Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression
Abstract
1. Introduction
2. Materials and Methods
2.1. P. lutzii Growth Conditions
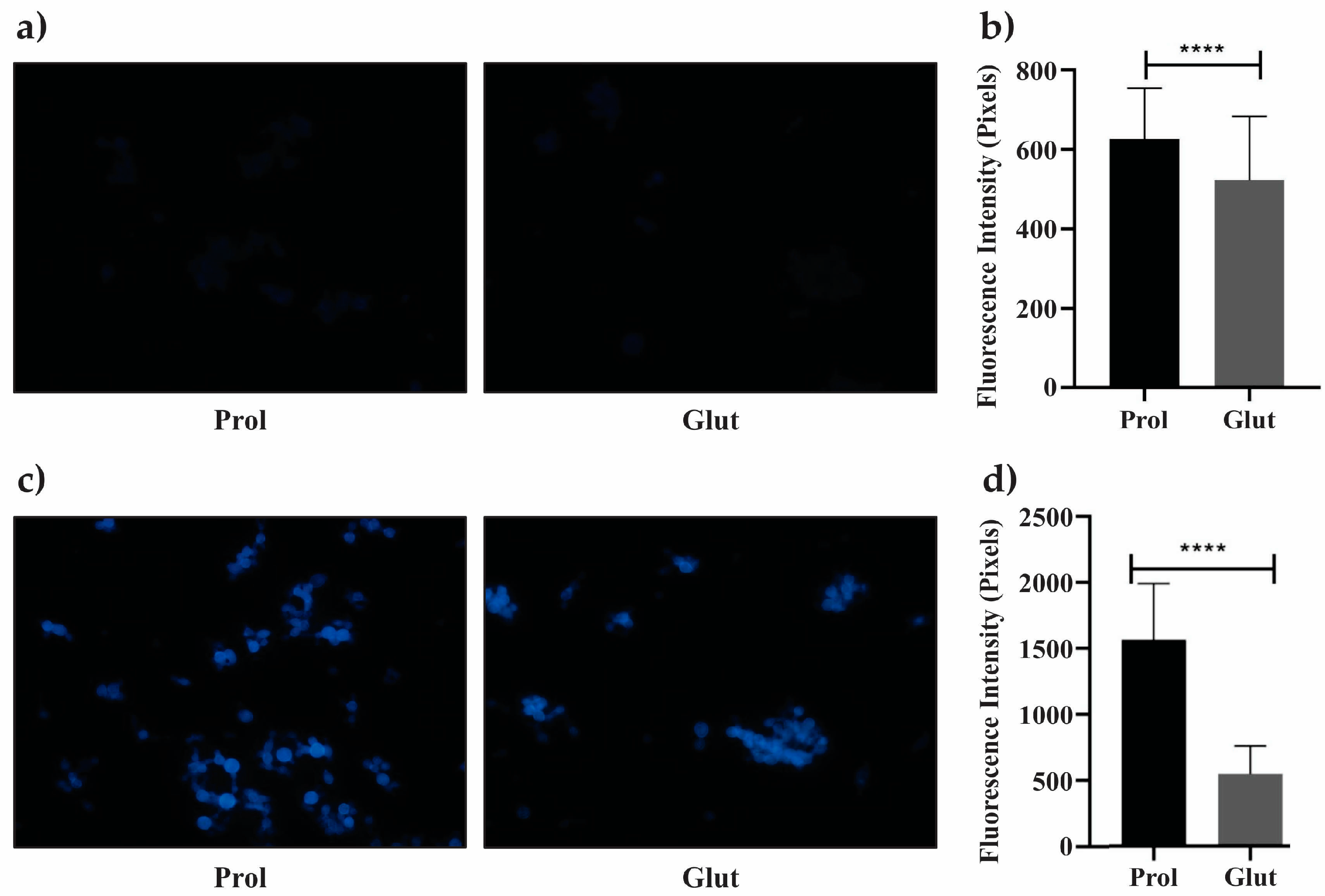
2.2. Viability Analysis of P. lutzii under NCR Conditions
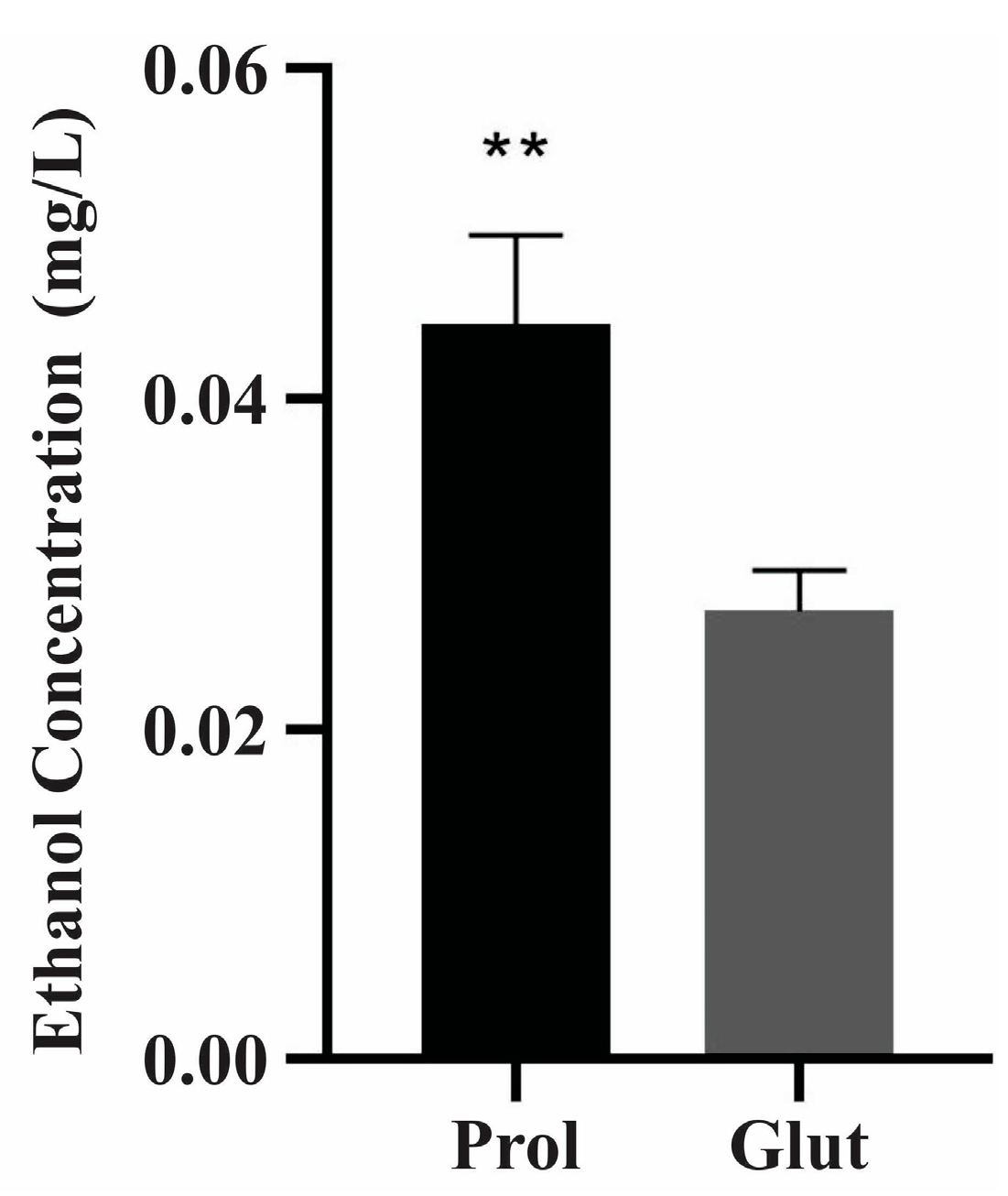
2.3. Ethanol Measurement Assay
2.4. RNA Extraction and Quantitative Real-Time RT-qPCR Analysis
2.5. P. lutzii Protein Extraction in NCR Conditions
2.6. P. lutzii nanoUPLC-MSE Analysis under NCR Conditions
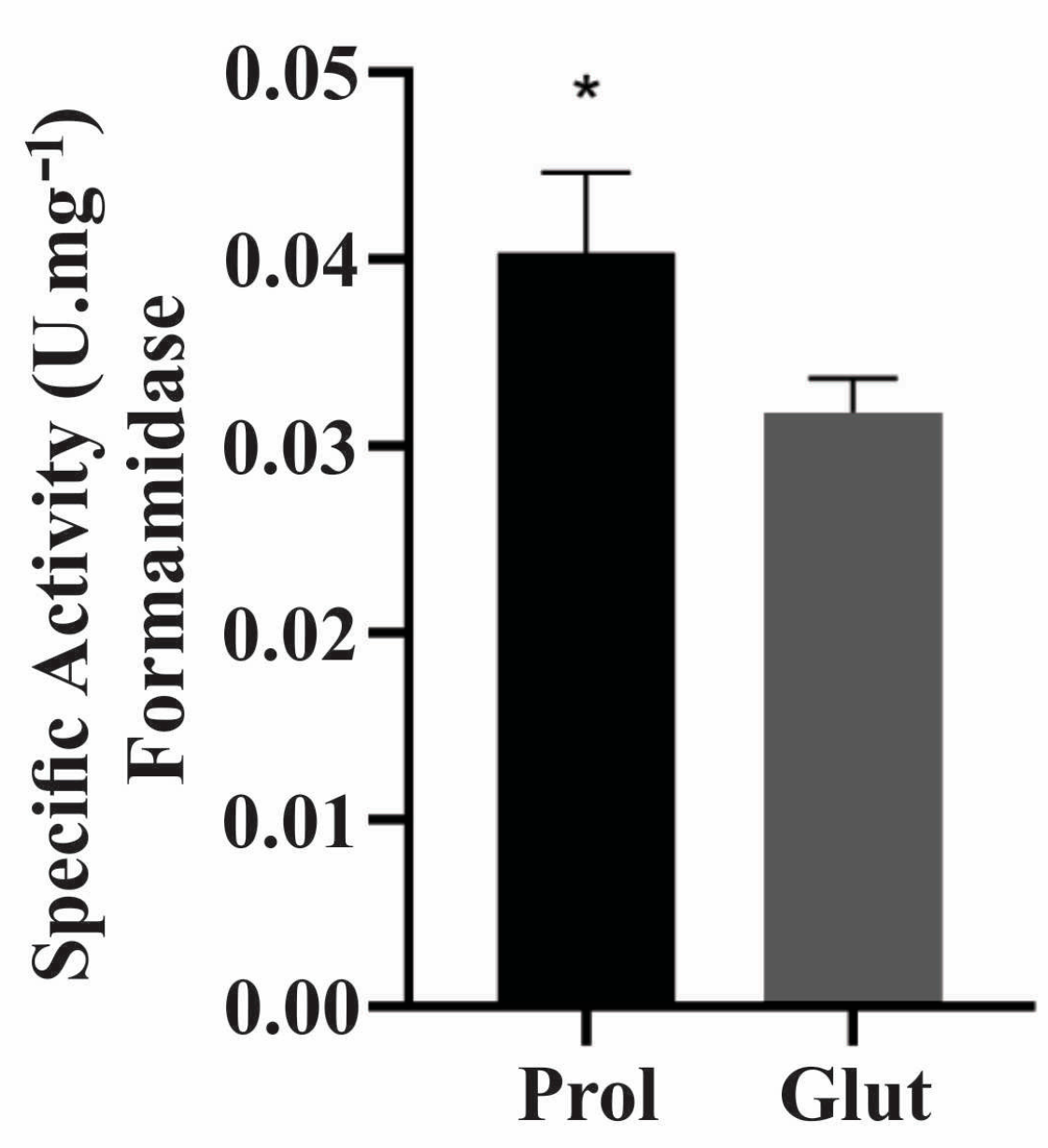
2.7. Enzymatic Activities under NCR Conditions
2.8. Estimation of Cell Wall Components in P. lutzii
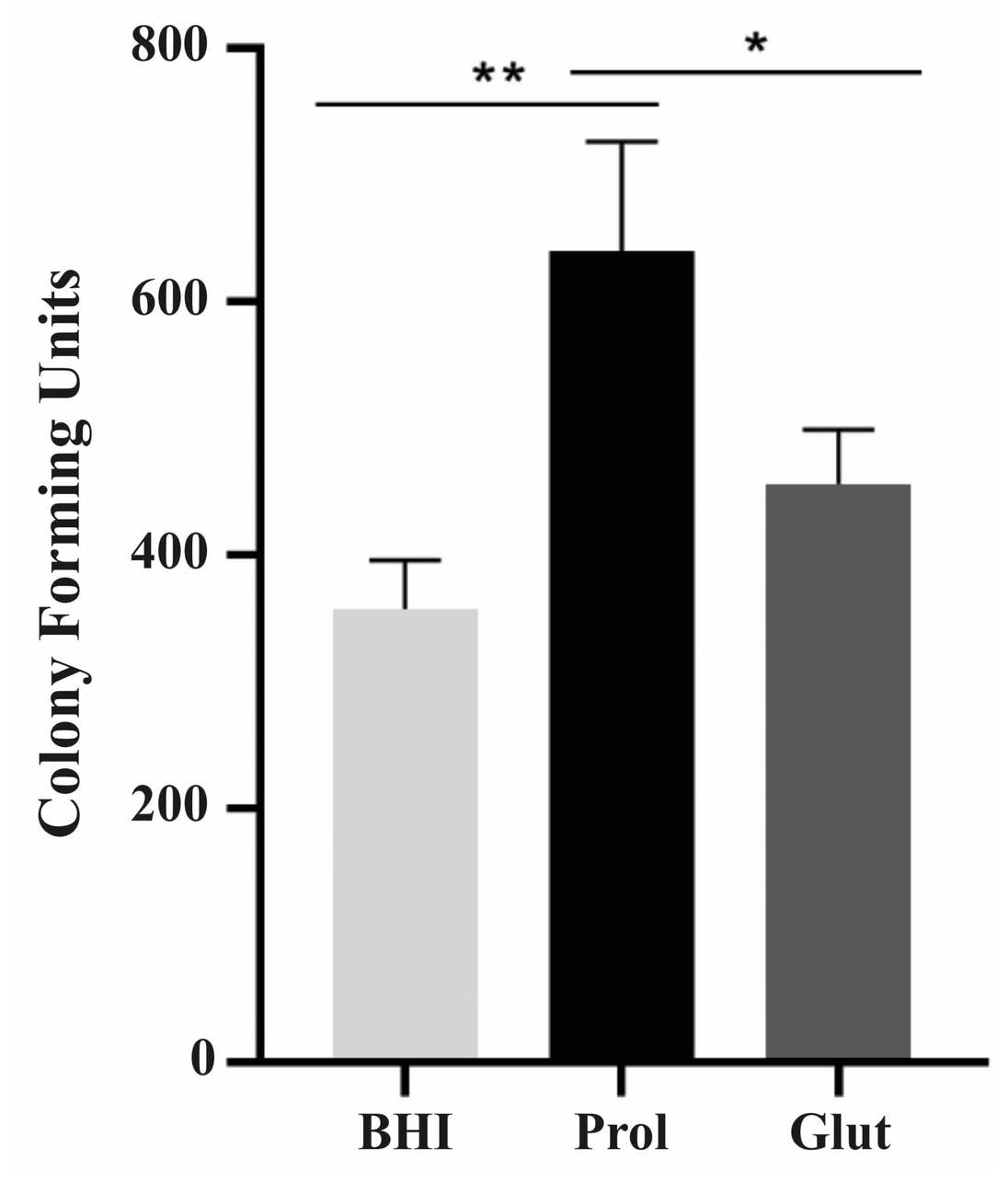
2.9. Ex Vivo Model of Infection in NCR Conditions
3. Results
3.1. P. lutzii Behavior under NCR Conditions
3.2. Proteomic Data Quality Analysis
3.3. Proteomic Analysis
3.4. An Overview of P. lutzii Metabolism under NCR Conditions
3.5. Proteins Potentially Involved in Adhesion during the NCR Response
3.6. Regulation of Cell Wall Metabolism in P. lutzii
3.7. Transcriptional Level Analysis of NCR-Related Genes
3.8. Protein Level Confirmation Analysis under NCR Conditions
3.9. Influence of NCR Training in P. lutzii Survival Inside Macrophages
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martinez, R. New trends in paracoccidioidomycosis epidemiology. J. Fungi 2017, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Turissini, D.A.; Gomez, O.M.; Teixeira, M.M.; McEwen, J.G.; Matute, D.R. Species boundaries in the human pathogen Paracoccidioides. Fungal Genet. Biol. 2017, 106, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Brummer, E.; Castaneda, E.; Restrepo, A. Paracoccidioidomycosis: An update. Clin. Microbiol. Rev. 1993, 6, 89–117. [Google Scholar] [CrossRef] [PubMed]
- San-Blas, G.; Niño-Vega, G.; Iturriaga, T. Paracoccidioides brasiliensis and paracoccidioidomycosis: Molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med. Mycol. 2002, 40, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Shikanai-Yasuda, M.A.; Mendes, R.P.; Colombo, A.L.; Queiroz-Telles, F.d.; Kono, A.S.G.; Paniago, A.M.; Nathan, A.; Valle, A.C.F.d.; Bagagli, E.; Benard, G. Brazilian guidelines for the clinical management of paracoccidioidomycosis. Rev. Soc. Bras. Med. Trop. 2017, 50, 715–740. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef]
- Lee, I.R.; Morrow, C.A.; Fraser, J.A. Nitrogen regulation of virulence in clinically prevalent fungal pathogens. FEMS Microbiol. Lett. 2013, 345, 77–84. [Google Scholar] [CrossRef]
- Gimeno, C.J.; Ljungdahl, P.O.; Styles, C.A.; Fink, G.R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell 1992, 68, 1077–1090. [Google Scholar] [CrossRef]
- Marini, A.-M.; Soussi-Boudekou, S.; Vissers, S.; Andre, B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997, 17, 4282–4293. [Google Scholar] [CrossRef]
- Brock, M. Fungal metabolism in host niches. Curr. Opin. Microbiol. 2009, 12, 371–376. [Google Scholar] [CrossRef]
- Fleck, C.B.; Schöbel, F.; Brock, M. Nutrient acquisition by pathogenic fungi: Nutrient availability, pathway regulation, and differences in substrate utilization. Int. J. Med. Microbiol. 2011, 301, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Magasanik, B.; Kaiser, C.A. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002, 290, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.J.; Zeh, C.; Saur, A.; Sonnewald, S.; Sonnewald, U.; Voll, L.M. The Ustilago maydis Nit2 homolog regulates nitrogen utilization and is required for efficient induction of filamentous growth. Eukaryot. Cell 2012, 11, 368–380. [Google Scholar] [CrossRef]
