SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans
Abstract
:1. Introduction
2. Materials & Methods
2.1. Bioinformatic Analyses
2.2. Media and STRAINS
2.3. Cloning
2.4. Creating HFI1 Gene Deletion and Complementation Strains
2.5. Phenotype and Virulence Assays
2.6. Murine Inhalation Model of Virulence
2.7. Protein Extraction
2.8. Western Blot Analyses
2.9. RNA Extraction and Reverse Transcription-Quantitative PCR
3. Results
3.1. Identification of the Gene-Encoding Hfi1 in C. neoformans
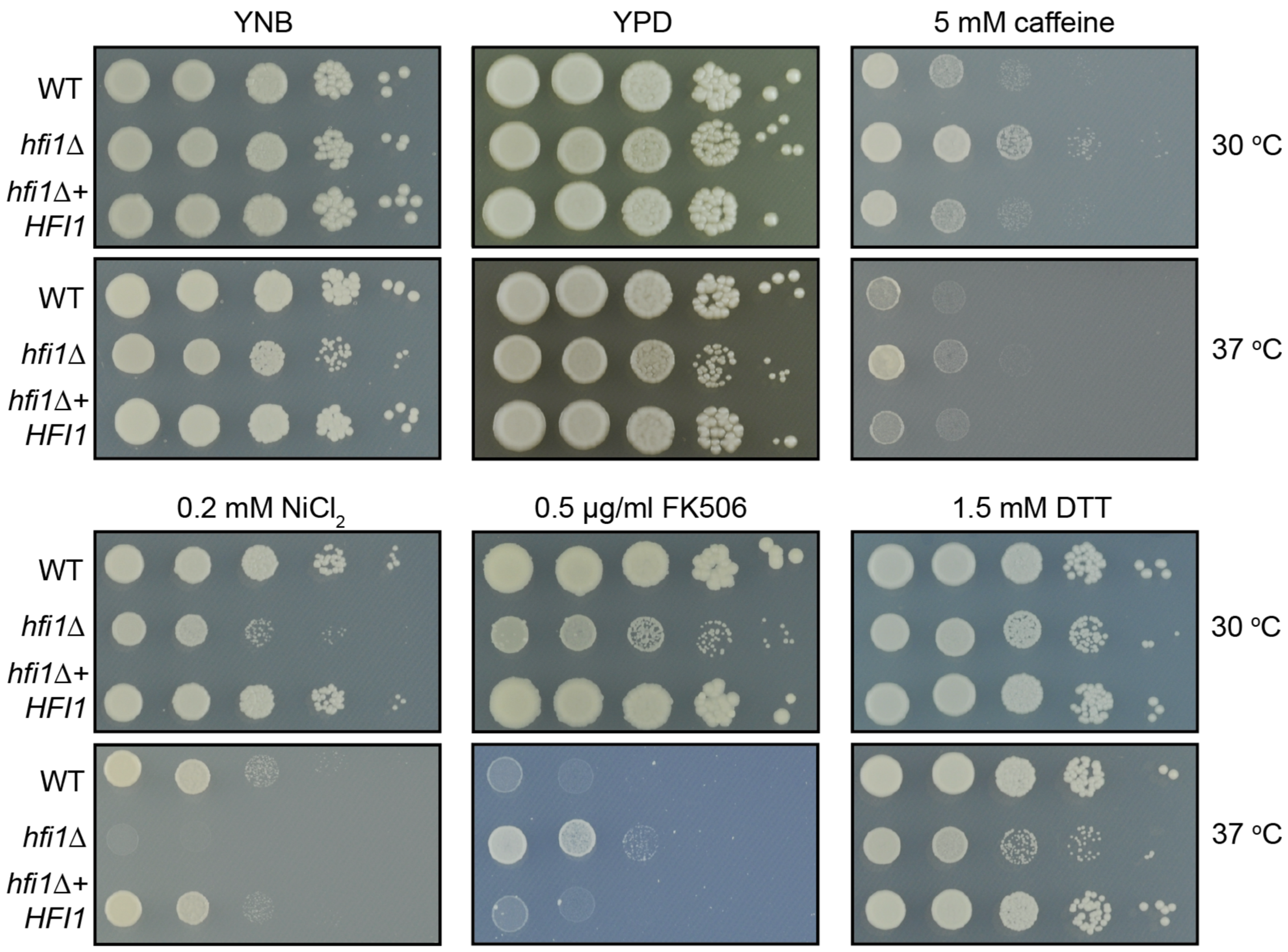
3.2. Hfi1 Is Essential for C. neoformans Growth during Stress
3.3. Deletion of HFI1 Impacts C. neoformans Virulence Traits In Vitro
3.4. HFI1 Is Important for C. neoformans Virulence in a Murine Inhalation Model of Infection
3.5. Hfi1 Is Required for SAGA to Perform a Subset of Its Post-Transcriptional Modification Functions
3.6. Deletion of Hfi1 Influences the Expression of Other SAGA Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gibney, E.R.; Nolan, C.M. Epigenetics and Gene Expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.D. The Quantitative Separation of Purines, Pyrimidines, and Nucleosides by Paper Chromatography. J. Biol. Chem. 1948, 175, 315–332. [Google Scholar] [CrossRef] [PubMed]
- Li, E. Chromatin Modification and Epigenetic Reprogramming in Mammalian Development. Nat. Rev. Genet. 2002, 3, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Smith, E.; Shilatifard, A. The Language of Histone Crosstalk. Cell 2010, 142, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Nagy, Z.; Tora, L. Distinct GCN5/PCAF-Containing Complexes Function as Co-Activators and are Involved in Transcription Factor and Global Histone Acetylation. Oncogene 2007, 26, 5341–5357. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Liu, Z.Z.; Wei, L.; Zhou, J.X.; Cai, X.W.; Su, Y.N.; Li, L.; Chen, S.; He, X.J. Three Functionally Redundant Plant-Specific Paralogs are Core Subunits of the SAGA Histone Acetyltransferase Complex in Arabidopsis. Mol. Plant. 2021, 14, 1071–1087. [Google Scholar] [CrossRef] [PubMed]
- Herbst, D.A.; Esbin, M.N.; Louder, R.K.; Dugast-Darzacq, C.; Dailey, G.M.; Fang, Q.; Darzacq, X.; Tjian, R.; Nogales, E. Structure of the Human SAGA Coactivator Complex: The Divergent Architecture of Human SAGA Allows Modular Coordination of Transcription Activation and Co-Transcriptional Splicing. Nat. Struct. Mol. Biol. 2021, 28, 989–996. [Google Scholar] [CrossRef]
- Helmlinger, D.; Tora, L. Sharing the SAGA. Trends Biochem. Sci. 2017, 42, 850–861. [Google Scholar] [CrossRef]
