Molecular Characterization and Pathogenicity of Alternaria spp. Associated with Black Rot of Sweet Cherries in Italy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Isolation
2.2. Micro and Macro-Morphological Characteristics
2.3. DNA Extraction and PCR Amplification
2.4. Phylogenetic Analysis
2.5. Pathogenicity Assay
2.6. Statistical Analysis
3. Results
3.1. Fungal Isolation, Identification, and Morphological Characterization
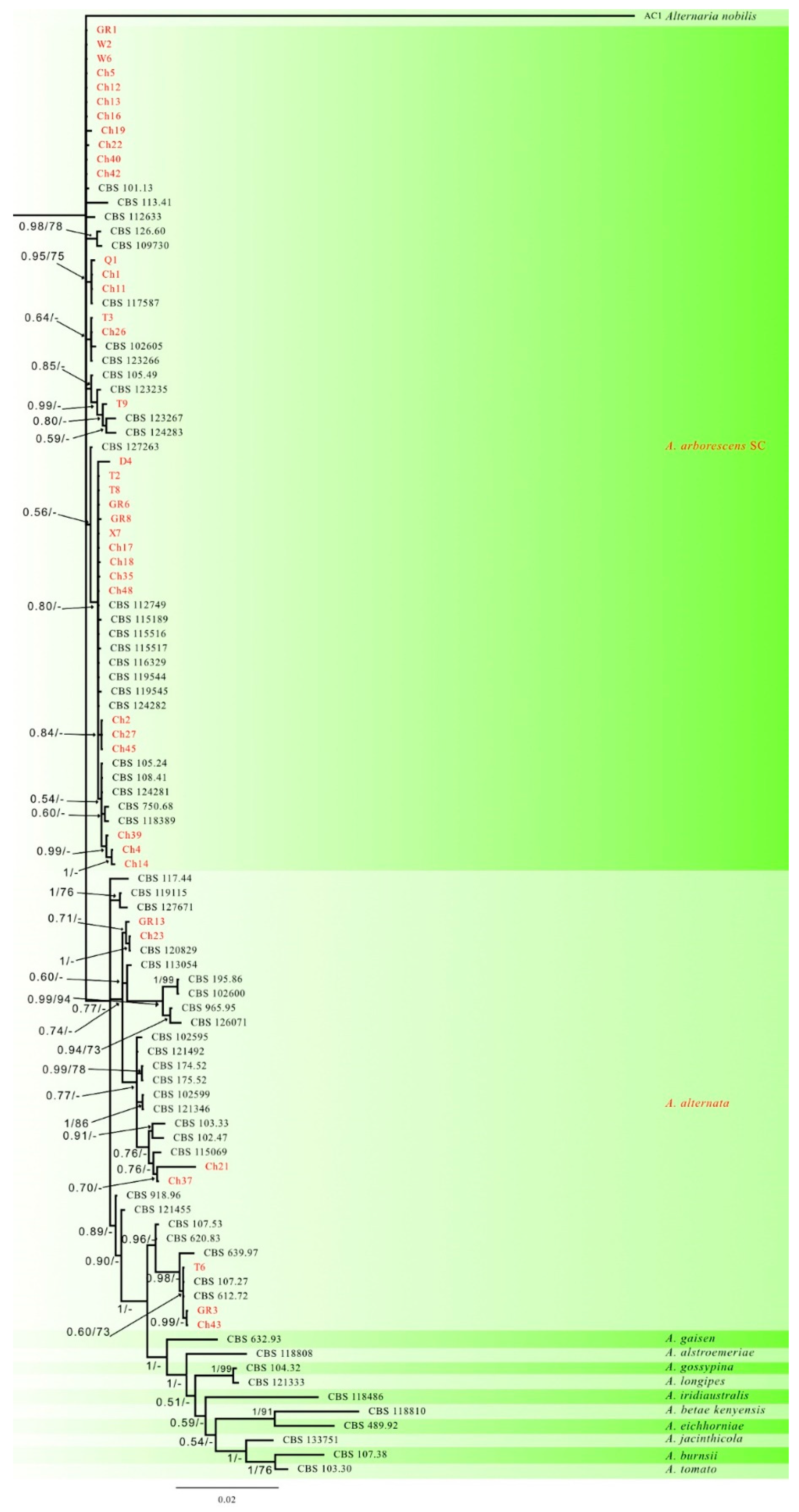
3.2. Phylogenetic Analysis
3.3. Pathogenicity Assay
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 27 February 2023).