- ter Schure, E.G.; van Riel, N.A.; Verrips, C.T. The role of ammonia metabolism in nitrogen catabolite repression in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2000, 24, 67–83. [Google Scholar] [CrossRef]
- Sieg, A.G.; Trotter, P.J. Differential contribution of the proline and glutamine pathways to glutamate biosynthesis and nitrogen assimilation in yeast lacking glutamate dehydrogenase. Microbiol. Res. 2014, 169, 709–716. [Google Scholar] [CrossRef]
- Zaman, S.; Lippman, S.I.; Zhao, X.; Broach, J.R. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 2008, 42, 27–81. [Google Scholar] [CrossRef]
- Wong, K.H.; Hynes, M.J.; Davis, M.A. Recent advances in nitrogen regulation: A comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot. Cell 2008, 7, 917–925. [Google Scholar] [CrossRef]
- Coffman, J.A.; Rai, R.; Loprete, D.M.; Cunningham, T.; Svetlov, V.; Cooper, T.G. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 3416–3429. [Google Scholar] [CrossRef]
- Hofman-Bang, J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 1999, 12, 35–71. [Google Scholar] [CrossRef]
- Yoo, H.-S.; Genbauffe, F.S.; Cooper, T.G. Identification of the ureidoglycolate hydrolase gene in the DAL gene cluster of Saccharomyces cerevisiae. Mol. Cell. Biol. 1985, 5, 2279–2288. [Google Scholar] [CrossRef] [PubMed]
- Genbauffe, F.S.; Cooper, T.G. Induction and repression of the urea amidolyase gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1986, 6, 3954–3964. [Google Scholar] [CrossRef]
- Courchesne, W.E.; Magasanik, B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: Roles of the URE2 and GLN3 genes. J. Bacteriol. 1988, 170, 708–713. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.M.; Magasanik, B. Role of the complex upstream region of the GDH2 gene in nitrogen regulation of the NAD-linked glutamate dehydrogenase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 6229–6247. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, J.; Rai, R.; el Berry, H.M.; Cooper, T.G. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 1993, 175, 64–73. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arst, H.N.; Cove, D.J. Nitrogen metabolite repression in Aspergillus nidulans. Mol. Gen. Genet. MGG 1973, 126, 111–141. [Google Scholar] [CrossRef]
- Fu, Y.-H.; Marzluf, G.A. Characterization of nit-2, the major nitrogen regulatory gene of Neurospora crassa. Mol. Cell. Biol. 1987, 7, 1691–1696. [Google Scholar] [CrossRef]
- Fu, Y.H.; Marzluf, G.A. nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc. Natl. Acad. Sci. USA 1990, 87, 5331–5335. [Google Scholar] [CrossRef]
- Kudla, B.; Caddick, M.; Langdon, T.; Martinez-Rossi, N.M.; Bennett, C.; Sibley, S.; Davies, R.; Arst, H., Jr. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990, 9, 1355–1364. [Google Scholar] [CrossRef]
- Ramachandra, S.; Linde, J.; Brock, M.; Guthke, R.; Hube, B.; Brunke, S. Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. PLoS ONE 2014, 9, e92734. [Google Scholar] [CrossRef]
- Godard, P.; Urrestarazu, A.; Vissers, S.; Kontos, K.; Bontempi, G.; Van Helden, J.; André, B. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3065–3086. [Google Scholar] [CrossRef]
- Scherens, B.; Feller, A.; Vierendeels, F.; Messenguy, F.; Dubois, E. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEMS Yeast Res. 2006, 6, 777–791. [Google Scholar] [CrossRef]