- Koutelou, E.; Hirsch, C.L.; Dent, S.Y.R. Multiple Faces of the SAGA Complex. Curr. Opin. Cell Biol. 2010, 22, 374–382. [Google Scholar] [CrossRef]
- Brown, C.E.; Howe, L.; Sousa, K.; Alley, S.C.; Carrozza, M.J.; Tan, S.; Workman, J.L. Recruitment of HAT Complexes by Direct Activator Interactions with the ATM-Related Tra1 Subunit. Science 2001, 292, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Paduch, M.; Kim, S.A.; Kramer, R.M.; Barrios, A.F.; Lu, V.; Luke, J.; Usatyuk, S.; Kossiakoff, A.A.; Tan, S. Structural Basis for Activation of SAGA Histone Acetyltransferase Gcn5 by Partner Subunit Ada2. Proc. Natl. Acad. Sci. USA 2018, 115, 10010–10015. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Dai, W.; Xiao, D.; Xiong, Q.; Liu, C.; Hu, J.; Ge, F.; Yu, X.; Li, S. Acetylation-Dependent SAGA Complex Dimerization Promotes Nucleosome Acetylation and Gene Transcription. Nat. Struct. Mol. Biol. 2022, 29, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Bian, C.; Xu, C.; Ruan, J.; Lee, K.K.; Burke, T.L.; Tempel, W.; Barsyte, D.; Li, J.; Wu, M.; Zhou, B.O.; et al. Sgf29 Binds Histone H3k4me2/3 and is Required for SAGA Complex Recruitment and Histone H3 Acetylation. EMBO J. 2011, 30, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, R.; Pray-Grant, M.G.; Selleck, W.; Grant, P.A.; Tan, S. Role of the Ada2 and Ada3 Transcriptional Coactivators in Histone Acetylation. J. Biol. Chem. 2002, 277, 7989–7995. [Google Scholar] [CrossRef] [PubMed]
- García-Molinero, V.; García-Martínez, J.; Reja, R.; Furió-Tarí, P.; Antúnez, O.; Vinayachandran, V.; Conesa, A.; Pugh, B.F.; Pérez-Ortín, J.E.; Rodríguez-Navarro, S. The SAGA/TREX-2 Subunit Sus1 Binds Widely to Transcribed Genes and Affects mRNA Turnover Globally. Epigenetics Chromatin 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wolberger, C. Uncovering the Role of Sgf73 in Maintaining SAGA Deubiquitinating Module Structure and Activity. J. Mol. Biol. 2015, 427, 1765–1778. [Google Scholar] [CrossRef]
- Lee, K.K.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Workman, J.L. The Deubiquitylation Activity of Ubp8 is Dependent Upon Sgf11 and its Association with the SAGA Complex. Mol. Cell. Biol. 2005, 25, 1173–1182. [Google Scholar] [CrossRef]
- Baker, S.P.; Grant, P.A. The SAGA Continues: Expanding the Cellular Role of a Transcriptional Co-Activator Complex. Oncogene 2007, 26, 5329–5340. [Google Scholar] [CrossRef]
- Rodríguez-Navarro, S.; Fischer, T.; Luo, M.J.; Antúnez, O.; Brettschneider, S.; Lechner, J.; Pérez-Ortín, J.E.; Reed, R.; Hurt, E. Sus1, a Functional Component of the SAGA Histone Acetylase Complex and the Nuclear Pore-Associated mRNA Export Machinery. Cell 2004, 116, 75–86. [Google Scholar] [CrossRef]