- ISTAT. Available online: http://dati.istat.it/Index.aspx?QueryId=33705&lang=en (accessed on 14 March 2023).
- Padilla-Zakour, O.I.; Ryona, I.; Cooley, H.J.; Robinson, T.L.; Osborne, J.; Freer, J. Shelf-Life Extension of Sweet Cherries by Field Management, Post-Harvest Treatments and Modified Atmosphere Packaging. N. Y. Fruit. Q. 2007, 15, 3–6. [Google Scholar]
- Chiabrando, V.; Garavaglia, L.; Giacalone, G. The Postharvest Quality of Fresh Sweet Cherries and Strawberries with an Active Packaging System. Foods 2019, 8, 335. [Google Scholar] [CrossRef]
- Sehirli, S.; Karabulut, O.A.; Ilhan, K.; Sehirli, A. Use and Efficiency of Disinfectants within a Hydrocooler System for Postharvest Disease Control in Sweet Cherry. Int. J. Fruit. Sci. 2020, 20, S1590–S1606. [Google Scholar] [CrossRef]
- Maghenzani, M.; Chiabrando, V.; Santoro, K.; Spadaro, D.; Giacalone, G. Essential oil vapour treatment (Thymus vulgaris and Satureja montana) on postharvest quality of sweet cherry (cv Ferrovia). J. Food Nutr. Res. 2018, 57, 161–169. [Google Scholar]
- Mari, M.; Spadaro, D.; Casals, C.; Collina, M.; De Cal, A.; Usall, J. Postharvest Diseases of Stone Fruits. In Postharvest Pathology of Fresh Horticultural Produce; Palou, L., Smilanick, J.L., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 111–140. ISBN 9781138630833. [Google Scholar]
- Ahmad, T.; Liu, Y.; Shujian, H.; Moosa, A. First Record of Alternaria alternata Causing Postharvest Fruit Rot of Sweet Cherry (Prunus avium) in China. Plant Dis. 2020, 104, 2030. [Google Scholar] [CrossRef]
- Cancino, S.; Lolas, M.A.; Galdós, L.; Hernández, Y.; Ferrada, E.; Riveros, P.; Blanco-Ulate, B.; Díaz, G.A. Occurrence of Alternaria alternata and A. tenuissima Causing Black Rot in Cherry Fruits (Prunus avium) in Central Chile. Plant Dis. 2023. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.Z.; Liu, Z.H. First Report of Black Spot Disease Caused by Alternaria alternata on Cherry Fruits in China. Plant Dis. 2012, 96, 1580. [Google Scholar] [CrossRef] [PubMed]
- Latinović, N.; Radišek, S.; Latinović, J. First Report of Alternaria alternata Causing Fruit Rot on Fig (Ficus carica) in Montenegro. Plant Dis. 2014, 98, 424. [Google Scholar] [CrossRef]
- Xiang, M.L.; Li, S.C.; Wu, F.; Zhao, X.Y.; Wang, Y.B.; An, X.X.; Zhang, Y.N.; Chen, M. First Report of Alternaria alternata Causing Fruit Rot on Tetradium ruticarpum in China. Plant Dis. 2021, 105, 1194. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The Sections of Alternaria: Formalizing Species-Group Concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef]
- Matić, S.; Tabone, G.; Garibaldi, A.; Gullino, M.L. Alternaria Leaf Spot Caused by Alternaria Species: An Emerging Problem on Ornamental Plants in Italy. Plant Dis. 2020, 104, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J. AlternariasSpp.: From General Saprophyte to Specific Parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria Redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.-J.; Zheng, X.-R.; Li, H.; Chen, F.-M. Alternaria alternata, the Causal Agent of a New Needle Blight Disease on Pinus Bungeana. J. Fungi 2023, 9, 71. [Google Scholar] [CrossRef] [PubMed]
