Effects of MAT1-2 Spore Ratios on Fruiting Body Formation and Degeneration in the Heterothallic Fungus Cordyceps militaris
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains
2.2. Media for Fungal Cultivation
2.3. Isolation of Ascospores from Fruiting Bodies
2.4. Total DNA Extraction
2.5. Identification of the Mating-Type Genes by PCR
2.6. Assays for the Fruiting Body Formation through Successive Culturing Generations
2.7. Assays for Effects of Spore Ratios on Fruiting Body Formation
2.8. Maintaining Fruiting Body Formation in the Heterokaryotic Strains
2.9. Statistical Analysis
3. Results and Discussion
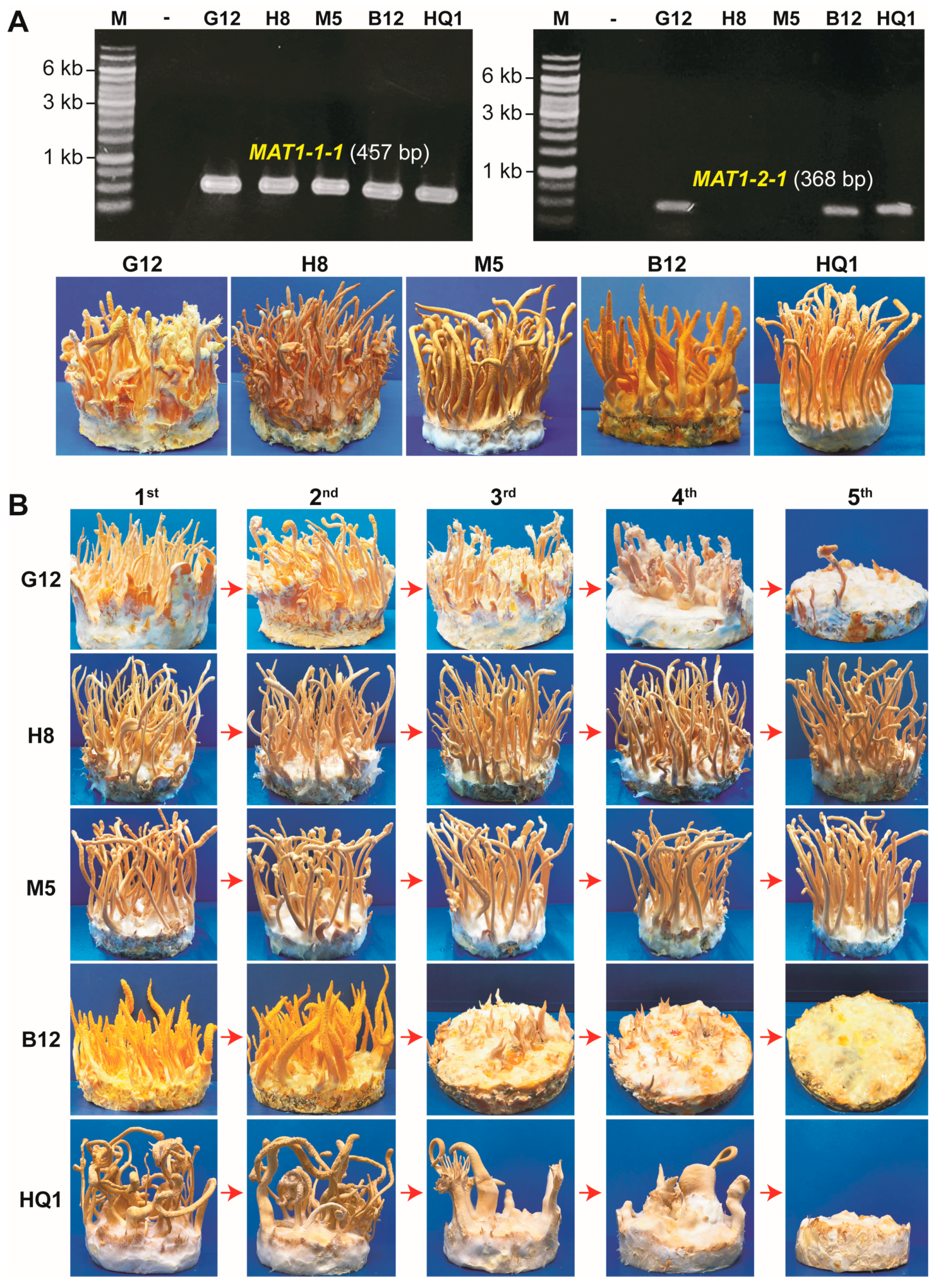
3.1. Heterokaryotic C. militaris Strains Carrying Both Mating-Type Loci Degenerate through Consecutive Culturing Generations
3.2. Both the Mating-Type Loci, MAT1-1 and MAT1-2, Are Required for Fruiting Body Formation in the Heterokaryotic C. militaris Strains
3.3. Excessive Increase in MAT1-2 Spore Ratios Caused the Degeneration in the Heterokaryotic Strains
3.4. Preserving the Monokaryotic Isolates Separately before Mating Mitigates the Degeneration in the Heterokaryotic Strains Caused by Successive Culturing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, G.-H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.B.; Wang, H.X. Pharmacological actions of Cordyceps, a prized folk medicine. J. Pharm. Pharmacol. 2005, 57, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Shin, H.-S.; Leyva-Gómez, G.; Prado-Audelo, M.L.D.; Cortes, H.; Singh, Y.D.; Panda, M.K.; Mishra, A.P.; Nigam, M.; Saklani, S.; et al. Cordyceps spp.: A review on its immune-stimulatory and other biological potentials. Front. Pharmacol. 2021, 11, 2250. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yan, H.; Zeng, B.; Hu, Z. Research progress on cordycepin synthesis and methods for enhancement of cordycepin production in Cordyceps militaris. Bioengineering 2022, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Phull, A.-R.; Ahmed, M.; Park, H.-J. Cordyceps militaris as a bio functional food source: Pharmacological potential, anti-inflammatory actions and related molecular mechanisms. Microorganisms 2022, 10, 405. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, Q.-Y.; Luo, R.; Fan, Q.; Duan, D.-E.; Dao, V.-M.; Wang, Y.-B.; Yu, H. Molecular phylogeny and morphology reveal cryptic species in the Cordyceps militaris complex from Vietnam. J. Fungi 2023, 9, 676. [Google Scholar] [CrossRef]
- Shweta; Abdullah, S.; Komal; Kumar, A. A brief review on the medicinal uses of Cordyceps militaris. Pharmacol. Res. Mod. Chin. Med. 2023, 7, 100228. [Google Scholar] [CrossRef]
- Krishna, K.V.; Ulhas, R.S.; Malaviya, A. Bioactive compounds from Cordyceps and their therapeutic potential. Crit. Rev. Biotechnol. 2023, 1–21. [Google Scholar] [CrossRef]