- Han, Y.; Luo, J.; Ranish, J.; Hahn, S. Architecture of the Saccharomyces cerevisiae SAGA Transcription Coactivator Complex. EMBO J. 2014, 33, 2534–2546. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zheng, X.; Guan, H.; Cao, Y.; Qu, H.; Kang, J.; Ren, X.; Lei, J.; Dong, M.-Q.; Li, X.; et al. Architecture of Saccharomyces cerevisiae SAGA Complex. Cell Discov. 2019, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.B.; Greber, B.J.; Nogales, E. Recent Insights into the Structure of TFIID, its Assembly, and its Binding to Core Promoter. Curr. Opin. Struct. Biol. 2020, 61, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.A.; Schieltz, D.; Pray-Grant, M.G.; Steger, D.J.; Reese, J.C.; Yates, J.R.; Workman, J.L. A Subset of TAFIIs are Integral Components of the SAGA Complex Required for Nucleosome Acetylation and Transcriptional Stimulation. Cell 1998, 94, 45–53. [Google Scholar] [CrossRef]
- Morgan, M.T.; Haj-Yahya, M.; Ringel, A.E.; Bandi, P.; Brik, A.; Wolberger, C. Structural Basis for Histone H2B Deubiquitination by the SAGA DUB Module. Science 2016, 351, 725–728. [Google Scholar] [CrossRef]
- Pascual-García, P.; Govind, C.K.; Queralt, E.; Cuenca-Bono, B.; Llopis, A.; Chavez, S.; Hinnebusch, A.G.; Rodríguez-Navarro, S. Sus1 Is Recruited to Coding Regions and Functions during Transcription Elongation in Association with SAGA and TREX2. Genes Dev. 2008, 22, 2811–2822. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, T.R.; Hay, C.; Price, M.S.; Giles, S.; Alspaugh, J.A. Cryptococcus neoformans Histone Acetyltransferase Gcn5 Regulates Fungal Adaptation to the Host. Eukaryot. Cell 2010, 9, 1193–1202. [Google Scholar] [CrossRef]
- Rösler, S.M.; Kramer, K.; Finkemeier, I.; Humpf, H.-U.; Tudzynski, B. The SAGA Complex in the Rice Pathogen Fusarium fujikuroi: Structure and Functional Characterization. Mol. Microbiol. 2016, 102, 951–974. [Google Scholar] [CrossRef]
- Gu, Q.; Wang, Y.; Zhao, X.; Yuan, B.; Zhang, M.; Tan, Z.; Zhang, X.; Chen, Y.; Wu, H.; Luo, Y.; et al. Inhibition of Histone Acetyltransferase GCN5 by a Transcription Factor FgPacC Controls Fungal Adaption to Host-Derived Iron Stress. Nucleic Acids Res. 2022, 50, 6190–6210. [Google Scholar] [CrossRef]
- Zhu, W.; Fan, X.; Zhao, Q.; Xu, Y.; Wang, X.; Chen, J. Bre1 and Ubp8 Regulate H2B Mono-Ubiquitination and the Reversible Yeast-Hyphae Transition in Candida albicans. Mol. Microbiol. 2021, 115, 332–343. [Google Scholar] [CrossRef]