- Abata, L.K.; Paz, I.A.; Viera, W.; Flores, F.J. First Report of Alternaria Rot Caused by Alternaria alternata on Peach in Ecuador. Plant Dis. 2016, 100, 2323. [Google Scholar] [CrossRef]
- Dogan, A.; Cat, A.; Catal, M.; Erkan, M. First Report of Alternaria alternata Causing Postharvest Decay in Fig (Ficus carica L. Cv. Bursa Siyahi) Fruit in Turkey. J. Biotechnol. 2018, 280, S84. [Google Scholar] [CrossRef]
- Gur, L.; Reuveni, M.; Cohen, Y. Occurrence and Etiology of Alternaria Leaf Blotch and Fruit Spot of Apple Caused by Alternaria alternata f. sp. mali on cv. Pink Lady in Israel. Eur. J. Plant Pathol. 2017, 147, 695–708. [Google Scholar] [CrossRef]
- Moslemi, A.; Ades, P.K.; Groom, T.; Nicolas, M.E.; Taylor, P.W.J. Alternaria infectoria and Stemphylium herbarum, Two New Pathogens of Pyrethrum (Tanacetum cinerariifolium) in Australia. Australas. Plant Pathol. 2017, 46, 91–101. [Google Scholar] [CrossRef]
- Munhuweyi, K.; Lennox, C.L.; Meitz-Hopkins, J.C.; Caleb, O.J.; Opara, U.L. Major Diseases of Pomegranate (Punica granatum L.), Their Causes and Management—A Review. Sci. Hortic. 2016, 211, 126–139. [Google Scholar] [CrossRef]
- Ustun, R.; Cat, A.; Uzun, B.; Catal, M. First Report of Alternaria Alternata Causing Leaf Spot Disease on Soybean (Glycine max) in Antalya Province of Turkey. Plant Dis. 2019, 103, 3284. [Google Scholar] [CrossRef]
- Şimşek, A.; Dinler, H.; Uysal Morca, A. Identification and Pathogenicity of Alternaria Alternata Causing Leaf Spot Disease on Sweet Cherry in Province of Turkey. J. Plant Dis. Prot. 2022, 129, 1355–1366. [Google Scholar] [CrossRef]
- Thomidis, T.; Tsipouridis, C. First Report of Alternaria Leaf Spot on Cherry Trees in Greece. Plant Dis. 2006, 90, 680. [Google Scholar] [CrossRef]
- Van de Perre, E.; Jacxsens, L.; Liu, C.; Devlieghere, F.; De Meulenaer, B. Climate impact on Alternaria moulds and their mycotoxins in fresh produce: The case of the tomato chain. Food Res. Int. 2015, 68, 41–46. [Google Scholar] [CrossRef]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007; ISBN 9070351684. [Google Scholar]
- Andrew, M.; Peever, T.L.; Pryor, B.M. An Expanded Multilocus Phylogeny Does Not Resolve Morphological Species within the Small-Spored Alternaria Species Complex. Mycologia 2009, 101, 95–109. [Google Scholar] [CrossRef]
- Ma, G.; Bao, S.; Zhao, J.; Sui, Y.; Wu, X. Morphological and Molecular Characterization of Alternaria Species Causing Leaf Blight on Watermelon in China. Plant Dis. 2021, 105, 60–70. [Google Scholar] [CrossRef]
- Somma, S.; Pose, G.; Pardo, A.; Mulè, G.; Pinto, V.F.; Moretti, A.; Logrieco, A.F. AFLP Variability, Toxin Production, and Pathogenicity of Alternaria Species from Argentinean Tomato Fruits and Puree. Int. J. Food Microbiol. 2011, 145, 414–419. [Google Scholar] [CrossRef]
- Stewart, J.E.; Andrew, M.; Bao, X.; Chilvers, M.I.; Carris, L.M.; Peever, T.L. Development of Sequence Characterized Amplified Genomic Regions (SCAR) for Fungal Systematics: Proof of Principle Using Alternaria, Ascochyta and Tilletia. Mycologia 2013, 105, 1077–1086. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria Section Alternaria: Species, Formae Speciales or Pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Rotondo, F.; Gannibal, P.B. Biodiversity and Taxonomy of the Pleomorphic Genus Alternaria. Mycol. Prog. 2016, 15, 3. [Google Scholar] [CrossRef]
