Coronatine-Induced Maize Defense against Gibberella Stalk Rot by Activating Antioxidants and Phytohormone Signaling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Disease Phenotype Investigation and Maize Growth
2.3. Assay of Activities of Antioxidant Enzymes, Defense Enzymes, and MDA Content
2.4. Measurement of Stomatal Density and Aperture
2.5. Assay of H2O2 Accumulation
2.6. Quantification of Phytohormones
2.7. RNA-Seq Transcriptome Analysis and qRT-PCR Validation
2.8. WGCNA for Identification of Key Candidate Genes Involved in Maize Defense Associated with COR
2.9. Detection and Analysis of Metabolites
3. Results
3.1. Disease Phenotype Investigation and Maize Growth
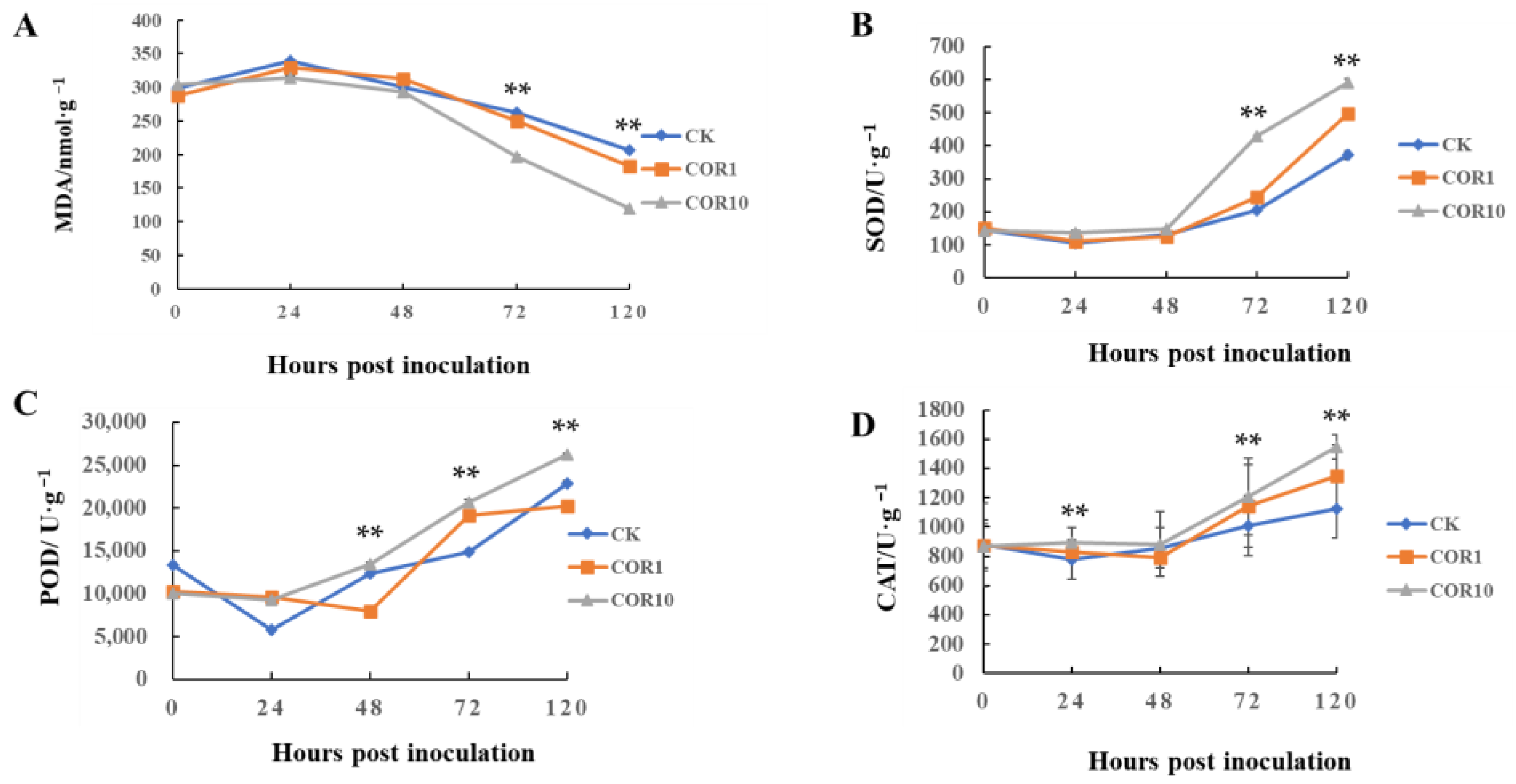
3.2. Effects of COR on MDA Accumulation, Activities of Antioxidants and Defense-Related Enzymes
3.3. COR Regulates Stomatal Closure
3.4. Effects of COR on H2O2 Accumulation
3.5. COR Enhanced the Jasmonate (JA-Ile and Me-JA) Concentration after Fusarium graminearum Infection
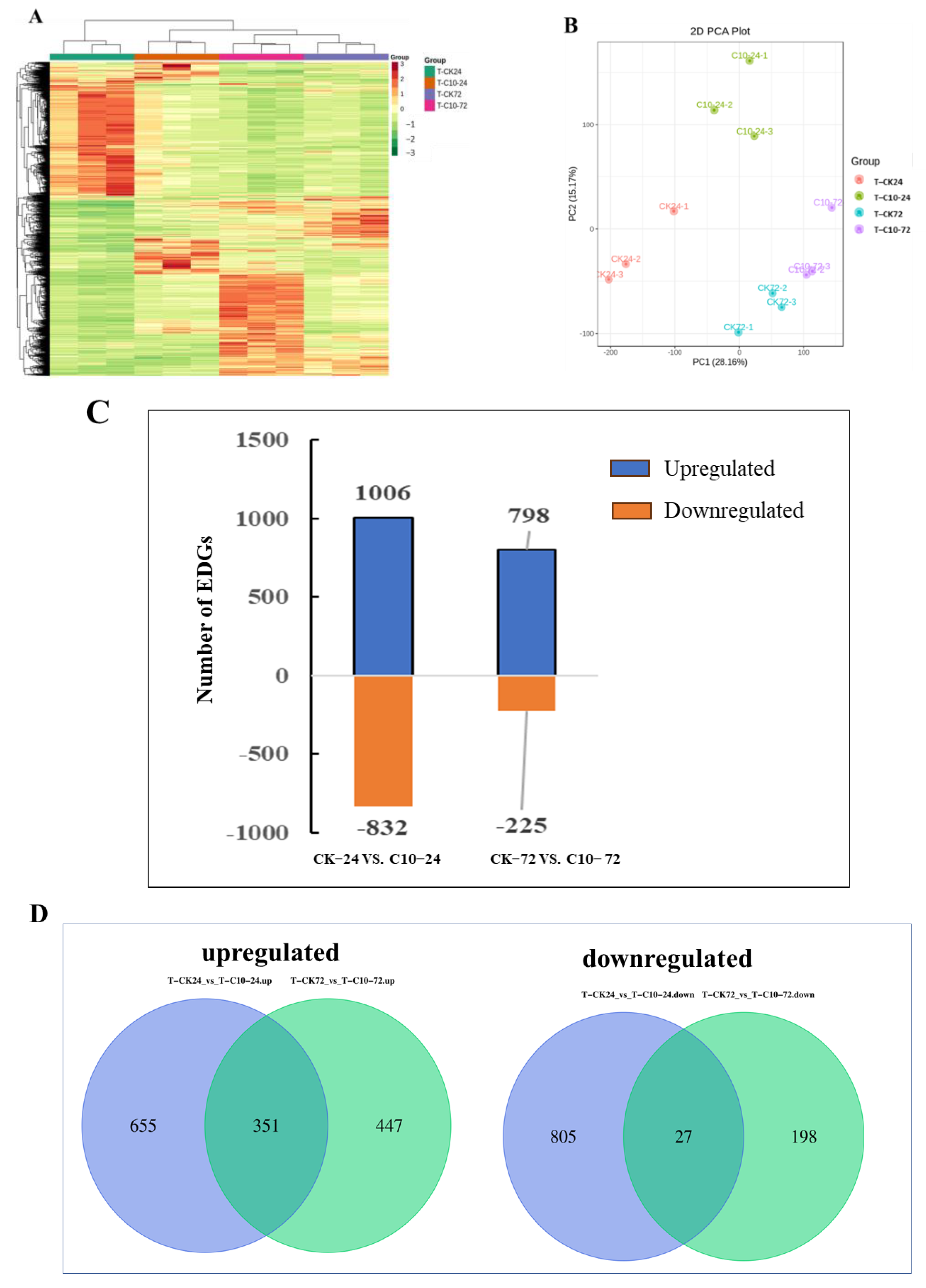
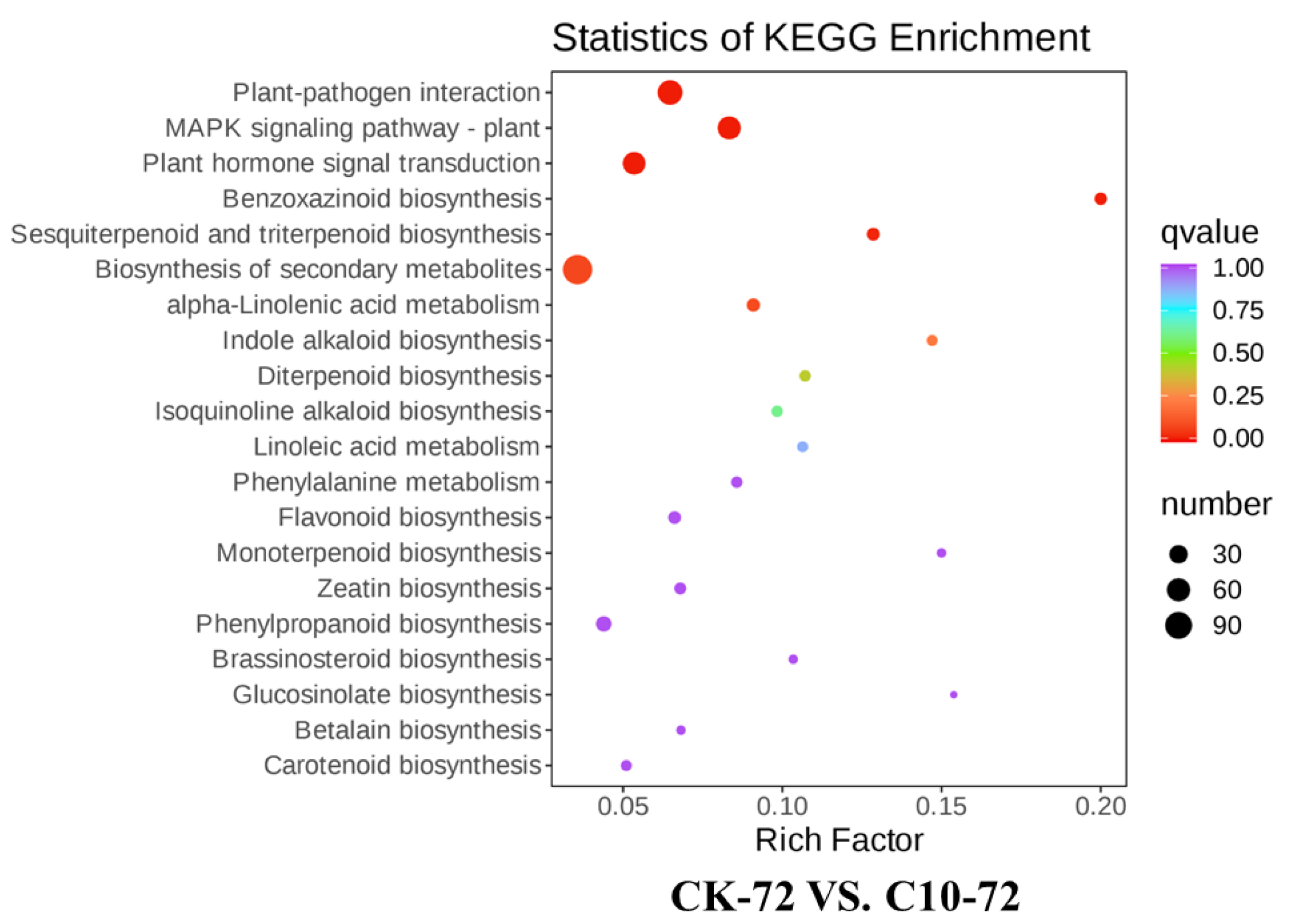
3.6. Transcriptome Responses Triggered by COR and Validation by qRT-PCR
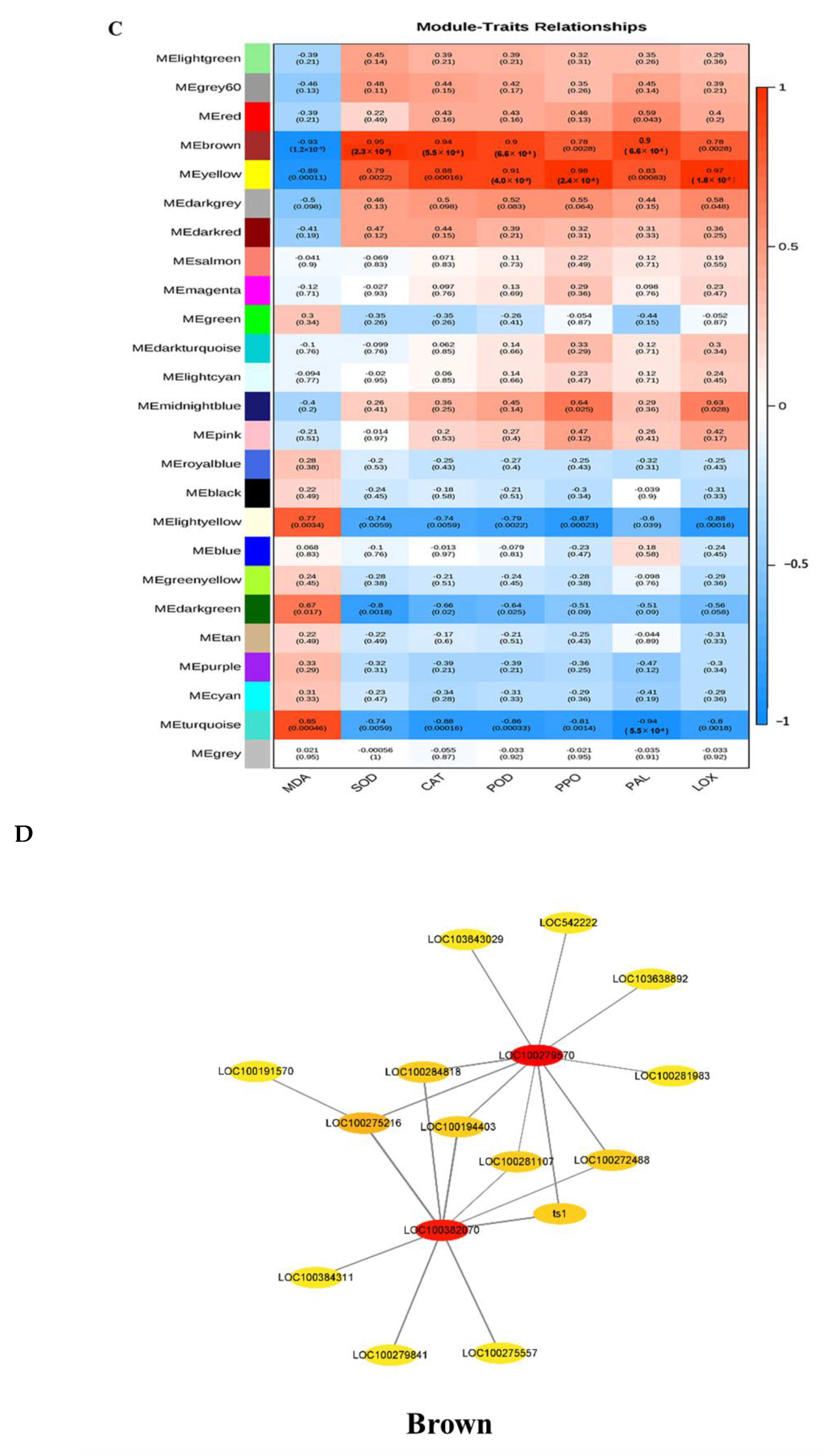
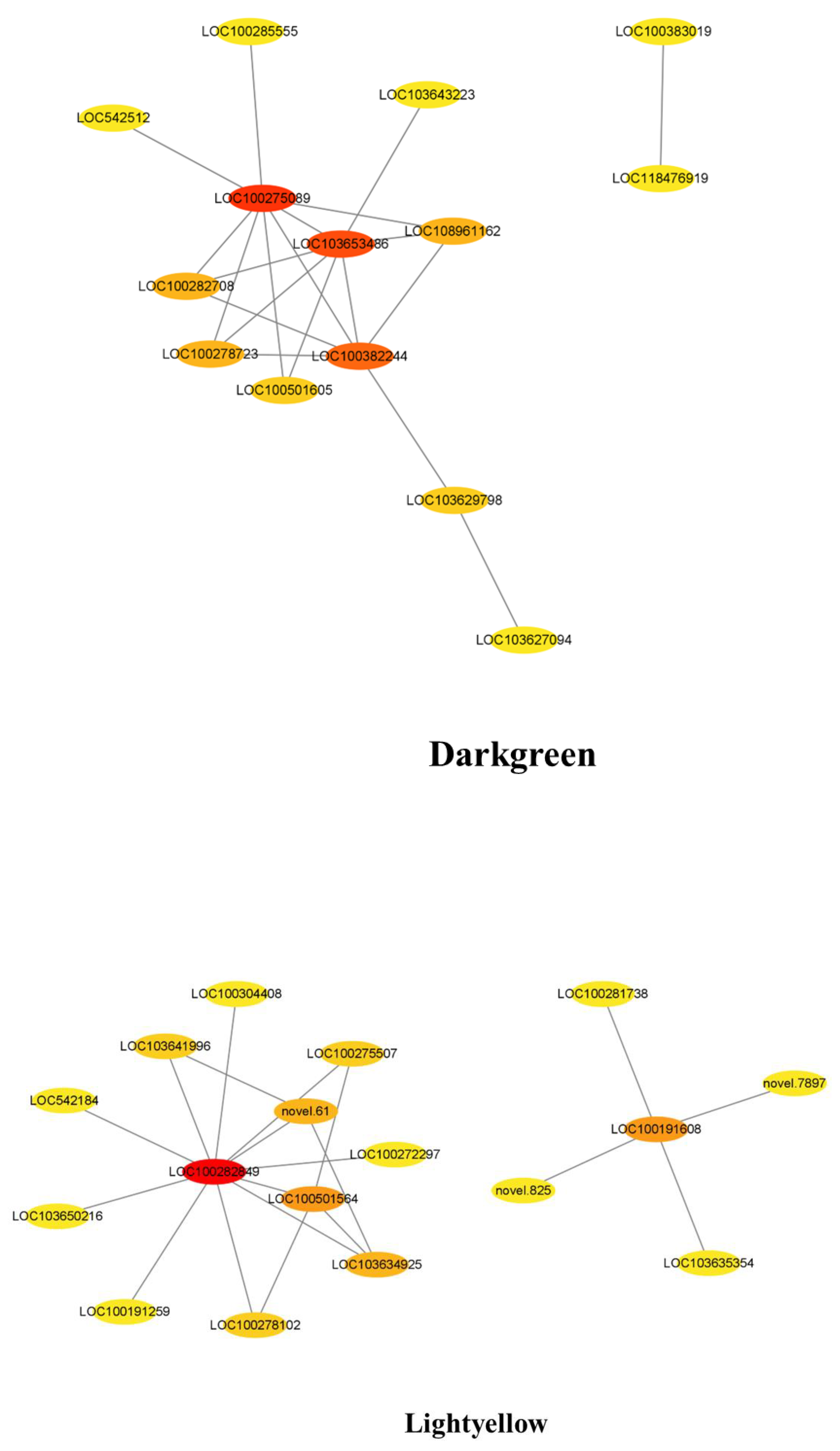
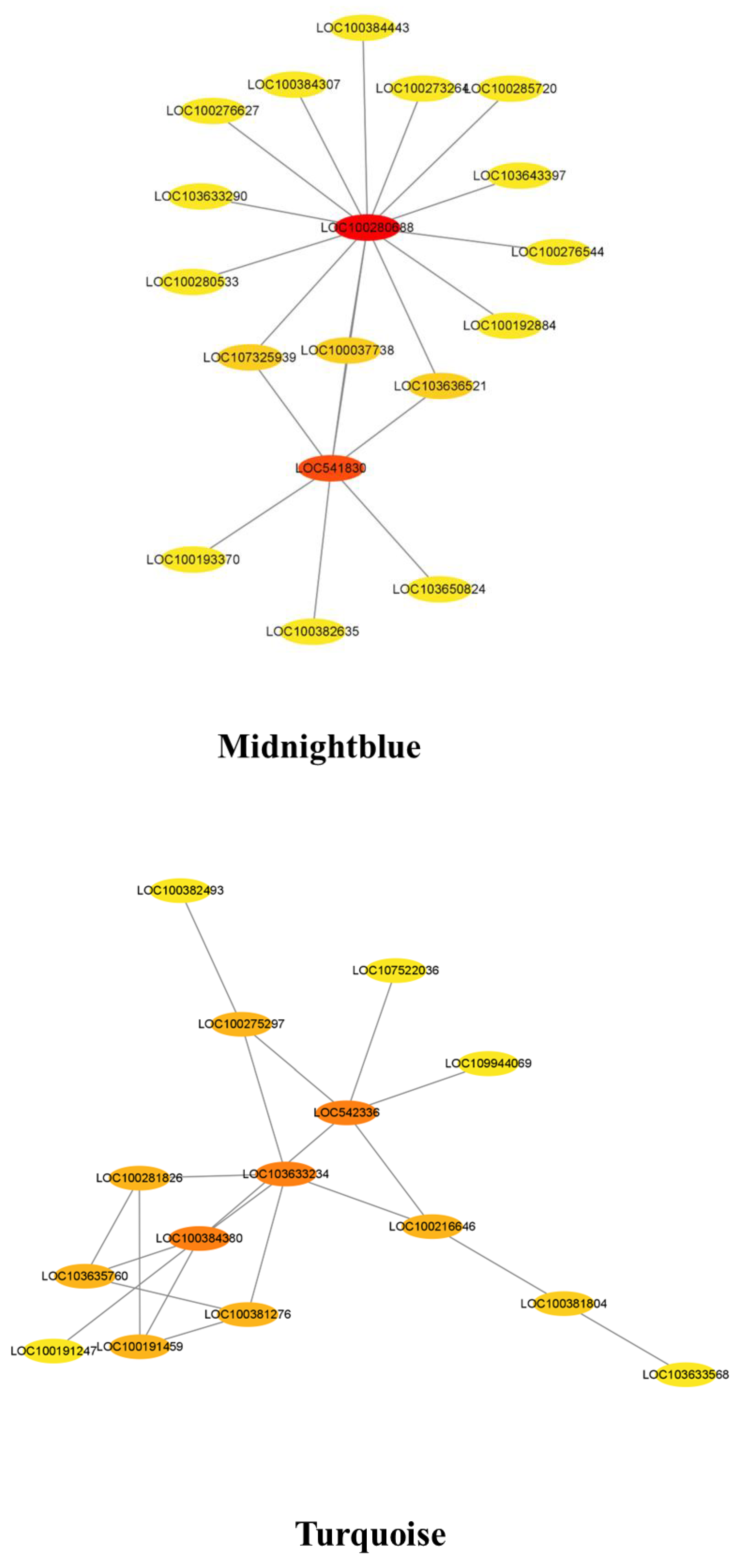
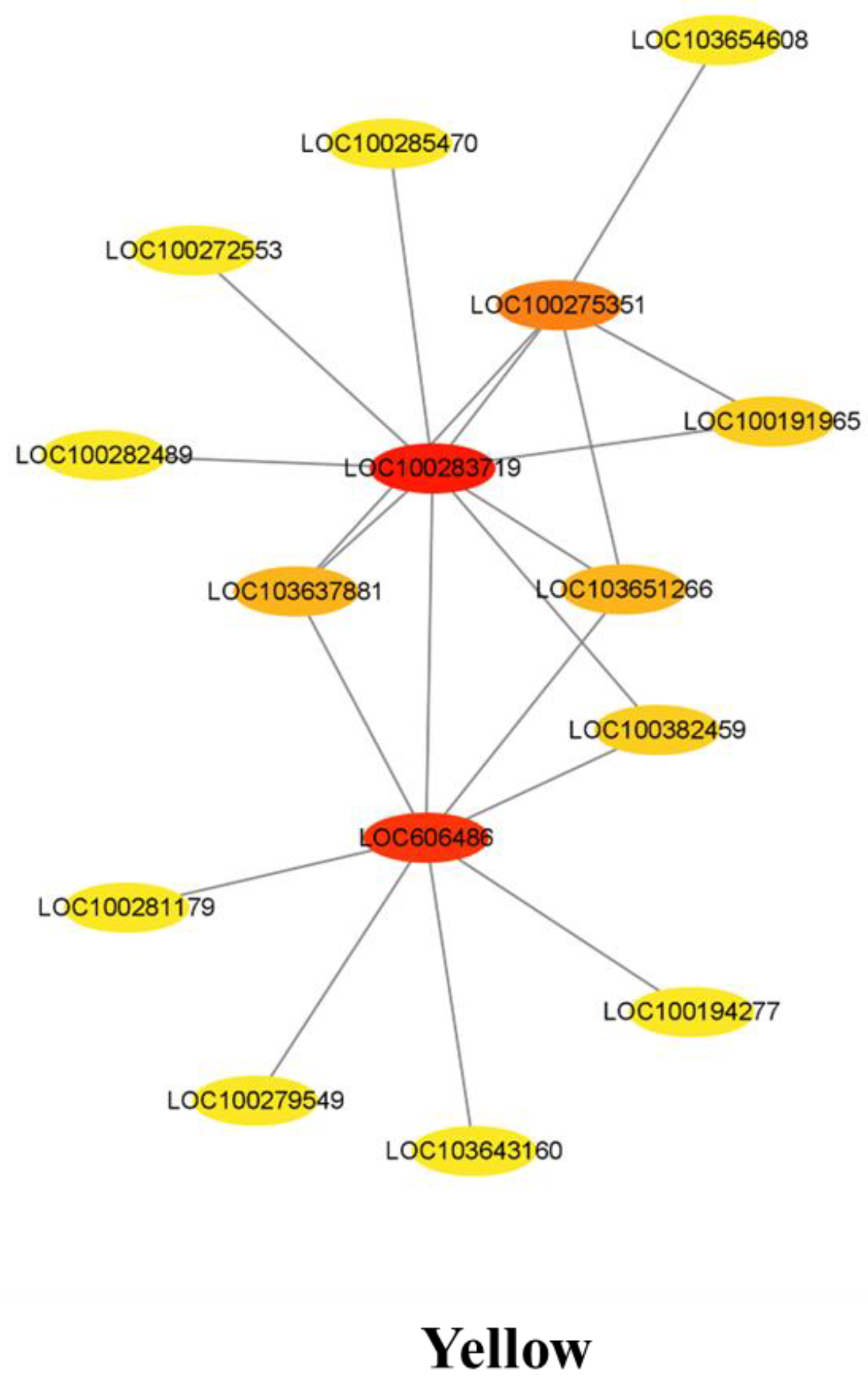
3.7. Co-Expression Network Analysis for the Candidate Hub Genes Involved in Maize Defense
3.8. COR Enhanced the Expression of Key Candidate Genes in Maize Defense Response to F. graminearum
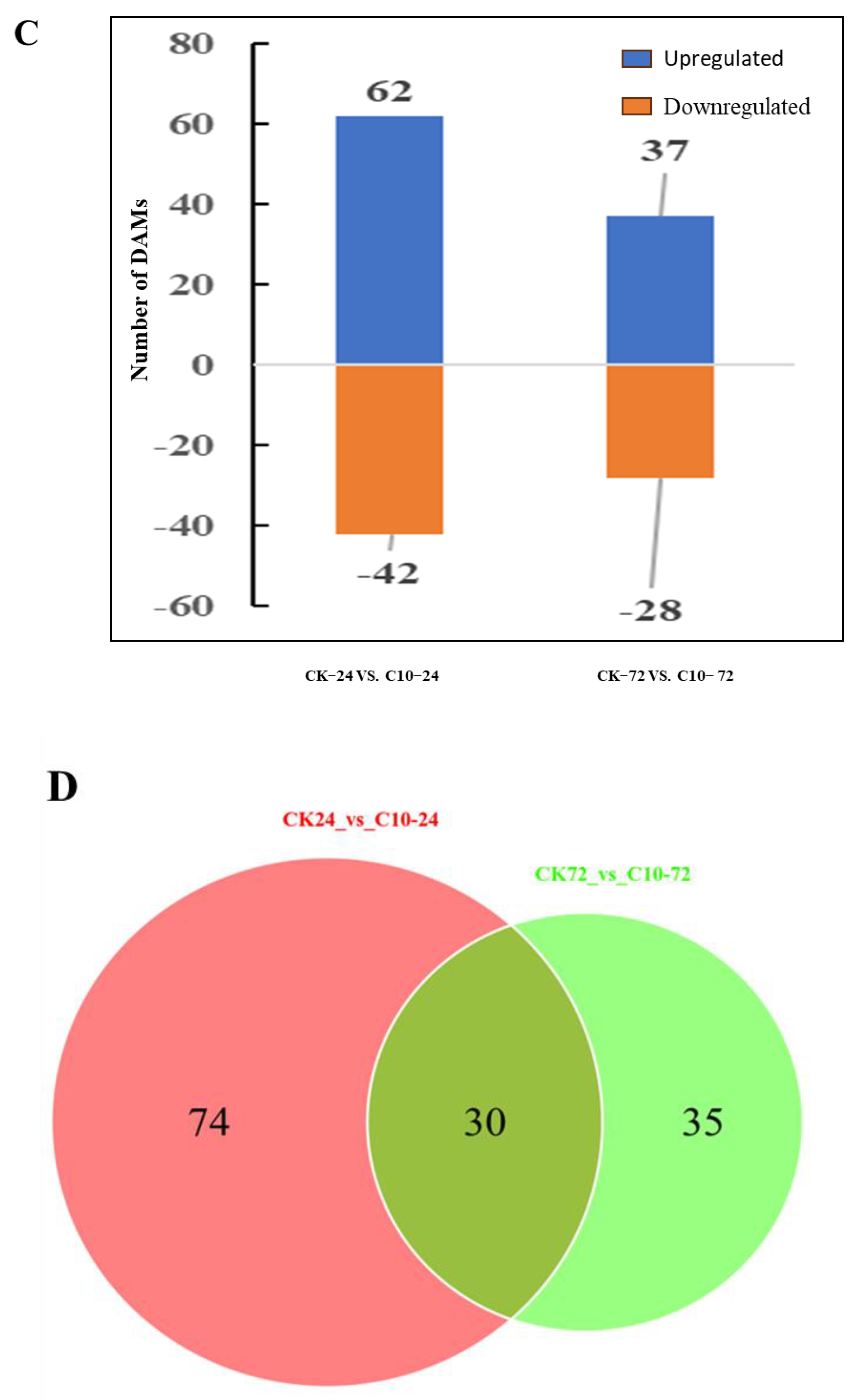
3.9. Metabolite Analysis of Maize in Response to COR
4. Discussion
4.1. Effects of COR on Hub Genes in the F. graminearum Associated Molecular Signaling Responses
4.2. Effects of COR on the Activity of Identified Candidate Genes in Phytohormone Signaling and Biosynthesis Pathways
4.3. COR Enhances Maize Resistance to GSR by Alpha-Linolenic Acid Metabolism and Flavonoid Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.; Beddington, I.R.; Crute, I.; Haddad, D.; Lawrence, D.; Muir, J.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- FAO Statistical Databases. Available online: http://faostat.fao.org/site/377/default.aspx#anco (accessed on 1 October 2021).
- Batool, R.; Umer, M.J.; Wang, Y.; He, K.; Zhang, T.; Bai, S.; Zhi, Y.; Chen, J.; Wang, Z. Synergistic effect of Beauveria bassiana and Trichoderma asperellum to induce maize (Zea mays L.) defense against the Asian corn borer, Ostrinia furnacalis (Lepidoptera, Crambidae) and larval immune response. Int. J. Mol. Sci. 2020, 21, 8215. [Google Scholar] [CrossRef]
- De Leon, C.; Jeffers, D. Maize Diseases: A Guide for Field Identification; Mexico FAO Statistical Databases; CIMMYT: México-Veracruz, Mexico, 2004. [Google Scholar]