- Zhao, S.; Zhao, X.; Zou, H.; Fu, J.; Du, G.; Zhou, J.; Chen, J. Comparative proteomic analysis of Saccharomyces cerevisiae under different nitrogen sources. J. Proteom. 2014, 101, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Robertson, S.L.; Parker, J.; Muddiman, D.C.; Dean, R.A. Comparative proteomic analysis between nitrogen supplemented and starved conditions in Magnaporthe oryzae. Proteome Sci. 2017, 15, 20. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S.; Rispail, N.; Prados-Rosales, R.C.; Di Pietro, A. A nitrogen response pathway regulates virulence functions in Fusarium oxysporum via the protein kinase TOR and the bZIP protein MeaB. Plant Cell 2010, 22, 2459–2475. [Google Scholar] [CrossRef]
- López-Berges, M.S.; Schäfer, K.; Hera, C.; Di Pietro, A. Combinatorial function of velvet and AreA in transcriptional regulation of nitrate utilization and secondary metabolism. Fungal Genet. Biol. 2014, 62, 78–84. [Google Scholar] [CrossRef]
- Kim, H.; Woloshuk, C. Role of AREA, a regulator of nitrogen metabolism, during colonization of maize kernels and fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 2008, 45, 947–953. [Google Scholar] [CrossRef]
- Frazzitta, A.E.; Vora, H.; Price, M.S.; Tenor, J.L.; Betancourt-Quiroz, M.; Toffaletti, D.L.; Cheng, N.; Perfect, J.R. Nitrogen source-dependent capsule induction in human-pathogenic Cryptococcus species. Eukaryot. Cell 2013, 12, 1439–1450. [Google Scholar] [CrossRef]
- Lee, I.R.; Chow, E.W.; Morrow, C.A.; Djordjevic, J.T.; Fraser, J.A. Nitrogen metabolite repression of metabolism and virulence in the human fungal pathogen Cryptococcus neoformans. Genetics 2011, 188, 309–323. [Google Scholar] [CrossRef]
- Lee, I.R.; Lim, J.W.; Ormerod, K.L.; Morrow, C.A.; Fraser, J.A. Characterization of an Nmr homolog that modulates GATA factor-mediated nitrogen metabolite repression in Cryptococcus neoformans. PLoS ONE 2012, 7, e32585. [Google Scholar] [CrossRef]
- Liao, W.-L.; Ramón, A.M.; Fonzi, W.A. GLN3 encodes a global regulator of nitrogen metabolism and virulence of C. albicans. Fungal Genet. Biol. 2008, 45, 514–526. [Google Scholar] [CrossRef]
- Cruz-Leite, V.R.M.; Salem-Izacc, S.M.; Novaes, E.; Neves, B.J.; de Almeida Brito, W.; Silva, L.O.H.S.; Paccez, J.D.; Parente-Rocha, J.A.; Pereira, M.; de Almeida Soares, C.M. Nitrogen Catabolite Repression in members of Paracoccidioides complex. Microb. Pathog. 2020, 149, 104281. [Google Scholar] [CrossRef]
- Fava-Neto, C. Estudos quantitativos sobre a fixacao do complemento na blastomicose sulamericana com antigenos polissacaridicos. Arq. Cir. Clin. Exp. 1955, 18, 197–254. [Google Scholar]
- Bookout, A.L.; Jeong, Y.; Downes, M.; Ruth, T.Y.; Evans, R.M.; Mangelsdorf, D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006, 126, 789–799. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Murad, A.M.; Rech, E.L. NanoUPLC-MSE proteomic data assessment of soybean seeds using the Uniprot database. BMC Biotechnol. 2012, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Parente-Rocha, J.A.; Parente, A.F.; Baeza, L.C.; Bonfim, S.M.; Hernandez, O.; McEwen, J.G.; Bailao, A.M.; Taborda, C.P.; Borges, C.L.; Soares, C.M. Macrophage Interaction with Paracoccidioides brasiliensis Yeast Cells Modulates Fungal Metabolism and Generates a Response to Oxidative Stress. PLoS ONE 2015, 10, e0137619. [Google Scholar] [CrossRef] [PubMed]
- Ramana, J.; Gupta, D. FaaPred: A SVM-based prediction method for fungal adhesins and adhesin-like proteins. PLoS ONE 2010, 5, e9695. [Google Scholar] [CrossRef]