- Ozkilinc, H.; Rotondo, F.; Pryor, B.M.; Peever, T.L. Contrasting Species Boundaries between Sections Alternaria and Porri of the Genus Alternaria. Plant Pathol. 2018, 67, 303–314. [Google Scholar] [CrossRef]
- Peever, T.L.; Ibañez, A.; Akimitsu, K.; Timmer, L.W. Worldwide Phylogeography of the Citrus Brown Spot Pathogen, Alternaria alternata. Phytopathology 2002, 92, 794–802. [Google Scholar] [CrossRef]
- Peever, T.L.; Su, G.; Carpenter-Boggs, L.; Timmer, L.W. Molecular Systematics of Citrus-Associated Alternaria Species. Mycologia 2004, 96, 119. [Google Scholar] [CrossRef]
- Pryor, B.M.; Michailides, T.J. Morphological, Pathogenic, and Molecular Characterization of Alternaria Isolates Associated with Alternaria Late Blight of Pistachio. Phytopathology 2002, 92, 406–416. [Google Scholar] [CrossRef]
- Hong, S.G.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt a 1 Allergen Homologs from Alternaria and Related Taxa: Analysis of Phylogenetic Content and Secondary Structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, A.; Akimitsu, K.; Yamamoto, M.; Yamamoto, H. Endopolygalacturonase Is Essential for Citrus Black Rot Caused by Alternaria Citri but Not Brown Spot Caused by Alternaria alternata. Mol. Plant Microbe Interact. 2001, 14, 749–757. [Google Scholar] [CrossRef] [PubMed]
- Isshiki, A.; Akimitsu, K.; Nishio, K.; Tsukamoto, M.; Yamamoto, H. Purification and Characterization of an Endopolygalacturonase from the Rough Lemon Pathotype of Alternaria alternata, the Cause of Citrus Brown Spot Disease. Physiol. Mol. Plant Pathol. 1997, 51, 155–167. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A Multi-Gene Phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of Localized Incongruence Using a Combinational Bootstrap Approach. Mol. Phylogenet Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Prencipe, S.; Meloni, G.R.; Nari, L.; Schiavon, G.; Spadaro, D. Pathogenicity, Molecular Characterization and Mycotoxigenic Potential of Alternaria Spp. Agents of Black Spots on Fruit and Leaves of Pyrus Communis in Italy. Phytopathology 2022, 113, 309–320. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-Spored Alternaria Pathogens in Section Porri Disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest Version 2. Program Distributed by the Author. Evolutionary Biology Centre, Uppsala University, Uppsala.-References-Scientific Research Publishing. 2004. Available online: https://www.scirp.org/(S(i43dyn45teexjx455qlt3d2q))/reference/ReferencesPapers.aspx?ReferenceID=1276217 (accessed on 29 March 2023).
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods). Version 4. Sinauer Associates, Sunderland.-References-Scientific Research Publishing. 2003. Available online: https://www.scirp.org/(S(czeh2tfqyw2orz553k1w0r45))/reference/ReferencesPapers.aspx?ReferenceID=1085917 (accessed on 29 March 2023).