- Gatch, E.W.; Hellmich, R.L.; Munkvold, G.P. A comparison of maize stalk rot occurrence in Bt and non-Bt hybrids. Plant Dis. 2002, 86, 1149–1155. [Google Scholar] [CrossRef]
- Ledencan, T.; Simic, D.; Brkic, I.; Jambrovic, A.; Zdunic, Z. Resistance of maize inbreds and, their hybrids to Fusarium stalk rot. Czech J. Genet. Plant Breed 2003, 39, 15–20. [Google Scholar] [CrossRef]
- Stephens, A.E.; Gardiner, D.M.; White, R.G.; Munn, A.L.; Manners, J.M. Phases of infection and gene expression of Fusarium graminearum during crown rot disease of wheat during crown rot disease of wheat. Mol. Plant-Microbe Interact. 2008, 21, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Kazan, K.; Gardiner, D.M.; Manners, J.M. On the trail of a cereal killer: Recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, L.; Zhang, Y.; Jiang, G.; Li, X.; Zhang, D.; Tang, W. In planta stage-specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell 2012, 13, 399–413. [Google Scholar] [CrossRef]
- Pieterse, C.J.; Leon-Reyes, A.; Ent, S.; Wees, S.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Chetouhi, C.; Bonhomme, L.; Lasserre-Zuber, P.; Cambon, F.; Pelletier, S.; Renou, J.P.; Langin, T. Transcriptome dynamics of a susceptible wheat upon Fusarium head blight reveals that molecular responses to Fusarium graminearum infection fit over the grain development processes. Funct. Integr. Genomics. 2016, 16, 183–201. [Google Scholar] [CrossRef]
- Ma, C.; Ma, X.; Yao, L.; Liu, Y.; Du, F.; Yang, X.; XU, M. qRfg3, a novel quantitative resistance locus against Gibberella stalk rot in maize. Theor. Appl. Genet. 2017, 130, 1723–1734. [Google Scholar] [CrossRef] [PubMed]
- Onrubia, M.; Moyano, E.; Bonfill, M.; Cusidó, R.M.; Goossens, A.; Palazón, J. Coronatine, a more powerful elicitor for inducing taxane biosynthesis in Taxus media cell cultures than methyl jasmonate. J. Plant Physiol. 2013, 170, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Svoboda, J.; Boland, W. Plant defense elicitors: Analogues of jasmonoyl–isoleucine conjugate. Phytochemistry 2010, 71, 1445–1449. [Google Scholar] [CrossRef]
- Chini, A.; Boter, M.; Solano, R. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS J. 2009, 276, 4682–4692. [Google Scholar] [CrossRef] [PubMed]
- Christensen, S.A.; Nemchenko, A.; Park, Y.S.; Borrego, E.; Huang, P.C.; Schmelz, E.A.; Kunze, S.; Feussner, I.; Yalpani, N.; Meeley, R.; et al. The novel monocot-specific 9-Lipoxygenase ZmLOX12 is required to mount an effective Jasmonate-Mediatedefense against Fusarium verticillioides in Maize. Mol. Plant-Microbe Interact. 