- Yang, J.; Chen, D.; Matar, K.A.O.; Zheng, T.; Zhao, Q.; Xie, Y.; Gao, X.; Li, M.; Wang, B.; Lu, G.-d. The Deubiquitinating Enzyme MoUbp8 is Required for Infection-Related Development, Pathogenicity, and Carbon Catabolite Repression in Magnaporthe oryzae. Appl. Microbiol. Biotechnol. 2020, 104, 5081–5094. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.; Correia-Mesquita, T.O.; Godoy, P.; Omran, R.P.; Whiteway, M. SAGA Complex Subunits in Candida albicans Differentially Regulate Filamentation, Invasiveness, and Biofilm Formation. Front. Cell. Infect. Microbiol. 2022, 12, 764711. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Stewart, J.I.P.; Liu, S.; Sheppard, D.C.; Lu, L.; Zhang, S.; Atomi, H. Spt20, a Structural Subunit of the SAGA Complex, Regulates Aspergillus fumigatus Biofilm Formation, Asexual Development, and Virulence. Appl. Environ. Microbiol. 2022, 88, e01535-21. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Ruhlmann, C.; Winston, F.; Schultz, P. Molecular Architecture of the S. cerevisiae SAGA Complex. Mol. Cell 2004, 15, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Grant, P.A.; Roberts, S.M.; Duggan, L.J.; Belotserkovskaya, R.; Pacella, L.A.; Winston, F.; Workman, J.L.; Berger, S.L. Functional Organization of the Yeast SAGA Complex: Distinct Components Involved in Structural Integrity, Nucleosome Acetylation, and TATA-Binding Protein Interaction. Mol. Cell. Biol. 1999, 19, 86–98. [Google Scholar] [CrossRef]
- Horiuchi, J.; Silverman, N.; Piña, B.; Marcus, G.A.; Guarente, L. Ada1, a Novel Component of the ADA/GCN5 Complex, has Broader Effects than GCN5, ADA2, or ADA3. Mol. Cell. Biol. 1997, 17, 3220–3228. [Google Scholar] [CrossRef]
- Sánchez-Gaya, V.; Casaní-Galdón, S.; Ugidos, M.; Kuang, Z.; Mellor, J.; Conesa, A.; Tarazona, S. Elucidating the Role of Chromatin State and Transcription Factors on the Regulation of the Yeast Metabolic Cycle: A Multi-Omic Integrative Approach. Front. Genet. 2018, 9, 578. [Google Scholar] [CrossRef]
- Nuño-Cabanes, C.; García-Molinero, V.; Martín-Expósito, M.; Gas, M.-E.; Oliete-Calvo, P.; García-Oliver, E.; de la Iglesia-Vayá, M.; Rodríguez-Navarro, S. SAGA–CORE Subunit Spt7 is Required for Correct Ubp8 Localization, Chromatin Association and Deubiquitinase Activity. Epigenetics Chromatin 2020, 13, 46. [Google Scholar] [CrossRef]
- World Health Organisation. WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organisation: Geneva, Switzerland, 2022; ISBN 978-92-4-006024-1. Available online: https://www.who.int/publications/i/item/9789240060241 (accessed on 25 October 2022).