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Wang, F.; Saito, S.; Michailides, T.J.; Xiao, C.L. Phylogenetic, Morphological, and Pathogenic Characterization of Alternaria Species Associated with Fruit Rot of Mandarin in California. Plant Dis. 2021, 105, 1555–1567. [Google Scholar] [CrossRef]
- Zhu, X.Q.; Xiao, C.L. Phylogenetic, Morphological, and Pathogenic Characterization of Alternaria Species Associated with Fruit Rot of Blueberry in California. Phytopathology 2015, 105, 1555–1567. [Google Scholar] [CrossRef]
- Kanetis, L.; Testempasis, S.; Goulas, V.; Samuel, S.; Myresiotis, C.; Karaoglanidis, G.S. Identification and Mycotoxigenic Capacity of Fungi Associated with Pre- and Postharvest Fruit Rots of Pomegranates in Greece and Cyprus. Int. J. Food Microbiol. 2015, 208, 84–92. [Google Scholar] [CrossRef]
- Riquelme, D.; Zuniga, C.; Tapia, E. First Report of Fruit Rot of Sweet Cultivars of Japanese Plum Caused by Alternaria alternata, A. arborescens, and A. tenuissima in Chile. Plant Dis. 2021, 105, 4167. [Google Scholar] [CrossRef]
- Ntasiou, P.; Myresiotis, C.; Konstantinou, S.; Papadopoulou-Mourkidou, E.; Karaoglanidis, G.S. Identification, Characterization and Mycotoxigenic Ability of Alternaria spp. Causing Core Rot of Apple Fruit in Greece. Int. J. Food Microbiol. 2015, 197, 22–29. [Google Scholar] [CrossRef]
- Harteveld, D.O.C.; Akinsanmi, O.A.; Drenth, A. Multiple Alternaria Species Groups Are Associated with Leaf Blotch and Fruit Spot Diseases of Apple in Australia. Plant Pathol. 2013, 62, 289–297. [Google Scholar] [CrossRef]
- Aiello, D.; Guarnaccia, V.; Azzaro, A.; Polizzi, G. Alternaria Brown Spot on New Clones of Sweet Orange and Lemon in Italy. Phytopathol. Mediterr. 2020, 59, 131–145. [Google Scholar] [CrossRef]
- Yang, J.; Chen, Y.Z.; Yu-Xuan, W.; Tao, L.; Zhang, Y.D.; Wang, S.R.; Zhang, G.C.; Zhang, J. Inhibitory Effects and Mechanisms of Vanillin on Gray Mold and Black Rot of Cherry Tomatoes. Pestic. Biochem. Physiol. 2021, 175, 104859. [Google Scholar] [CrossRef] [PubMed]
- Al-Rahbi, B.A.A.; Al-Sadi, A.M.; Al-Mahmooli, I.H.; Al-Maawali, S.S.; Al-Mahruqi, N.M.T.; Velazhahan, R. Meyerozyma guilliermondii SQUCC-33Y Suppresses Postharvest Fruit Rot of Strawberry Caused by Alternaria Alternata. Australas. Plant Pathol. 2021, 50, 349–352. [Google Scholar] [CrossRef]