- Lu, Y.; Xia, Y.; Luo, F.; Dong, C.; Wang, C. Functional convergence and divergence of mating-type genes fulfilling in Cordyceps militaris. Fungal Genet. Biol. 2016, 88, 35–43. [Google Scholar] [CrossRef]
- Shrestha, B.; Kim, H.-K.; Sung, G.-H.; Spatafora, J.W.; Sung, J.-M. Bipolar heterothallism, a principal mating system of Cordyceps militaris in vitro. Biotechnol. Bioprocess Eng. 2004, 9, 440–446. [Google Scholar] [CrossRef]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, S. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2012, 12, R116. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, E.; Yamagishi, K.; Hara, A. Heterothallism in Cordyceps takaomontana. FEMS Microbiol. Lett. 2005, 250, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.-H.; Shrestha, B.; Han, S.-K.; Kim, S.-Y.; Sung, J.-M. Heterothallic type of mating system for Cordyceps cardinalis. Mycobiology 2010, 38, 282. [Google Scholar] [CrossRef] [PubMed]
- Danner, C.; Mach, R.L.; Mach-Aigner, A.R. The phenomenon of strain degeneration in biotechnologically relevant fungi. Appl. Microbiol. Biotechnol. 2023, 107, 4745–4758. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Z.; Liu, X.; Cui, B.; Miao, W.; Cheng, W.; Zhao, F. Characteristics analysis reveals the progress of Volvariella volvacea mycelium subculture degeneration. Front. Microbiol. 2019, 10, 2045. [Google Scholar] [CrossRef]
- Pérez, G.; Lopez-Moya, F.; Chuina, E.; Ibañez-Vea, M.; Garde, E.; López-Llorca, L.V.; Pisabarro, A.G.; Ramírez, L. Strain Degeneration in Pleurotus ostreatus: A genotype dependent oxidative stress process which triggers oxidative stress, cellular detoxifying and cell wall reshaping genes. J. Fungi 2021, 7, 862. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, X.; Chen, C.; Cheng, Z.; Wang, W.; Yun, J. Successive mycelial subculturing decreased lignocellulase activity and increased ROS accumulation in Volvariella volvacea. Front. Microbiol. 2022, 13, 997485. [Google Scholar] [CrossRef]
- Du, X.-H.; Zhao, Q.; Xia, E.-H.; Gao, L.-Z.; Richard, F.; Yang, Z.L. Mixed-reproductive strategies, competitive mating-type distribution and life cycle of fourteen black morel species. Sci. Rep. 2017, 7, 1493. [Google Scholar] [CrossRef]
- Shrestha, B.; Zhang, W.; Zhang, Y.; Liu, X. The medicinal fungus Cordyceps militaris: Research and development. Mycol. Prog. 2012, 11, 599–614. [Google Scholar] [CrossRef]
- Sun, S.-J.; Deng, C.-H.; Zhang, L.-Y.; Hu, K.-H. Molecular analysis and biochemical characteristics of degenerated strains of Cordyceps militaris. Arch. Microbiol. 2017, 199, 939–944. [Google Scholar] [CrossRef]
- Sung, J.-M.; Park, Y.-J.; Lee, J.-O.; Han, S.-K.; Lee, W.-H.; Choi, S.-K.; Shrestha, B. Effect of preservation periods and subcultures on fruiting body formation of Cordyceps militaris in vitro. Mycobiology 2006, 34, 196. [Google Scholar] [CrossRef] [PubMed]