- Borges, C.L.; Pereira, M.; Felipe, M.S.; de Faria, F.P.; Gomez, F.J.; Deepe Jr, G.S.; Soares, C.M. The antigenic and catalytically active formamidase of Paracoccidioides brasiliensis: Protein characterization, cDNA and gene cloning, heterologous expression and functional analysis of the recombinant protein. Microbes Infect. 2005, 7, 66–77. [Google Scholar] [CrossRef]
- Silber, P.M.; Gandolfi, A.J.; Brendel, K. Adaptation of a γ-glutamyl transpeptidase assay to microtiter plates. Anal. Biochem. 1986, 158, 68–71. [Google Scholar] [CrossRef]
- Lima, P.d.S.; Casaletti, L.; Bailão, A.M.; Vasconcelos, A.T.R.d.; Fernandes, G.d.R.; Soares, C.M.d.A. Transcriptional and proteomic responses to carbon starvation in Paracoccidioides. PLoS Neglected Trop. Dis. 2014, 8, e2855. [Google Scholar] [CrossRef]
- Moreira, A.L.E.; Cruz-Leite, V.R.M.; Silva, L.O.H.S.; Parente, A.F.A.; Bailão, A.M.; de Almeida Soares, C.M.; Parente-Rocha, J.A.; Ruiz, O.H.; Borges, C.L. Proteome characterization of Paracoccidioides lutzii conidia by using nanoUPLC-MSE. Fungal Biol. 2020, 124, 766–780. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Arenas, E.; Cabezón, V.; Bermejo, C.; Arroyo, J.; Nombela, C.; Diez-Orejas, R.; Gil, C. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell. Proteom. 2007, 6, 460–478. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.J.; Cooper, T.G. Stress-responsive Gln3 localization in Saccharomyces cerevisiae is separable from and can overwhelm nitrogen source regulation. J. Biol. Chem. 2007, 282, 18467–18480. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Tate, J.J.; Shanmuganatham, K.; Howe, M.M.; Nelson, D.; Cooper, T.G. Nuclear Gln3 import is regulated by nitrogen catabolite repression whereas export is specifically regulated by glutamine. Genetics 2015, 201, 989–1016. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Airoldi, E.M.; Miller, D.; Athanasiadou, R.; Brandt, N.; Abdul-Rahman, F.; Neymotin, B.; Hashimoto, T.; Bahmani, T.; Gresham, D. Steady-state and dynamic gene expression programs in Saccharomyces cerevisiae in response to variation in environmental nitrogen. Mol. Biol. Cell 2016, 27, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Brosnan, J.T. Interorgan amino acid transport and its regulation. J. Nutr. 2003, 133, 2068S–2072S. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Bender, J.A.; Fink, G.R. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 2004, 3, 1076–1087. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot. Cell 2002, 1, 657–662. [Google Scholar] [CrossRef]
- Lorenz, M.C.; Fink, G.R. The glyoxylate cycle is required for fungal virulence. Nature 2001, 412, 83–86. [Google Scholar] [CrossRef]
- Barelle, C.J.; Priest, C.L.; MacCallum, D.M.; Gow, N.A.; Odds, F.C.; Brown, A.J. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 2006, 8, 961–971. [Google Scholar] [CrossRef]
- Derengowski, L.; Tavares, A.; Silva, S.; Procópio, L.; Felipe, M.; Silva-Pereira, I. Upregulation of glyoxylate cycle genes upon Paracoccidioides brasiliensis internalization by murine macrophages and in vitro nutritional stress condition. Med. Mycol. 2008, 46, 125–134. [Google Scholar] [CrossRef]
- Baeza, L.C.; Da Mata, F.R.; Pigosso, L.L.; Pereira, M.; De Souza, G.H.; Coelho, A.S.; Soares, C.M.d.A. Differential metabolism of a two-carbon substrate by members of the Paracoccidioides genus. Front. Microbiol. 2017, 8, 2308. [Google Scholar] [CrossRef] [PubMed]