- Ezra, D.; Shulhani, R.; Bar Ya’Akov, I.; Harel-Beja, R.; Holland, D.; Shtienberg, D. Factors Affecting the Response of Pomegranate Fruit to Alternaria alternata, the Causal Agent of Heart Rot. Plant Dis. 2019, 103, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Jia, M.S.; Li, S.C.; Xiao, L.H.; Wang, Y.B.; Peng, W.W.; Chen, J.Y.; Xiang, M.L. First Report of Postharvest Fruit Rot in Solanum muricatum Caused by Alternaria alternata in Southwest China. Plant Dis. 2022, 106, 2520. [Google Scholar] [CrossRef]
- Guo, L.W.; He, S.H.; Gao, Z.R.; Wu, Z.T.; Duan, R.Q.; Yang, K.Z.; Wei, Y.J.; He, X.H. First Report of Fruit Rot in Opium Poppy (Papaver somniferum) Caused by Alternaria alternata in China. Plant Dis. 2020, 104, 3264. [Google Scholar] [CrossRef]
- Basım, E.; Basım, H.; Abdulai, M.; Baki, D.; Öztürk, N. Identification and Characterization of Alternaria alternata Causing Leaf Spot of Olive Tree (Olea europaea) in Turkey. Crop Prot. 2017, 92, 79–88. [Google Scholar] [CrossRef]
- Serdani, M.; Kang, J.C.; Andersen, B.; Crous, P.W. Characterisation of Alternaria Species-Groups Associated with Core Rot of Apples in South Africa. Mycol. Res. 2002, 106, 561–569. [Google Scholar] [CrossRef]
- Pryor, B.M.; Gilbertson, R.L. Molecular Phylogenetic Relationships amongst Alternaria Species and Related Fungi Based upon Analysis of Nuclear ITS and Mt SSU RDNA Sequences. Mycol. Res. 2000, 104, 1312–1321. [Google Scholar] [CrossRef]
- Elfar, K.; Zoffoli, J.P.; Latorre, B.A. Identification and Characterization of Alternaria Species Associated with Moldy Core of Apple in Chile. Plant Dis. 2018, 102, 2158–2169. [Google Scholar] [CrossRef]
- Prusky, D. Pathogen Quiescence in Postharvest Diseases. Annu. Rev. Phytopathol. 1996, 34, 413–434. [Google Scholar] [CrossRef]
- Rotem, J. The Genus Alternaria: Biology, Epidemiology, and Pathogenicity; The American Phytopathological Society: Saint Paul, MN, USA, 1994; ISBN 978-0-89054-152-4. [Google Scholar]
- Mmbaga, M.T.; Shi, A.; Kim, M.-S. Identification of Alternaria alternata as a Causal Agent for Leaf Blight in Syringa Species. Plant Pathol J 2011, 27, 120–127. [Google Scholar] [CrossRef]
- Fontaine, K.; Fourrier-Jeandel, C.; Armitage, A.D.; Boutigny, A.L.; Crépet, M.; Caffier, V.; Gnide, D.C.; Shiller, J.; Le Cam, B.; Giraud, M.; et al. Identification and Pathogenicity of Alternaria Species Associated with Leaf Blotch Disease and Premature Defoliation in French Apple Orchards. PeerJ 2021, 9, e12496. [Google Scholar] [CrossRef]