- Serna-Espinosa, B.N.; Guzmán-Sanabria, D.; Forero-Castro, M.; Escandón, P.; Sánchez-Quitian, Z.A. Environmental Status of Cryptococcus neoformans and Cryptococcus gattii in Colombia. J. Fungi 2021, 7, 410. [Google Scholar] [CrossRef]
- Chatterjee, S.; Prados-Rosales, R.; Itin, B.; Casadevall, A.; Stark, R.E. Solid-State NMR Reveals the Carbon-Based Molecular Architecture of Cryptococcus neoformans Fungal Eumelanins in the Cell Wall. J. Biol. Chem. 2015, 290, 13779–13790. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chrétien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal Cell Morphology Affects Host Cell Interactions and Pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Almeida, F.; Wolf, J.M.; Casadevall, A. Virulence-Associated Enzymes of Cryptococcus neoformans. Eukaryot. Cell 2015, 14, 1173–1185. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.L.; Nakayasu, E.S.; Oliveira, D.L.; Nimrichter, L.; Nosanchuk, J.D.; Almeida, I.C.; Casadevall, A. Extracellular Vesicles Produced by Cryptococcus neoformans Contain Protein Components Associated with Virulence. Eukaryot. Cell 2008, 7, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Arras, S.D.M.; Ormerod, K.L.; Erpf, P.E.; Espinosa, M.I.; Carpenter, A.C.; Blundell, R.D.; Stowasser, S.R.; Schulz, B.L.; Tanurdzic, M.; Fraser, J.A. Convergent Microevolution of Cryptococcus neoformans Hypervirulence in the Laboratory and the Clinic. Sci. Rep. 2017, 7, 17918. [Google Scholar] [CrossRef] [PubMed]
- Haynes, B.C.; Skowyra, M.L.; Spencer, S.J.; Gish, S.R.; Williams, M.; Held, E.P.; Brent, M.R.; Doering, T.L. Toward an Integrated Model of Capsule Regulation in Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002411. [Google Scholar] [CrossRef] [PubMed]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J., 3rd; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the Genome and Transcriptome of Cryptococcus neoformans var. grubii Reveals Complex RNA Expression and Microevolution Leading to Virulence Attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar]
- Fraser, J.A.; Subaran, R.L.; Nichols, C.B.; Heitman, J. Recapitulation of the Sexual Cycle of the Primary Fungal Pathogen Cryptococcus neoformans var. gattii: Implications for an Outbreak on Vancouver Island, Canada. Eukaryot. Cell 2003, 2, 1036–1045. [Google Scholar] [PubMed]
- Arras, S.D.M.; Chitty, J.L.; Blake, K.L.; Schulz, B.L.; Fraser, J.A. A Genomic Safe Haven for Mutant Complementation in Cryptococcus neoformans. PLoS ONE 2015, 10, e0122916. [Google Scholar] [CrossRef]
- Arras, S.D.; Fraser, J.A. Chemical Inhibitors of Non-Homologous End Joining Increase Targeted Construct Integration in Cryptococcus neoformans. PLoS ONE 2016, 11, e0163049. [Google Scholar] [CrossRef]
- Pitkin, J.W.; Panaccione, D.G.; Walton, J.D. A Putative Cyclic Peptide Efflux Pump Encoded by the TOXA Gene of the Plant-Pathogenic Fungus Cochliobolus carbonum. Microbiology 1996, 142, 1557–1565. [Google Scholar] [CrossRef]