- Xiong, C.; Xia, Y.; Zheng, P.; Wang, C. Increasing oxidative stress tolerance and subculturing stability of Cordyceps militaris by overexpression of a glutathione peroxidase gene. Appl. Microbiol. Biotechnol. 2012, 97, 2009–2015. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Xin, X.; Weng, Y.; Gui, Z. Transcriptome-wide analysis reveals the progress of Cordyceps militaris subculture degeneration. PLoS ONE 2017, 12, e0186279. [Google Scholar] [CrossRef] [PubMed]
- Lou, H.; Lin, J.; Guo, L.; Wang, X.; Tian, S.; Liu, C.; Zhao, Y.; Zhao, R. Advances in research on Cordyceps militaris degeneration. Appl. Microbiol. Biotechnol. 2019, 103, 7835–7841. [Google Scholar] [CrossRef]
- Kang, N.; Lee, H.-H.; Park, I.; Seo, Y.-S. Development of high cordycepin-producing Cordyceps militaris strains. Mycobiology 2017, 45, 31–38. [Google Scholar] [CrossRef]
- Tran, V.T.; Do, T.B.X.L.; Nguyen, T.K.; Vu, X.T.; Dao, B.N.; Nguyen, H.H. A simple, efficient and universal method for the extraction of genomic DNA from bacteria, yeasts, molds and microalgae suitable for PCR-based applications. Vietnam. J. Sci. Technol. Eng. 2017, 59, 66–74. [Google Scholar] [CrossRef]
- Yokoyama, E.; Arakawa, M.; Yamagishi, K.; Hara, A. Phylogenetic and structural analyses of the mating-type loci in Clavicipitaceae. FEMS Microbiol. Lett. 2006, 264, 182–191. [Google Scholar] [CrossRef]
- Wellham, P.A.D.; Hafeez, A.; Gregori, A.; Brock, M.; Kim, D.-H.; Chandler, D.; de Moor, C.H. Culture degeneration reduces sex-related gene expression, alters metabolite production and reduces insect pathogenic response in Cordyceps militaris. Microorganisms 2021, 9, 1559. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, T.X.; Thai, H.-D.; Dinh, B.-H.T.; Nguyen, H.T.; Tran, H.T.P.; Bui, K.-L.T.; Tran, T.B.; Pham, H.T.; Mai, L.T.D.; Le, D.H.; et al. Effects of MAT1-2 Spore Ratios on Fruiting Body Formation and Degeneration in the Heterothallic Fungus Cordyceps militaris. J. Fungi 2023, 9, 971. https://doi.org/10.3390/jof9100971
Vu TX, Thai H-D, Dinh B-HT, Nguyen HT, Tran HTP, Bui K-LT, Tran TB, Pham HT, Mai LTD, Le DH, et al. Effects of MAT1-2 Spore Ratios on Fruiting Body Formation and Degeneration in the Heterothallic Fungus Cordyceps militaris. Journal of Fungi. 2023; 9(10):971. https://doi.org/10.3390/jof9100971
Chicago/Turabian StyleVu, Tao Xuan, Hanh-Dung Thai, Bich-Hang Thi Dinh, Huong Thi Nguyen, Huyen Thi Phuong Tran, Khanh-Linh Thi Bui, Tram Bao Tran, Hien Thanh Pham, Linh Thi Dam Mai, Diep Hong Le, and et al. 2023. "Effects of MAT1-2 Spore Ratios on Fruiting Body Formation and Degeneration in the Heterothallic Fungus Cordyceps militaris" Journal of Fungi 9, no. 10: 971. https://doi.org/10.3390/jof9100971





