. Ital. Dermatol. Venereol. 2010, 145, 415–424. [Google Scholar]
- Shiao, S.L.; Kershaw, K.M.; Limon, J.J.; You, S.; Yoon, J.; Ko, E.Y.; Guarnerio, J.; Potdar, A.A.; McGovern, D.P.B.; Bose, S.; et al. Commensal bacteria and fungi differentially regulate tumor responses to radiation therapy. Cancer Cell 2021, 39, 1202–1213.e6. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.M.; Kachur, S.; Dwan, M.G.; Abraham, A.G.; Aziz, M.; Hsueh, P.R.; Huang, Y.T.; Busch, J.D.; Lamit, L.J.; Gehring, C.A.; et al. FungiQuant: A broad-coverage fungal quantitative real-time PCR assay. BMC Microbiol. 2012, 12, 255. [Google Scholar] [CrossRef]
- He, X.X.; Li, Y.H.; Yan, P.G.; Meng, X.C.; Chen, C.Y.; Li, K.M.; Li, J.N. Relationship between clinical features and intestinal microbiota in Chinese patients with ulcerative colitis. World J. Gastroenterol. 2021, 27, 4722–4737. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H.; et al. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Bhakat, B.; Pal, J.; Das, S.; Charaborty, S.K.; SircarMedical, N.R. A Prospective Study to Evaluate the Possible Role of Cholecalciferol Supplementation on Autoimmunity in Hashimoto’s Thyroiditis. J. Assoc. Physicians India 2023, 71, 1. [Google Scholar]
- Ouyang, J.; Yan, J.; Zhou, X.; Isnard, S.; Harypursat, V.; Cui, H.; Routy, J.P.; Chen, Y. Relevance of biomarkers indicating gut damage and microbial translocation in people living with HIV. Front. Immunol. 2023, 14, 1173956. [Google Scholar] [CrossRef]
- Gupta, A.K.; Daigle, D.; Foley, K.A. The prevalence of culture-confirmed toenail onychomycosis in at-risk patient populations. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1039–1044. [Google Scholar] [CrossRef]
- Gupta, A.K.; Versteeg, S.G.; Shear, N.H. Onychomycosis in the 21st Century: An Update on Diagnosis, Epidemiology, and Treatment. J. Cutan. Med. Surg. 2017, 21, 525–539. [Google Scholar] [CrossRef]
- Moreno-Coutino, G.; Arenas, R.; Reyes-Teran, G. Improvement in onychomycosis after initiation of combined antiretroviral therapy. Int. J. Dermatol. 2013, 52, 311–313. [Google Scholar] [CrossRef]
- Salzer, H.J.F.; Schafer, G.; Hoenigl, M.; Gunther, G.; Hoffmann, C.; Kalsdorf, B.; Alanio, A.; Lange, C. Clinical, Diagnostic, and Treatment Disparities between HIV-Infected and Non-HIV-Infected Immunocompromised Patients with Pneumocystis jirovecii Pneumonia. Respiration 2018, 96, 52–65. [Google Scholar] [CrossRef]