2014, 27, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Gorman, Z.; Christensen, S.A.; Yan, Y.; He, Y.; Borrego, E.; Kolomiets, M.V. Green leaf volatiles and jasmonic acid enhance susceptibility to anthracnose diseases caused by Colletotrichum graminicola in maize. Mol. Plant Pathol. 2020, 21, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; Xiao, J.; Jia, X.P.; Ke, P.B.; He, L.Q.; Cao, A.Z.; Wang, H.Y.; Wu, Y.F.; Gao, X.Q.; Wang, X. The role of wheat jasmonic acid and ethylene pathways in response to Fusarium graminearum infection. Plant Grow. Regul. 2016, 80, 69–77. [Google Scholar] [CrossRef]
- Wang, K.D.; Gorman, Z.; Huang, P.C.; Kenerley, C.M.; Kolomiets, M.V. Trichoderma virens colonization of maize roots triggers rapid accumulation of 12-oxophytodienoate and twoγ—Ketols in leaves as priming agents of induced systemic resistance. Plant Signal. Behav. 2020, 15, 1792187. [Google Scholar] [CrossRef]
- Ma, L.; Sun, Y.; Ruan, X.; Huang, P.; Wang, S.; Li, S.; Zhou, Y.; Wang, F.; Cao, Y.; Wang, Q.; et al. Genome-Wide characterization of Jasmonates signaling components reveals the essential role of ZmCOI1a-ZmJAZ15 action module in regulating maize immunity to Gibberella Stalk Rot. Int. J. Mol. Sci. 2021, 22, 870. [Google Scholar] [CrossRef]
- Liu, L.; Zhai, Z.; Duan, L. Study on resistance of cabbage seedlings to black rot induced by coronatine treatment and its biochemical and physiological mechanism. Chin. J. Pestic. Sci. 2012, 14, 30–34. [Google Scholar]
- Wang, N.; Wu, F. Role of coronatine in inducing infection and improving resistance to Pseudomonas syringae pv. mori in mulberry seedlings. Agri. Biotech. 2017, 6, 23–30. [Google Scholar]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M.; et al. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl–isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ruan, X.; Ma, L.; Wang, F.; Gao, X. Rapid screening and evaluation of maize seedling resistance to stalk rot caused by Fusarium spp. Bio-Protocol 2018, 8, e2859. [Google Scholar] [CrossRef]
- Shao, J.; Song, Y.; Zhou, Y.; Wan, Z.; Li, R.; Yu, J. Diagnostic value of fluorescein-labeled chitinase staining in formalin-fixed and paraffin-embedded tissues of fungal disease. Med. Mycol. 2019, 58, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Kanagaraj, A.; Verma, D.; Lange, T.; Daniell, H. Release of hormones from conjugates: Chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol. 2011, 155, 222–235. [Google Scholar] [CrossRef]
- Zhang, W.; Corwin, J.A.; Copeland, D.; Feusier, J.; Eshbaugh, R.; Chen, F.; Atwell, S.; Kliebenstein, D. Plastic transcriptomes stabilize immunity to pathogen diversity: The Jasmonic acid and salicylic acid networks within the Arabidopsis/Botrytis pathosystem. Plant Cell 2017, 29, 2727–2752. [Google Scholar] [CrossRef]