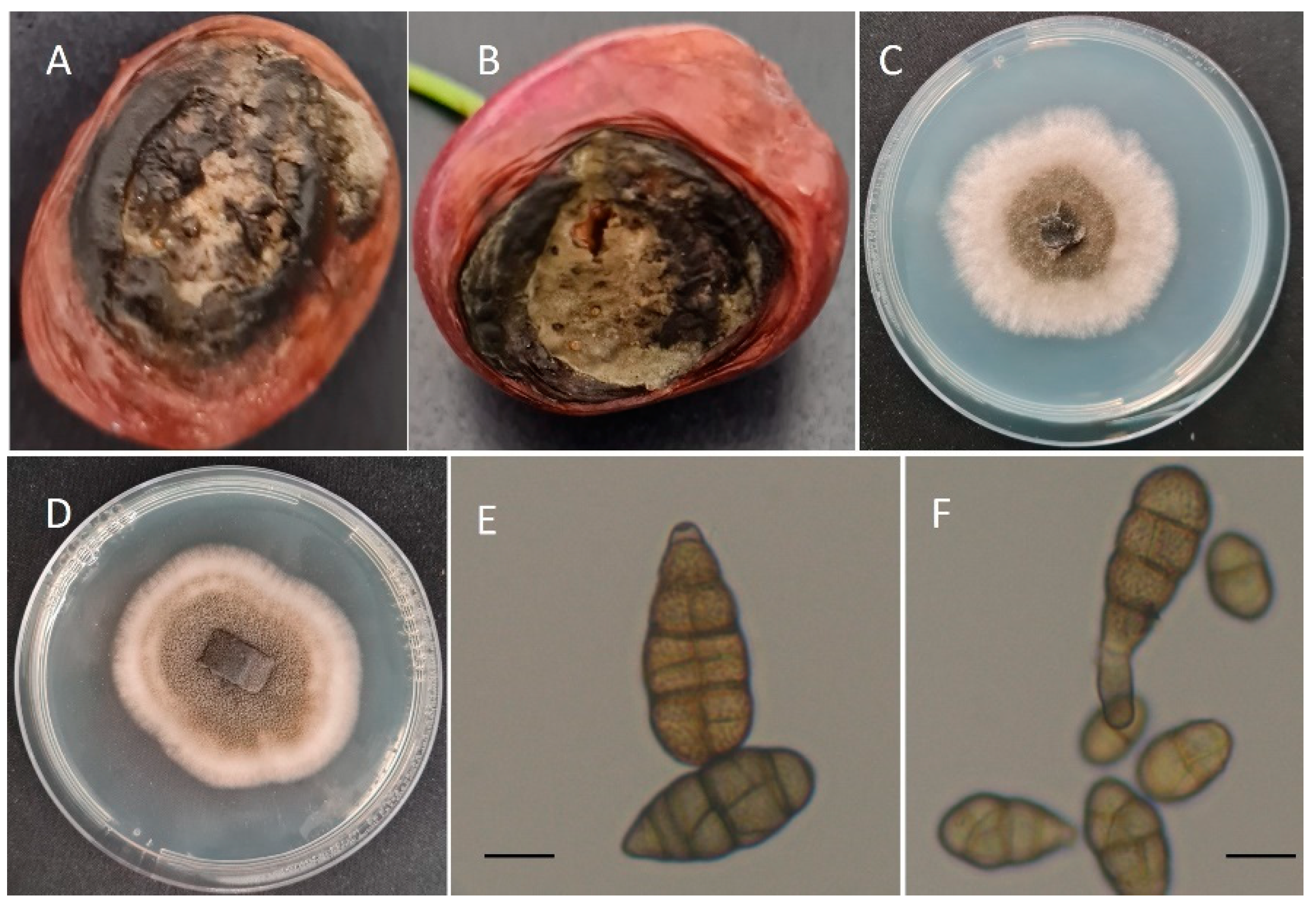

| Isolate Name | Fungal Species | Year of Isolation | Cherry Cultivar | Colony Growth (cm ± SD) | Size of Conidia (Length × Width; μm) | Rot Diameter (mm ± SD) 1 |
|---|---|---|---|---|---|---|
| D4 | AASC | 2020 | Kordia | 4.94 ± 0.59 | 21.73 × 11.31 | 12.06 b–d ± 1.52 |
| T2 | AASC | 2020 | Ferrovia | 5.14 ± 0.28 | 20.13 × 11.19 | 9.56 bc ± 1.22 |
| T3 | AASC | 2020 | Ferrovia | 4.98 ± 0.55 | 21.68 × 11.23 | 12.21 bc ± 1.66 |
| T6 | A. alternata | 2020 | Ferrovia | 4.90 ± 0.59 | 20.64 × 11.18 | 13.81 b–d ± 1.82 |
| T8 | AASC | 2020 | Ferrovia | 4.96 ± 0.55 | 20.57 × 11.10 | 10.65 b–d ± 1.86 |
| T9 | AASC | 2020 | Ferrovia | 5.08 ± 0.35 | 21.53 × 11.27 | 10.75 bc ± 2.31 |
| GR1 | AASC | 2020 | Regina | 4.90 ± 0.58 | 21.73 × 11.24 | 11.39 b–d ± 1.88 |
| GR3 | A. alternata | 2020 | Regina | 4.70 ± 0.51 | 20.83 × 11.12 | 9.06 bc ± 1.81 |
| GR6 | AASC | 2020 | Regina | 4.8 8± 0.53 | 20.89 × 10.99 | 14.94 b–d ± 2.77 |
| GR8 | AASC | 2020 | Regina | 4.94 ± 0.59 | 20.94 × 11.20 | 11.94 b–d ± 1.68 |
| GR13 | A. alternata | 2020 | Regina | 4.82 ± 0.51 | 20.93 × 11.21 | 9.06 bc ± 1.45 |
| W2 | AASC | 2020 | Kordia | 5.02 ± 0.33 | 21.07 × 11.16 | 11.06 b–d ± 1.53 |
| W6 | AASC | 2020 | Kordia | 5.30 ± 0.41 | 20.43 × 11.12 | 11.40 b–d ± 2.29 |
| Q1 | AASC | 2020 | Kordia | 5.00 ± 0.78 | 19.76 × 11.64 | 14.43 b–d ± 2.64 |
| X7 | AASC | 2020 | Kordia | 4.96 ± 0.62 | 20.84 × 11.31 | 15.25 cd ± 2.94 |
| Ch1 | AASC | 2021 | Regina | 4.70 ± 0.70 | 21.25 × 11.11 | 9.38 bc ± 1.98 |
| Ch2 | AASC | 2021 | Regina | 5.16 ± 0.15 | 20.59 × 10.86 | 10.94 bc ± 2.59 |
| Ch4 | AASC | 2021 | Regina | 4.94 ± 0.55 | 21.30 × 11.21 | 8.43 ab ± 2.03 |
| Ch5 | AASC | 2021 | Regina | 4.76 ± 0.92 | 21.32 × 11.15 | 13.93 b–d ± 1.94 |
| Ch11 | AASC | 2021 | Regina | 5.12 ± 0.13 | 20.75 × 11.18 | 9.11 bc ± 1.61 |
| Ch12 | AASC | 2021 | Regina | 4.96 ± 0.55 | 20.88 × 11.02 | 10.71 bc ± 3.54 |
| Ch13 | AASC | 2021 | Regina | 4.94 ± 0.59 | 21.16 × 11.17 | 9.12 bc ± 1.66 |
| Ch14 | AASC | 2021 | Regina | 5.26 ± 0.49 | 20.20 × 10.84 | 10.06 b–d ± 1.59 |
| Ch16 | AASC | 2021 | Regina | 5.12 ± 0.77 | 21.40 × 11.31 | 11.75 b–d ± 1.88 |
| Ch17 | AASC | 2021 | Regina | 5.08 ± 0.74 | 21.32 × 11.14 | 17.00 d ± 2.13 |