- Brock, M.; Fischer, R.; Linder, D.; Buckel, W. Methylcitrate synthase from Aspergillus nidulans: Implications for propionate as an antifungal agent. Mol. Microbiol. Orig. Artic. 2000, 35, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Domin, N.; Wilson, D.; Brock, M. Methylcitrate cycle activation during adaptation of Fusarium solani and Fusarium verticillioides to propionyl-CoA-generating carbon sources. Microbiology 2009, 155, 3903–3912. [Google Scholar] [CrossRef]
- Santos, L.P.A.; Assunção, L.d.P.; Lima, P.d.S.; Tristão, G.B.; Brock, M.; Borges, C.L.; Silva-Bailão, M.G.; Soares, C.M.d.A.; Bailão, A.M. Propionate metabolism in a human pathogenic fungus: Proteomic and biochemical analyses. IMA Fungus 2020, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Grahl, N.; Puttikamonkul, S.; Macdonald, J.M.; Gamcsik, M.P.; Ngo, L.Y.; Hohl, T.M.; Cramer, R.A. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathog. 2011, 7, e1002145. [Google Scholar] [CrossRef]
- Lima, P.d.S.; Chung, D.; Bailão, A.M.; Cramer, R.A.; Soares, C.M.d.A. Characterization of the Paracoccidioides hypoxia response reveals new insights into pathogenesis mechanisms of this important human pathogenic fungus. PLoS Neglected Trop. Dis. 2015, 9, e0004282. [Google Scholar] [CrossRef]
- Parente, A.F.A.; de Rezende, T.C.V.; De Castro, K.P.; Bailão, A.M.; Parente, J.A.; Borges, C.L.; Silva, L.P.; Soares, C.M.d.A. A proteomic view of the response of Paracoccidioides yeast cells to zinc deprivation. Fungal Biol. 2013, 117, 399–410. [Google Scholar] [CrossRef]
- Parente, A.F.; Bailao, A.M.; Borges, C.L.; Parente, J.A.; Magalhaes, A.D.; Ricart, C.A.; Soares, C.M.d.A. Proteomic analysis reveals that iron availability alters the metabolic status of the pathogenic fungus Paracoccidioides brasiliensis. PLoS ONE 2011, 6, e22810. [Google Scholar] [CrossRef]
- de Arruda Grossklaus, D.; Bailão, A.M.; Rezende, T.C.V.; Borges, C.L.; de Oliveira, M.A.P.; Parente, J.A.; Soares, C.M.d.A. Response to oxidative stress in Paracoccidioides yeast cells as determined by proteomic analysis. Microbes Infect. 2013, 15, 347–364. [Google Scholar] [CrossRef]
- da Silva Rodrigues, L.N.; de Almeida Brito, W.; Parente, A.F.A.; Weber, S.S.; Bailão, A.M.; Casaletti, L.; Borges, C.L.; Soares, C.M.d.A. Osmotic stress adaptation of Paracoccidioides lutzii, Pb01, monitored by proteomics. Fungal Genet. Biol. 2016, 95, 13–23. [Google Scholar] [CrossRef]
- Campos, É.G.; Jesuino, R.; Dantas Ada, S.; Brigido Mde, M.; Felipe, M. Oxidative stress response in Paracoccidioides brasiliensis. Genet. Mol. Res. GMR 2005, 4, 409–429. [Google Scholar]
- Youseff, B.H.; Holbrook, E.D.; Smolnycki, K.A.; Rappleye, C.A. Extracellular superoxide dismutase protects Histoplasma yeast cells from host-derived oxidative stress. PLoS Pathog. 2012, 8, e1002713. [Google Scholar] [CrossRef]
- Penninckx, M.J. An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2002, 2, 295–305. [Google Scholar]
- Springael, J.-Y.; Penninckx, M.J. Nitrogen-source regulation of yeast gamma-glutamyl transpeptidase synthesis involves the regulatory network including the GATA zinc-finger factors Gln3, Nil1/Gat1 and Gzf3. Biochem. J. 2003, 371, 589–595. [Google Scholar] [CrossRef]
- Cleare, L.G.; Zamith-Miranda, D.; Nosanchuk, J.D. Heat shock proteins in Histoplasma and Paracoccidioides. Clin. Vaccine Immunol. 2017, 24, e00221-17. [Google Scholar] [CrossRef]
- Tiwari, S.; Thakur, R.; Shankar, J. Role of Heat-Shock Proteins in Cellular Function and in the Biology of Fungi. Biotechnol. Res. Int. 2015, 2015, 132635. [Google Scholar] [CrossRef]