- Chen, K.; Du, K.; Shi, Y.; Yin, L.; Shen, W.H.; Yu, Y.; Liu, B.; Dong, A. H3K36 methyltransferase SDG708 enhances drought tolerance by promoting abscisic acid biosynthesis in rice. New Phytol. 2021, 230, 1967–1984. [Google Scholar] [CrossRef]
- Hu, Y.; Wu, Q.; Peng, Z.; Sprague, S.A.; Wang, W.; Park, J.; Akhunov, E.; Jagadish, K.S.V.; Nakata, P.A.; Cheng, N.; et al. Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci. Rep. 2017, 7, 15950. [Google Scholar] [CrossRef]
- Šimura, J.; Antoniadi, I.; Široká, J.; Tarkowská, D.; Strnad, M.; Ljung, K.; Novák, O. Plant hormonomics: Multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 2018, 177, 476–489. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, 480–484. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Batool, R.; Umer, M.J.; Wang, Y.; He, K.; Shabbir, M.Z.; Zhang, T.; Bai, S.; Chen, J.; Wang, Z. Myco-Synergism boosts herbivory-induced maize defense by triggering antioxidants and phytohormone signaling. Front. Plant Sci. 2022, 13, 790504. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Yang, X.; Tzin, V.; Peng, Y.; Romeis, J.; Li, Y. Plant breeding involving genetic engineering does not result in unacceptable unintended effects in rice relative to conventional cross-breeding. Plant J. 2020, 103, 2236–2249. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yin, G.; Guo, Y.; Zhang, D.; Chen, S.; Xu, M. A major QTL for resistance to Gibberella stalk rot in maize. Theor. Appl. Genet. 2010, 121, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, Q.; Wang, W.; Li, Y.; Guo, Y.; Zhang, D.; Ma, X.; Song, W.; Zhao, J.; Xu, M. A transposon-directed epigenetic change in ZmCCT underlies quantitative resistance to Gibberella stalk rot in maize. New Phytol. 2017, 215, 1503–1515. [Google Scholar] [CrossRef]
- Thomma, B.P.; Nurnberger, T.; Joosten, M.H. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Si, H.; Zang, J.; Pang, X.; Yu, L.; Cao, H.; Xing, J.; Zhang, K.; Dong, J. Comparative proteomic analysis of the defense response to Gibberella stalk rot in maize and reveals that ZmWRKY83 is involved in plant disease resistance. Front. Plant Sci. 2021, 12, 694973. [Google Scholar] [CrossRef]
- Sun, Y.; Ruan, X.; Wang, Q.; Zhou, Y.; Wang, F.; Ma, L.; Wang, Z.; Gao, X. Integrated Gene Co-expression Analysis and metabolites profiling highlight the important role of ZmHIR3 in maize resistance to Gibberella stalk rot. Front. Plant Sci. 2021, 12, 664733. [Google Scholar] [CrossRef]
- Henkes, G.J.; Thorpe, M.R.; Minchin, P.E.H.; Schurr, U.; Rose, U.S.R. Jasmonic acid treatment to part of the root system is consistent with simulated leaf herbivory, diverting recently assimilated carbon towards untreated roots within an hour. Plant Cell Environ. 2008, 31, 1229–1236. [Google Scholar] [CrossRef]