- Southern, E. Southern Blotting. Nat. Protoc. 2006, 1, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Christensen, W.B. Urea Decomposition as a Means of Differentiating Proteus and Paracolon Cultures from Each Other and from Salmonella and Shigella Types. J. Bacteriol. 1946, 52, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Chaskes, S.; Tyndall, R.L. Pigment Production by Cryptococcus neoformans from Para- and Ortho-Diphenols: Effect of the Nitrogen Source. J. Clin. Microbiol. 1975, 1, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Wright, L.C.; Santangelo, R.T.; Muller, M.; Moran, V.R.; Kuchel, P.W.; Sorrell, T.C. Identification of Extracellular Phospholipase B, Lysophospholipase, and Acyltransferase Produced by Cryptococcus neoformans. Infect. Immun. 1997, 65, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.M.; Mukherjee, J.; Cole, G.T.; Casadevall, A.; Perfect, J.R. Urease as a Virulence Factor in Experimental Cryptococcosis. Infect. Immun. 2000, 68, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hommel, B.; Mukaremera, L.; Cordero, R.J.B.; Coelho, C.; Desjardins, C.A.; Sturny-Leclère, A.; Janbon, G.; Perfect, J.R.; Fraser, J.A.; Casadevall, A.; et al. Titan Cells Formation in Cryptococcus neoformans is Finely Tuned by Environmental Conditions and Modulated by Positive and Negative Genetic Regulators. PLoS Pathog. 2018, 14, e1006982. [Google Scholar] [CrossRef]
- Dambuza, I.M.; Drake, T.; Chapuis, A.; Zhou, X.; Correia, J.; Taylor-Smith, L.; LeGrave, N.; Rasmussen, T.; Fisher, M.C.; Bicanic, T.; et al. The Cryptococcus neoformans Titan Cell is an Inducible and Regulated Morphotype Underlying Pathogenesis. PLoS Pathog. 2018, 14, e1006978. [Google Scholar] [CrossRef]
- Kuo, M.H.; Brownell, J.E.; Sobel, R.E.; Ranalli, T.A.; Cook, R.G.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Transcription-Linked Acetylation by Gcn5p of Histones H3 and H4 at Specific Lysines. Nature 1996, 383, 269–272. [Google Scholar] [CrossRef]
- Li, B.; Sun, J.; Dong, Z.; Xue, P.; He, X.; Liao, L.; Yuan, L.; Jin, Y. GCN5 Modulates Osteogenic Differentiation of Periodontal Ligament Stem Cells through DKK1 Acetylation in Inflammatory Microenvironment. Sci. Rep. 2016, 6, 26542. [Google Scholar] [CrossRef]
- Grant, P.A.; Eberharter, A.; John, S.; Cook, R.G.; Turner, B.M.; Workman, J.L. Expanded Lysine Acetylation Specificity of Gcn5 in Native Complexes. J. Biol. Chem. 1999, 274, 5895–5900. [Google Scholar] [CrossRef] [PubMed]
- Riss, A.; Scheer, E.; Joint, M.; Trowitzsch, S.; Berger, I.; Tora, L. Subunits of Ada-Two-A-Containing (ATAC) or Spt-Ada-Gcn5-Acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J. Biol. Chem. 2015, 290, 28997–29009. [Google Scholar] [CrossRef] [PubMed]