- Pathakumari, B.; Liang, G.; Liu, W. Immune defence to invasive fungal infections: A comprehensive review. Biomed. Pharmacother. 2020, 130, 110550. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Lopez, P.; Moreno-Coutino, G.; Fernandez-Martinez, R.; Espinoza-Hernandez, J.; Rodriguez-Zulueta, P.; Reyes-Teran, G. Evaluation of improvement of onychomycosis in HIV-infected patients after initiation of combined antiretroviral therapy without antifungal treatment. Mycoses 2015, 58, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Vanden Bossche, H.; Dromer, F.; Improvisi, I.; Lozano-Chiu, M.; Rex, J.H.; Sanglard, D. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 1998, 36 (Suppl. 1), 119–128. [Google Scholar] [PubMed]
- Ryder, N.S. Terbinafine: Mode of action and properties of the squalene epoxidase inhibition. Br. J. Dermatol. 1992, 126 (Suppl. 39), 2–7. [Google Scholar] [CrossRef] [PubMed]
- Maskan Bermudez, N.; Rodriguez-Tamez, G.; Perez, S.; Tosti, A. Onychomycosis: Old and New. J. Fungi 2023, 9, 559. [Google Scholar] [CrossRef]
- Li, J.; Chen, D.; Yu, B.; He, J.; Zheng, P.; Mao, X.; Yu, J.; Luo, J.; Tian, G.; Huang, Z.; et al. Fungi in Gastrointestinal Tracts of Human and Mice: From Community to Functions. Microb. Ecol. 2018, 75, 821–829. [Google Scholar] [CrossRef]
- Kombrink, A.; Tayyrov, A.; Essig, A.; Stockli, M.; Micheller, S.; Hintze, J.; van Heuvel, Y.; Durig, N.; Lin, C.W.; Kallio, P.T.; et al. Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria. ISME J. 2019, 13, 588–602. [Google Scholar] [CrossRef]
- Fernandez de Ullivarri, M.; Arbulu, S.; Garcia-Gutierrez, E.; Cotter, P.D. Antifungal Peptides as Therapeutic Agents. Front. Cell Infect. Microbiol. 2020, 10, 105. [Google Scholar] [CrossRef]
- Noverr, M.C.; Huffnagle, G.B. Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 2004, 72, 6206–6210. [Google Scholar] [CrossRef]
- Garcia, C.; Tebbji, F.; Daigneault, M.; Liu, N.N.; Kohler, J.R.; Allen-Vercoe, E.; Sellam, A. The Human Gut Microbial Metabolome Modulates Fungal Growth via the TOR Signaling Pathway. mSphere 2017, 2, 10–1128. [Google Scholar] [CrossRef]
- Zhang, F.; Aschenbrenner, D.; Yoo, J.Y.; Zuo, T. The gut mycobiome in health, disease, and clinical applications in association with the gut bacterial microbiome assembly. Lancet Microbe 2022, 3, e969–e983. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Erb Downward, J.R.; Falkowski, N.R.; Young, V.B.; Kao, J.Y.; Huffnagle, G.B. Interplay between the gastric bacterial microbiota and Candida albicans during postantibiotic recolonization and gastritis. Infect. Immun. 2012, 80, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, F.; Yang, X.; Wu, N.; Jiang, W.; Li, X.; Li, X.; Liu, Y. Changes in the composition of intestinal fungi and their role in mice with dextran sulfate sodium-induced colitis. Sci. Rep. 2015, 5, 10416. [Google Scholar] [CrossRef] [PubMed]