- Gómez, K.; Quenguan, F.; Aristizabal, D.; Escobar, G.; Quiñones, W.; García-Beltrán, O.; Durango, D. Elicitation of isoflavonoids in Colombian edible legume plants with jasmonates and structurally related compounds. Heliyon 2022, 8, e08979. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Duan, L.; Tian, X.; Wang, B.; Eneji, A.E.; Li, Z. Coronatine alleviates salinity stress in cotton by improving the antioxidative defense system and radical-scavenging activity. Plant Physiol. 2008, 165, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Song, C.; Wang, B.; Zhou, J.; Kangasjarvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Yuan, H.; Zeng, X.; Yang, Q.; Xu, Q.; Wang, Y.; DunZhu, J.; Zha, S.; Tashi, N. Gene coexpression network analysis combined with metabonomics reveals the resistance responses to powdery mildew in Tibetan hulless barley. Sci. Rep. 2018, 8, 14928. [Google Scholar] [CrossRef] [PubMed]
- Vanderbeld, B.; Snedden, W.A. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 2007, 64, 683–697. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, K.; Hotta, C.; Gardner, M.; Braam, J.; Webb, A. The Arabidopsis thaliana calmodulin-like protein CML24 is a regulator of rhythmic Ca2+ signaling and flowering time. Comp. Biochem. Physiol. 2008, 150, 148–154. [Google Scholar] [CrossRef]
- Magnan, F.; Ranty, B.; Charpenteau, M.; Sotta, B.; Galaud, J.P.; Aldon, D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008, 56, 575–589. [Google Scholar] [CrossRef]
- Jyothilakshmi, V.; Michael, R.; Bettina, H.; Jonathan, G.; Wilhelm, B.; Axel, M. CML42-Mediated Calcium signaling coordinates responses to spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 2012, 159, 1159–1175. [Google Scholar]
- Shang, Y.; Yan, L.; Liu, Z.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.; Wang, X.; Du, S.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef]
- Zhao, W.; Liu, Y.; Zhou, J.; Zhao, S.; Zhang, X.; Min, D. Genome-wide analysis of the lectin receptor-like kinase family in foxtail millet (Setaria italica L.). Plant Cell Tiss. 2016, 127, 335–346. [Google Scholar] [CrossRef]
- Wang, C.; Ru, J.; Liu, Y.; Yang, J.; Li, M.; Xu, Z.; Fu, J. The maize WRKY transcription factor ZmWRKY40 confers drought resistance in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2580. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zhang, Y.; Duan, R.; Fan, J.; Jiao, P.; Sun, H.; Guan, S.; Liu, S. Overexpression of maize Glutathione S-Transferase ZmGST26 decreases drought resistance of Arabidopsis. Agronomy 2022, 12, 2948. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Yu, J.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef] [PubMed]