- Ciurciu, A.; Komonyi, O.; Pankotai, T.; Boros, I.M. The Drosophila Histone Acetyltransferase Gcn5 and Transcriptional Adaptor Ada2a are Involved in Nucleosomal Histone H4 Acetylation. Mol. Cell. Biol. 2006, 26, 9413–9423. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.M.; Laribee, R.N.; Strahl, B.D. Protein Modifications in Transcription Elongation. Biochim. Biophys. Acta 2009, 1789, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Schram, A.W.; Baas, R.; Jansen, P.W.; Riss, A.; Tora, L.; Vermeulen, M.; Timmers, H.T. A Dual Role for SAGA-Associated Factor 29 (SGF29) in ER Stress Survival by Coordination of both Histone H3 Acetylation and Histone H3 Lysine-4 Trimethylation. PLoS ONE 2013, 8, e70035. [Google Scholar] [CrossRef] [PubMed]
- Fischer, V.; Schumacher, K.; Tora, L.; Devys, D. Global Role for Coactivator Complexes in RNA Polymerase II Transcription. Transcription 2019, 10, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Varthi, M.; Sykes, S.M.; Phillips, C.; Warzecha, C.; Zhu, W.; Wyce, A.; Thorne, A.W.; Berger, S.L.; McMahon, S.B. The Putative Cancer Stem Cell Marker USP22 is a Subunit of the Human SAGA Complex Required for Activated Transcription and Cell-Cycle Progression. Mol. Cell 2008, 29, 102–111. [Google Scholar] [CrossRef]
- Geng, Q.; Li, H.; Wang, D.; Sheng, R.C.; Zhu, H.; Klosterman, S.J.; Subbarao, K.V.; Chen, J.Y.; Chen, F.M.; Zhang, D.D. The Verticillium dahliae Spt-Ada-Gcn5 Acetyltransferase Complex Subunit Ada1 Is Essential for Conidia and Microsclerotia Production and Contributes to Virulence. Front. Microbiol. 2022, 13, 852571. [Google Scholar] [CrossRef]
- Henry, K.W.; Wyce, A.; Lo, W.S.; Duggan, L.J.; Emre, N.T.; Kao, C.F.; Pillus, L.; Shilatifard, A.; Osley, M.A.; Berger, S.L. Transcriptional Activation via Sequential Histone H2B Ubiquitylation and Deubiquitylation, Mediated by SAGA-Associated Ubp8. Genes Dev. 2003, 17, 2648–2663. [Google Scholar] [CrossRef]
- Chen, M.; Liu, Y.; Liu, Z.; Su, L.; Yan, L.; Huang, Y.; Huang, Y.; Zhang, W.; Xu, X.; Zheng, F. Histone Acetyltransferase Gcn5-Mediated Histone H3 Acetylation Facilitates Cryptococcal Morphogenesis and Sexual Reproduction. mSphere 2023, e00299-23. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.K.; Stephenson, C.J.; Villamor, T.C.; Dyba, T.G.; Schulz, B.L.; Fraser, J.A. SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans. J. Fungi 2023, 9, 1198. https://doi.org/10.3390/jof9121198
Yu CK, Stephenson CJ, Villamor TC, Dyba TG, Schulz BL, Fraser JA. SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans. Journal of Fungi. 2023; 9(12):1198. https://doi.org/10.3390/jof9121198
Chicago/Turabian StyleYu, Chendi K., Christina J. Stephenson, Tristan C. Villamor, Taylor G. Dyba, Benjamin L. Schulz, and James A. Fraser. 2023. "SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans" Journal of Fungi 9, no. 12: 1198. https://doi.org/10.3390/jof9121198
APA StyleYu, C. K., Stephenson, C. J., Villamor, T. C., Dyba, T. G., Schulz, B. L., & Fraser, J. A. (2023). SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans. Journal of Fungi, 9(12), 1198. https://doi.org/10.3390/jof9121198