- Tamayo, D.; Muñoz, J.F.; Torres, I.; Almeida, A.J.; Restrepo, A.; McEwen, J.G.; Hernández, O. Involvement of the 90 kDa heat shock protein during adaptation of Paracoccidioides brasiliensis to different environmental conditions. Fungal Genet. Biol. 2013, 51, 34–41. [Google Scholar] [CrossRef]
- Tomazett, M.V.; Zanoelo, F.F.; Bailão, E.F.C.; Bailão, A.M.; Borges, C.L.; Soares, C.M.d.A. Molecular and biochemical characterization of carbonic anhydrases of Paracoccidioides. Genet. Mol. Biol. 2016, 39, 416–425. [Google Scholar] [CrossRef]
- Fraser, J.A.; Davis, M.A.; Hynes, M.J. The formamidase gene of Aspergillus nidulans: Regulation by nitrogen metabolite repression and transcriptional interference by an overlapping upstream gene. Genetics 2001, 157, 119–131. [Google Scholar] [CrossRef]
- Silva, L.O.S.; Moreira, T.R.; Goncales, R.A.; Tomazett, M.V.; Parente-Rocha, J.A.; Mattos, K.; Paccez, J.D.; Ruiz, O.H.; Pereira, M.; Soares, C.M.A.; et al. Paracoccidioides lutzii Formamidase Contributes to Fungal Survival in Macrophages. Microorganisms 2022, 10, 2011. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.; Ferguson, D.; Rai, R.; Bysani, N. The GLN3 gene product is required for transcriptional activation of allantoin system gene expression in Saccharomyces cerevisiae. J. Bacteriol. 1990, 172, 1014–1018. [Google Scholar] [CrossRef]
- McLean, R.J.; Nickel, J.C.; Cheng, K.J.; Costerton, J.W. The ecology and pathogenicity of urease-producing bacteria in the urinary tract. Crit. Rev. Microbiol. 1988, 16, 37–79. [Google Scholar] [CrossRef]
- Tavares, A.H.; Silva, S.S.; Bernardes, V.V.; Maranhao, A.Q.; Kyaw, C.M.; Pocas-Fonseca, M.; Silva-Pereira, I. Virulence insights from the Paracoccidioides brasiliensis transcriptome. Genet. Mol. Res. GMR 2005, 4, 372–389. [Google Scholar]
- Rappleye, C.A.; Goldman, W.E. Defining virulence genes in the dimorphic fungi. Annu. Rev. Microbiol. 2006, 60, 281–303. [Google Scholar] [CrossRef]
- Mora, D.; Arioli, S. Microbial urease in health and disease. PLoS Pathog. 2014, 10, e1004472. [Google Scholar] [CrossRef]
- Feder, V.; Kmetzsch, L.; Staats, C.C.; Vidal-Figueiredo, N.; Ligabue-Braun, R.; Carlini, C.R.; Vainstein, M.H. Cryptococcus gattii urease as a virulence factor and the relevance of enzymatic activity in cryptococcosis pathogenesis. FEBS J. 2015, 282, 1406–1418. [Google Scholar] [CrossRef]
- Tyvold, S.S.; Solligard, E.; Lyng, O.; Steinshamn, S.L.; Gunnes, S.; Aadahl, P. Continuous monitoring of the bronchial epithelial lining fluid by microdialysis. Respir. Res. 2007, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Mathys, V.; Ang, E.L.; Koh, V.H.; Martinez Gomez, J.M.; Ang, M.L.; Zainul Rahim, S.Z.; Tan, M.P.; Pethe, K.; Alonso, S. Urease activity represents an alternative pathway for Mycobacterium tuberculosis nitrogen metabolism. Infect. Immun. 2012, 80, 2771–2779. [Google Scholar] [CrossRef]
- Maier, R.J.; Benoit, S.L. Role of nickel in microbial pathogenesis. Inorganics 2019, 7, 80. [Google Scholar] [CrossRef]
- Hensel, M.; Arst, H.N., Jr.; Aufauvre-Brown, A.; Holden, D.W. The role of the Aspergillus fumigatus are A gene in invasive pulmonary aspergillosis. Mol. Gen. Genet. MGG 1998, 258, 553–557. [Google Scholar] [CrossRef]
- Xiong, Z.; Zhang, N.; Xu, L.; Deng, Z.; Limwachiranon, J.; Guo, Y.; Han, Y.; Yang, W.; Scharf, D.H. Urease of Aspergillus fumigatus Is Required for Survival in Macrophages and Virulence. Microbiol. Spectr. 2023, 11, e0350822. [Google Scholar] [CrossRef] [PubMed]