- Hafren, A.; Macia, A.J.; Love, J.J.; Milner, M.; Drucker, M.; Hofius, D. Selective autophagy limits cauliflower mosaic virus infection by NBR1- mediated targeting of viral capsid protein and particles. Proc. Natl. Acad. Sci. USA 2017, 114, 2026–2035. [Google Scholar] [CrossRef] [PubMed]
- Leong, J.X.; Raffeiner, M.; Spinti, D.; Langin, G.; Franz-Wachtel, M.; Guzman, A.R.; Kim, J.G.; Pandey, P.; Minina, A.E.; Macek, B.; et al. A bacterial effector counteracts host autophagy by promoting degradation of an autophagy component. EMBO J. 2022, 41, e110352. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Lukomska, J.; Moniuszko, G.; Gwozdecki, R.; Surowiecki, P.; Lewandowska, M.; Liszewska, F.; Wawrzyńska, A.; Sirko, A. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 2011, 7, 1145–1158. [Google Scholar] [CrossRef]
- Wang, T.; Li, Y.; Huang, Y.; Zhao, X.; Dong, Z.; Jin, W.; Huang, W. Amino acid permease 6 regulates grain protein content in maize. Crop J. 2022, 10, 1536–1544. [Google Scholar] [CrossRef]
- Markakis, M.N.; Cnodder, D.T.; Lewandowski, M.; Simon, D.; Boron, A.; Balcerowicz, D.; Doubbo, T.; Taconnat, L.; Renou, J.P.; Höfte, H.; et al. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biol. 2012, 12, 208. [Google Scholar] [CrossRef]
- Chen, G.; Wang, J.; Wang, H.; Wang, C.; Tang, X.; Li, J.; Zhang, L.; Song, J.; Hou, J.; Yuan, L. Genome-wide analysis of proline-rich extension-like receptor protein kinase (PERK) in Brassica rapa and its association with the pollen development. BMC Genom. 2020, 21, 401. [Google Scholar] [CrossRef]
- Bai, L.; Zhou, Y.; Song, C. Arabidopsis proline-rich extension-like receptor kinase 4 modulates the early event toward abscisic acid response in root tip growth. Plant Signal. 2009, 4, 107294. [Google Scholar]
- Wei, K.; Chen, J.; Wang, Y.; Chen, Y.; Chen, S.; Lin, Y.; Pan, S.; Zhong, X.; Xie, D. Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 2012, 19, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Creelman, R.A.; Mullet, J.E. Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Feussner, I. The Oxylipin Pathways: Biochemistry and Function. Annu. Rev. Plant Biol. 2018, 69, 363. [Google Scholar] [CrossRef] [PubMed]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Chen, M.; Lam, P.; Dini-Andreote, F.; Dai, L.; Wei, Z. Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 2022, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Siciliano, I.; Amaral, C.; Spadaro, D.; Garibaldi, A.; Gullino, M. Jasmonic acid, abscisic acid, and salicylic acid are involved in the phytoalexin responses of rice to Fusarium fujikuroi, a high gibberellin producer pathogen. J. Agric. Food Chem. 2015, 63, 8134–8142. [Google Scholar] [CrossRef]