| Ch18 | AASC | 2021 | Regina | 4.94 ± 0.47 | 21.71 × 11.23 | 14.25 b–d ± 1.89 |
| Ch19 | AASC | 2021 | Regina | 4.74 ± 0.81 | 21.10 × 11.22 | 12.64 b–d ± 1.61 |
| Ch21 | A. alternata | 2021 | Regina | 5.06 ± 0.34 | 21.02 × 10.88 | 11.00 bc ± 1.98 |
| Ch22 | AASC | 2021 | Regina | 5.00 ± 0.21 | 20.87 × 11.07 | 10.06 bc ± 1.66 |
| Ch23 | A. alternata | 2022 | Regina | 5.12 ± 0.53 | 20.90 × 11.03 | 11.50 b–d ± 1.90 |
| Ch26 | AASC | 2022 | Regina | 5.18 ± 0.48 | 21.04 × 11.12 | 11.94 b–d ± 1.87 |
| Ch27 | AASC | 2022 | Regina | 4.86 ± 0.77 | 21.12 ×11.09 | 9.25 bc ± 1.80 |
| Ch35 | AASC | 2022 | Regina | 4.98 ± 0.41 | 21.03 × 11.16 | 10.78 b–d ± 1.93 |
| Ch37 | A. alternata | 2022 | Regina | 4.52 ± 0.78 | 21.12 × 11.19 | 9.17 bc ± 1.52 |
| Ch39 | AASC | 2022 | Regina | 4.74 ± 0.57 | 21.07 × 10.98 | 11.50 bc ± 1.93 |
| Ch40 | AASC | 2022 | Regina | 5.18 ± 0.26 | 21.12 × 11.11 | 10.50 bc ± 1.74 |
| Ch42 | AASC | 2022 | Regina | 5.04 ± 0.27 | 20.94 × 11.03 | 12.06 b–d ± 1.52 |
| Ch43 | A. alternata | 2022 | Regina | 4.68 ± 0.68 | 21.05 × 11.01 | 10.88 bc ± 1.23 |
| Ch45 | AASC | 2022 | Regina | 4.92 ± 0.59 | 21.20 × 11.29 | 10.81 bc ± 1.94 |
| Ch48 | AASC | 2022 | Regina | 4.88 ± 0.50 | 21.11 × 11.05 | 9.33 bc ± 1.97 |
| Healthy control | - | - | Regina | - | - | 0 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waqas, M.; Prencipe, S.; Guarnaccia, V.; Spadaro, D. Molecular Characterization and Pathogenicity of Alternaria spp. Associated with Black Rot of Sweet Cherries in Italy. J. Fungi 2023, 9, 992. https://doi.org/10.3390/jof9100992
Waqas M, Prencipe S, Guarnaccia V, Spadaro D. Molecular Characterization and Pathogenicity of Alternaria spp. Associated with Black Rot of Sweet Cherries in Italy. Journal of Fungi. 2023; 9(10):992. https://doi.org/10.3390/jof9100992
Chicago/Turabian StyleWaqas, Muhammad, Simona Prencipe, Vladimiro Guarnaccia, and Davide Spadaro. 2023. "Molecular Characterization and Pathogenicity of Alternaria spp. Associated with Black Rot of Sweet Cherries in Italy" Journal of Fungi 9, no. 10: 992. https://doi.org/10.3390/jof9100992
APA StyleWaqas, M., Prencipe, S., Guarnaccia, V., & Spadaro, D. (2023). Molecular Characterization and Pathogenicity of Alternaria spp. Associated with Black Rot of Sweet Cherries in Italy. Journal of Fungi, 9(10), 992. https://doi.org/10.3390/jof9100992