- Coffman, J.A.; Rai, R.; Cooper, T.G. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J. Bacteriol. 1995, 177, 6910–6918. [Google Scholar] [CrossRef][Green Version]
- Stanbrough, M.; Magasanik, B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J. Bacteriol. 1995, 177, 94–102. [Google Scholar] [CrossRef]
- Xu, S.; Falvey, D.A.; Brandriss, M.C. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995, 15, 2321–2330. [Google Scholar] [CrossRef]
- Hopke, A.; Brown, A.J.; Hall, R.A.; Wheeler, R.T. Dynamic fungal cell wall architecture in stress adaptation and immune evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Dubey, L.K.; Moeller, J.B.; Schlosser, A.; Sorensen, G.L.; Holmskov, U. Induction of innate immunity by Aspergillus fumigatus cell wall polysaccharides is enhanced by the composite presentation of chitin and beta-glucan. Immunobiology 2014, 219, 179–188. [Google Scholar] [CrossRef]
- Brown, G.D.; Gordon, S. Fungal β-glucans and mammalian immunity. Immunity 2003, 19, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Felipe, M.S.S.; Andrade, R.V.; Arraes, F.c.B.; Nicola, A.M.; Maranhao, A.Q.; Torres, F.A.; Silva-Pereira, I.; Poças-Fonseca, M.J.; Campos, E.G.; Moraes, L.d.M. Transcriptional profiles of the human pathogenic fungus Paracoccidioides brasiliensis in mycelium and yeast cells. J. Biol. Chem. 2005, 280, 24706–24714. [Google Scholar] [CrossRef] [PubMed]
- Camacho, E.; Niño-Vega, G.A. Paracoccidioides spp.: Virulence factors and immune-evasion strategies. Mediat. Inflamm. 2017, 2017, 5313691. [Google Scholar] [CrossRef]
- Rappleye, C.A.; Eissenberg, L.G.; Goldman, W.E. Histoplasma capsulatum α-(1, 3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. USA 2007, 104, 1366–1370. [Google Scholar] [CrossRef] [PubMed]
- Damveld, R.A.; Franken, A.; Arentshorst, M.; Punt, P.J.; Klis, F.M.; van den Hondel, C.A.; Ram, A.F. A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 2008, 178, 873–881. [Google Scholar] [CrossRef] [PubMed]
- Schmalhorst, P.S.; Krappmann, S.; Vervecken, W.; Rohde, M.; Muller, M.; Braus, G.H.; Contreras, R.; Braun, A.; Bakker, H.; Routier, F.H. Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 2008, 7, 1268–1277. [Google Scholar] [CrossRef]
- Kizjakina, K.; Tanner, J.J.; Sobrado, P. Targeting UDP-galactopyranose mutases from eukaryotic human pathogens. Curr. Pharm. Des. 2013, 19, 2561–2573. [Google Scholar] [CrossRef]
- Crowe, J.D.; Sievwright, I.K.; Auld, G.C.; Moore, N.R.; Gow, N.A.; Booth, N.A. Candida albicans binds human plasminogen: Identification of eight plasminogen-binding proteins. Mol. Microbiol. 2003, 47, 1637–1651. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.; Nosanchuk, J.D.; Cordero, R.J.; Frases, S.; Casadevall, A.; Almeida, I.C. Characterization of yeast extracellular vesicles: Evidence for the participation of different pathways of cellular traffic in vesicle biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular transport in Histoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 2008, 10, 1695–1710. [Google Scholar] [CrossRef]
- Araújo, D.S.; de Sousa Lima, P.; Baeza, L.C.; Parente, A.F.A.; Bailão, A.M.; Borges, C.L.; Soares, C.M.d.A. Employing proteomic analysis to compare Paracoccidioides lutzii yeast and mycelium cell wall proteins. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 1304–1314. [Google Scholar] [CrossRef]