- Hasegawa, M.; Mitsuhara, I.; Seo, S.; Okada, K.; Yamane, H.; Iwai, T.; Ohashi, Y. Analysis on blast fungus-responsive characters of a flavonoid phytoalexin sakuranetin; accumulation in infected rice leaves, antifungal activity and detoxification by fungus. Molecules 2014, 19, 11404–11418. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; et al. Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 2021, 19, 671–688. [Google Scholar] [CrossRef]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Zhang, F.; Zhang, G.; Jiang, X.; Yu, H.; Hou, B. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.J.; Li, P.; Wang, T.; Zhang, F.; Huang, X.; Hou, B.K. The maize secondary metabolism glycosyltransferase UFGT2 modifies flavonols and contributes to plant acclimation to abiotic stresses. Ann. Bot. 2018, 122, 1203–1217. [Google Scholar] [CrossRef] [PubMed]








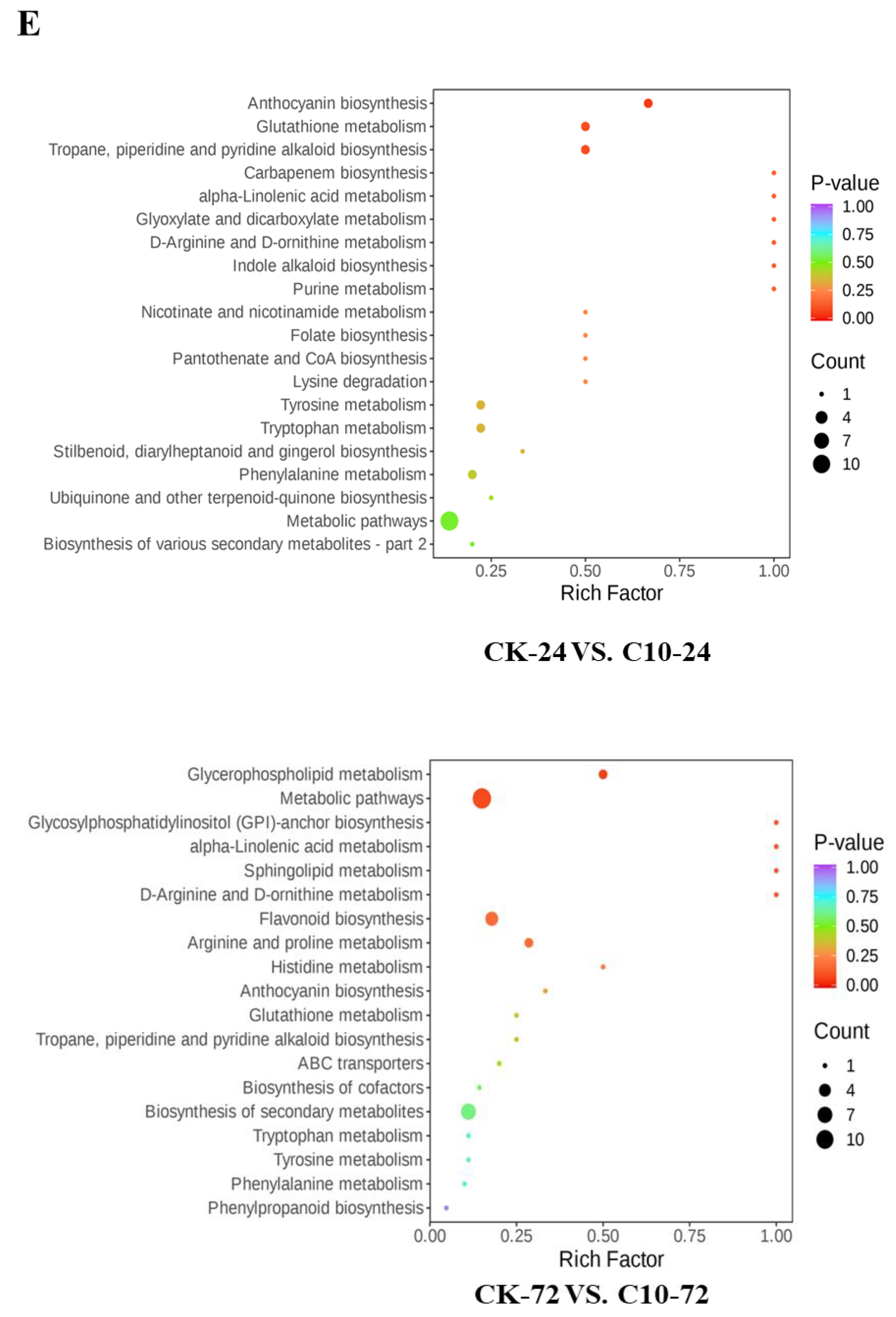
| Treatments | Plant Height (cm) | Stem Diameter (cm) | Leaf Length (cm) | Leaf Width (cm) | SPAD | Fresh Weight (g) | Dry Weight (g) |
|---|---|---|---|---|---|---|---|
| CK | 59.80 ± 6.42 a | 11.02 ± 1.20 a | 51.20 ± 2.29 b | 4.24 ± 0.16 a | 23.26 ± 1.11 a | 29.21 ± 3.05 a | 2.72 ± 0.36 a |
| COR1 | 56.90 ± 6.11 a | 9.40 ± 1.25 b | 60.90 ± 3.96 a | 3.60 ± 0.37 b | 22.60 ± 2.70 a | 27.30 ± 4.54 a | 2.10 ± 0.37 ab |
| COR10 | 50.13 ± 6.21 b | 9.45 ± 1.12 b | 45.93 ± 2.72 c | 3.91 ± 0.55 ab | 21.43 ± 2.45 a | 27.56 ± 3.35 a | 2.19 ± 0.36 ab |
| COR50 | 43.82 ± 3.39 b | 9.39 ± 1.28 b | 45.06 ± 2.64 c | 3.57 ± 0.46 b | 20.57 ± 3.17 a | 20.48 ± 4.13 b | 1.92 ± 0.57 b |
| Treatments | Stomatal Length (um) | Stomatal Width (um) | Aperture Length (um) | Aperture Width (um) | Stomatal Density (Number/mm2) | Stomatal Area (um2) | Aperture Area (um2) |
|---|---|---|---|---|---|---|---|
| CK | 51.59 ± 5.46 a | 25.92 ± 0.95 a | 51.13 ± 3.09 a | 8.51 ± 0.92 a | 67.20 ± 5.19 a | 1075.31 ± 51.36 a | 326.54 ± 17.73 a |
| COR10 | 43.58 ± 1.67 b | 21.05 ± 3.60 b | 34.74 ± 1.7 b | 7.79 ± 0.66 b | 65.28 ± 3.94 a | 904.16 ± 34.45 b | 217.93 ± 17.36 b |
| Module | Gene ID | Annotation | Identified as Key Candidate Gene |
|---|---|---|---|
| Black | LOC103641718 | Uncharacterized | × |
| Blue | LOC100382049 | K(+) efflux antiporter 3 chloroplastic | × |
| Brown | LOC100279570 | WRKY transcription factor 40 | √ |
| Brown | LOC100382070 | Calcium-binding protein CML42 Calcium-binding protein | √ |
| Cyan | LOC100127513 | ZCN2 protein | × |
| Dark green | LOC100275089 | NBR1-like protein | √ |
| Dark green | LOC100382244 | Amino acid permease 6 | √ |
| Dark grey | LOC100274778 | Uncharacterized | × |