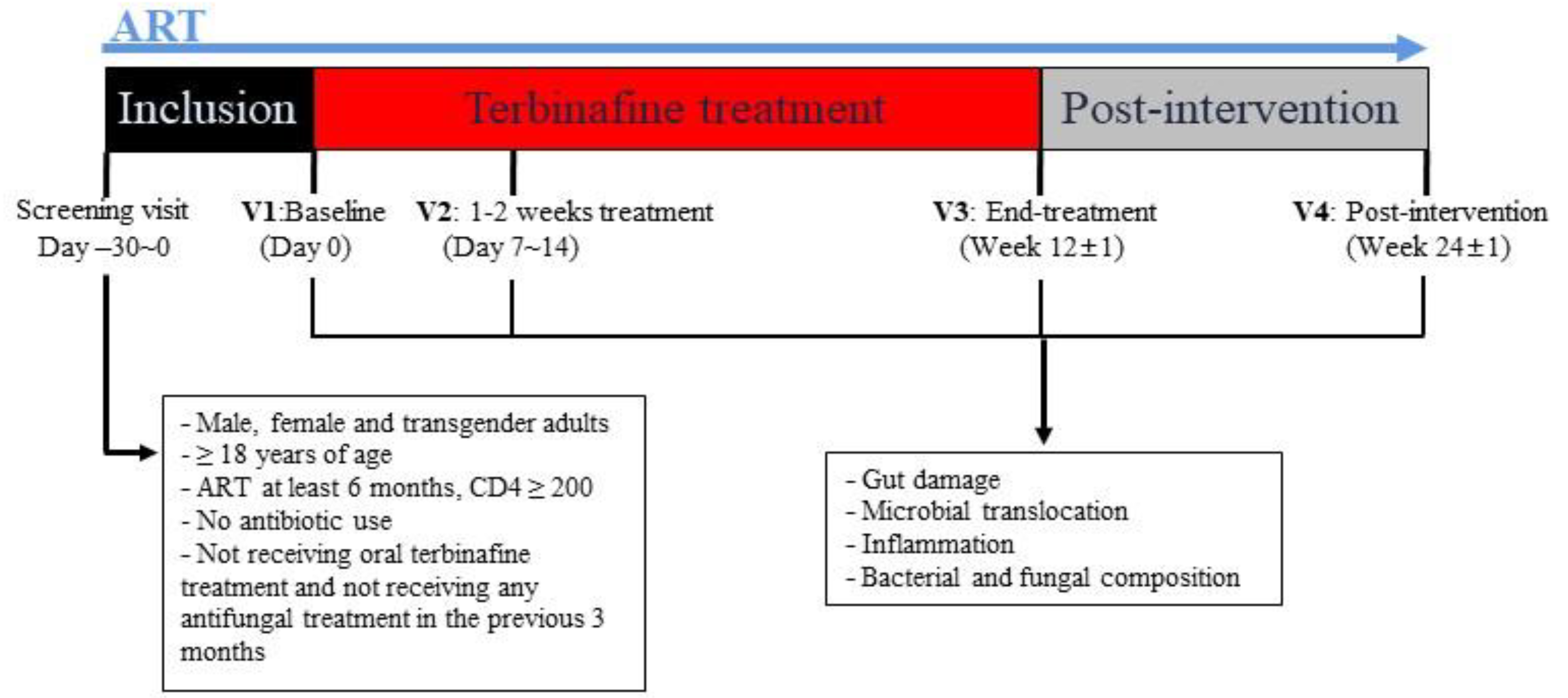



| Characteristics | All Participants (n = 23) | Participants Completing All Visits (n = 20) |
|---|---|---|
| Age (Years, mean ± SD) | 40.9 ± 12.3 | 41.7 ± 12.6 |
| Sex (Female/male) | 1/22 | 1/20 (5%) |
| Ethnicity (Chinese) | 23/23 | 20/20 (100%) |
| Sexual behavior | ||
| MSM | 9(39.1%) | 8/20 (40%) |
| Heterosexual | 13(56.6%) | 11/20 (55%) |
| Bisexual | 1(4.3%) | 1/20 (5%) |
| Time since HIV diagnosis (Years) | 3.91 (IQR 2.33–6.58) | 3.88 (IQR 2.40–8.15) |
| Duration of ART (Years) | 3.83 (IQR 2.08–6.58) | 3.88 (IQR 2.40–7.58) |
| Duration of onychomycosis infection (Years) | 6.41 ± 4.35 | 6.33 ± 4.03 |
| Viral load (HIV-1 RNA, copies/mL) | <50 | <50 |
| CD4+ T-cells median (cells/μL) | 326 (IQR 226–472) | 309 (IQR 225–456) |
| CD8+ T-cells median (cells/μL) | 648 (IQR 478–985) | 643 (IQR 449–897) |
| CD4+/CD8+ ratio | 0.5 (IQR 0.3–0.7) | 0.45 (IQR 0.35–0.72) |
| BMI (kg/m2, mean± SD) | 22.63 ± 3.81 | 22.18 ± 3.57 |
| Current smoker | 4/23(17.4%) | 3/20 (15%) |
| ART regimens | ||
| 3TC + TDF + EFV | 19(82.6%) | 16/20 (80%) |
| EVG/c/FTC/TAF | 3(13.1%) | 3/20 (15%) |
| Zidovudine and Lamivudine + LPV/r | 1(4.3%) | 1/20 (5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ouyang, J.; Yan, J.; Zhou, X.; Isnard, S.; Tang, S.; Costiniuk, C.T.; Chen, Y.; Routy, J.-P.; Chen, Y. The Influence of Oral Terbinafine on Gut Fungal Microbiome Composition and Microbial Translocation in People Living with HIV Treated for Onychomycosis. J. Fungi 2023, 9, 963. https://doi.org/10.3390/jof9100963
Ouyang J, Yan J, Zhou X, Isnard S, Tang S, Costiniuk CT, Chen Y, Routy J-P, Chen Y. The Influence of Oral Terbinafine on Gut Fungal Microbiome Composition and Microbial Translocation in People Living with HIV Treated for Onychomycosis. Journal of Fungi. 2023; 9(10):963. https://doi.org/10.3390/jof9100963
Chicago/Turabian StyleOuyang, Jing, Jiangyu Yan, Xin Zhou, Stéphane Isnard, Shengquan Tang, Cecilia T. Costiniuk, Yaling Chen, Jean-Pierre Routy, and Yaokai Chen. 2023. "The Influence of Oral Terbinafine on Gut Fungal Microbiome Composition and Microbial Translocation in People Living with HIV Treated for Onychomycosis" Journal of Fungi 9, no. 10: 963. https://doi.org/10.3390/jof9100963
APA StyleOuyang, J., Yan, J., Zhou, X., Isnard, S., Tang, S., Costiniuk, C. T., Chen, Y., Routy, J.-P., & Chen, Y. (2023). The Influence of Oral Terbinafine on Gut Fungal Microbiome Composition and Microbial Translocation in People Living with HIV Treated for Onychomycosis. Journal of Fungi, 9(10), 963. https://doi.org/10.3390/jof9100963