- Oliveira, A.R.d.; Oliveira, L.N.; Chaves, E.G.A.; Weber, S.S.; Bailão, A.M.; Parente-Rocha, J.A.; Baeza, L.C.; Soares, C.M.d.A.; Borges, C.L. Characterization of extracellular proteins in members of the Paracoccidioides complex. Fungal Biol. 2018, 122, 738–751. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Nakayasu, E.S.; Matsuo, A.L.; Sobreira, T.J.; Longo, L.V.; Ganiko, L.; Almeida, I.C.; Puccia, R. Vesicle and vesicle-free extracellular proteome of Paracoccidioides brasiliensis: Comparative analysis with other pathogenic fungi. J. Proteome Res. 2012, 11, 1676–1685. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Tomazett, M.V.; Baeza, L.C.; Paccez, J.D.; Parente-Rocha, J.A.; Ribeiro-Dias, F.; Soares, C.M.d.A. Identification and characterization of Paracoccidioides lutzii proteins interacting with macrophages. Microbes Infect. 2019, 21, 401–411. [Google Scholar] [CrossRef]
- Longo, L.V.; da Cunha, J.P.; Sobreira, T.J.; Puccia, R. Proteome of cell wall-extracts from pathogenic Paracoccidioides brasiliensis: Comparison among morphological phases, isolates, and reported fungal extracellular vesicle proteins. EuPA Open Proteom. 2014, 3, 216–228. [Google Scholar] [CrossRef]
- Chaves, E.G.A.; Parente-Rocha, J.A.; Baeza, L.C.; Araujo, D.S.; Borges, C.L.; de Oliveira, M.A.P.; Soares, C.M.A. Proteomic Analysis of Paracoccidioides brasiliensis During Infection of Alveolar Macrophages Primed or Not by Interferon-Gamma. Front. Microbiol. 2019, 10, 96. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.S.; Parente, A.F.A.; Borges, C.L.; Parente, J.A.; Bailão, A.M.; de Almeida Soares, C.M. Analysis of the secretomes of Paracoccidioides mycelia and yeast cells. PLoS ONE 2012, 7, e52470. [Google Scholar] [CrossRef] [PubMed]
- Chaves, E.G.A.; Weber, S.S.; Báo, S.N.; Pereira, L.A.; Bailão, A.M.; Borges, C.L.; Soares, C.M.d.A. Analysis of Paracoccidioides secreted proteins reveals fructose 1, 6-bisphosphate aldolase as a plasminogen-binding protein. BMC Microbiol. 2015, 15, 53. [Google Scholar] [CrossRef]
- Arvizu-Rubio, V.J.; García-Carnero, L.C.; Mora-Montes, H.M. Moonlighting proteins in medically relevant fungi. PeerJ 2022, 10, e14001. [Google Scholar] [CrossRef]
- Rødkær, S.V.; Færgeman, N.J. Glucose-and nitrogen sensing and regulatory mechanisms in Saccharomyces cerevisiae. FEMS Yeast Res. 2014, 14, 683–696. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz-Leite, V.R.M.; Moreira, A.L.E.; Silva, L.O.S.; Inácio, M.M.; Parente-Rocha, J.A.; Ruiz, O.H.; Weber, S.S.; Soares, C.M.d.A.; Borges, C.L. Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression. J. Fungi 2023, 9, 1102. https://doi.org/10.3390/jof9111102
Cruz-Leite VRM, Moreira ALE, Silva LOS, Inácio MM, Parente-Rocha JA, Ruiz OH, Weber SS, Soares CMdA, Borges CL. Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression. Journal of Fungi. 2023; 9(11):1102. https://doi.org/10.3390/jof9111102
Chicago/Turabian StyleCruz-Leite, Vanessa Rafaela Milhomem, André Luís Elias Moreira, Lana O’Hara Souza Silva, Moises Morais Inácio, Juliana Alves Parente-Rocha, Orville Hernandez Ruiz, Simone Schneider Weber, Célia Maria de Almeida Soares, and Clayton Luiz Borges. 2023. "Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression" Journal of Fungi 9, no. 11: 1102. https://doi.org/10.3390/jof9111102
APA StyleCruz-Leite, V. R. M., Moreira, A. L. E., Silva, L. O. S., Inácio, M. M., Parente-Rocha, J. A., Ruiz, O. H., Weber, S. S., Soares, C. M. d. A., & Borges, C. L. (2023). Proteomics of Paracoccidioides lutzii: Overview of Changes Triggered by Nitrogen Catabolite Repression. Journal of Fungi, 9(11), 1102. https://doi.org/10.3390/jof9111102