| Dark red | LOC100127520 | MFT2-Corn MFT-like protein | × |
| Dark turquoise | LOC118473652 | Photosystem I P700 chlorophyll a apoprotein A2 | × |
| Green | LOC103625941 | Receptor-like kinase TMK2 | × |
| Green yellow | LOC100272716 | Tryptophan aminotransferase | × |
| Grey60 | LOC100273423 | Phospholipase A1-IIgamma | × |
| Light cyan | LOC109945353 | Uncharacterized | × |
| Light green | BA1 | bHLH transcription factor | × |
| Light yellow | LOC100282849 | Alanine aminotransferase 2 | × |
| Light yellow | LOC100191608 | HXXXD-type acyl-transferase family protein | √ |
| Light yellow | LOC100501564 | Prolin-rich extensin-like receptor protein kinase | √ |
| Magenta | novel.8180 | Uncharacterized | × |
| Midnight blue | LOC100280688 | D-3-phosphoglycerate dehydrogenase 2 chloroplastic | × |
| Midnight blue | LOC541830 | Glutathione S-transferase | √ |
| Pink | LOC100273965 | Major facilitator superfamily defense 1 | × |
| Purple | LOC100127521 | ZCN10 protein | × |
| Red | LOC100279184 | TOM1-like protein 9 | × |
| Royal blue | LOC100136884 | PTO-like protein kinase | × |
| Salmon | LOC100193868 | Uncharacterized | × |
| Tan | LOC100193845 | Pollen proteins Ole e I like | × |
| Turquoise | LOC100384380 | AP2-like ethylene-responsive transcription factor | √ |
| Turquoise | LOC542336 | Cyclin 2 | × |
| Turquoise | LOC103633234 | Abnormal spindle-like microcephaly-associated protein homolog | × |
| Yellow | LOC100283719 | Uncharacterized | × |
| Yellow | LOC606486 | Glycosyltransferase | √ |
| Yellow | LOC100275351 | Basic leucine zipper 25 | √ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Sui, Y.; Yu, C.; Wang, X.; Zhang, W.; Wang, B.; Yan, J.; Duan, L. Coronatine-Induced Maize Defense against Gibberella Stalk Rot by Activating Antioxidants and Phytohormone Signaling. J. Fungi 2023, 9, 1155. https://doi.org/10.3390/jof9121155
Liu M, Sui Y, Yu C, Wang X, Zhang W, Wang B, Yan J, Duan L. Coronatine-Induced Maize Defense against Gibberella Stalk Rot by Activating Antioxidants and Phytohormone Signaling. Journal of Fungi. 2023; 9(12):1155. https://doi.org/10.3390/jof9121155
Chicago/Turabian StyleLiu, Mei, Yiping Sui, Chunxin Yu, Xuncheng Wang, Wei Zhang, Baomin Wang, Jiye Yan, and Liusheng Duan. 2023. "Coronatine-Induced Maize Defense against Gibberella Stalk Rot by Activating Antioxidants and Phytohormone Signaling" Journal of Fungi 9, no. 12: 1155. https://doi.org/10.3390/jof9121155
APA StyleLiu, M., Sui, Y., Yu, C., Wang, X., Zhang, W., Wang, B., Yan, J., & Duan, L. (2023). Coronatine-Induced Maize Defense against Gibberella Stalk Rot by Activating Antioxidants and Phytohormone Signaling. Journal of Fungi, 9(12), 1155. https://doi.org/10.3390/jof9121155






