Structure of the Mating-Type Genes and Mating Systems of Verpa bohemica and Verpa conica (Ascomycota, Pezizomycotina)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Cultivation and Ascosporic Isolates
2.2. DNA Extraction and Sequencing Scheme
2.3. Genome Assembly and Annotation
2.4. Determination of the MAT Locus Structures
2.5. Comparison of MAT Loci
2.6. Motif Identification of Secondary MAT Genes
2.7. Phylogenetic Analysis
2.8. Primer Design for MAT Genes
3. Results
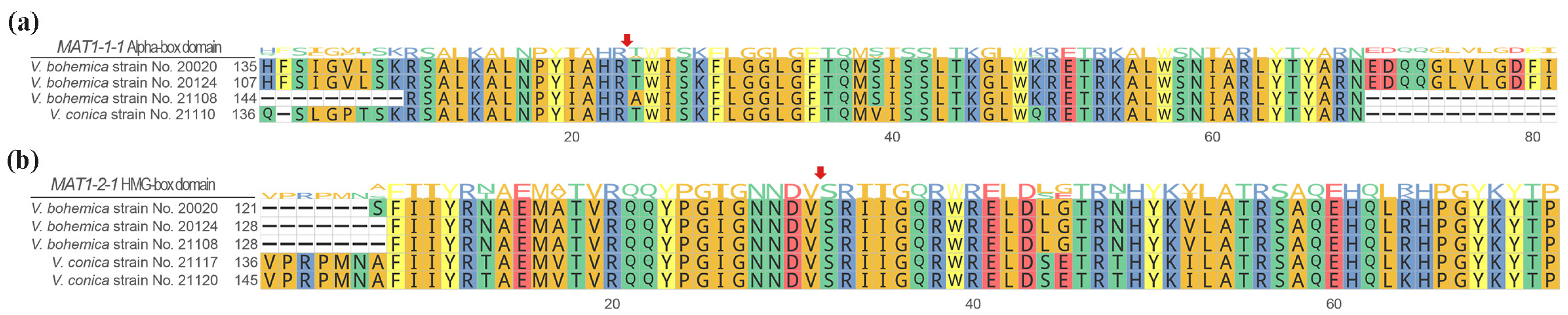
3.1. Structure of the MAT1–1-1 and MAT1–2-1 Genes of V. bohemica and V. conica
3.2. Structure of the MAT1–1-10 and MAT1-1-11 Genes of V. bohemica and V. conica
3.3. Phylogenetic Analysis of MAT Genes
3.4. Analysis of Conserved Motifs of MAT1-1-10 and MAT1-1-11 Proteins
3.5. MAT Locus Gene Content of Verpa
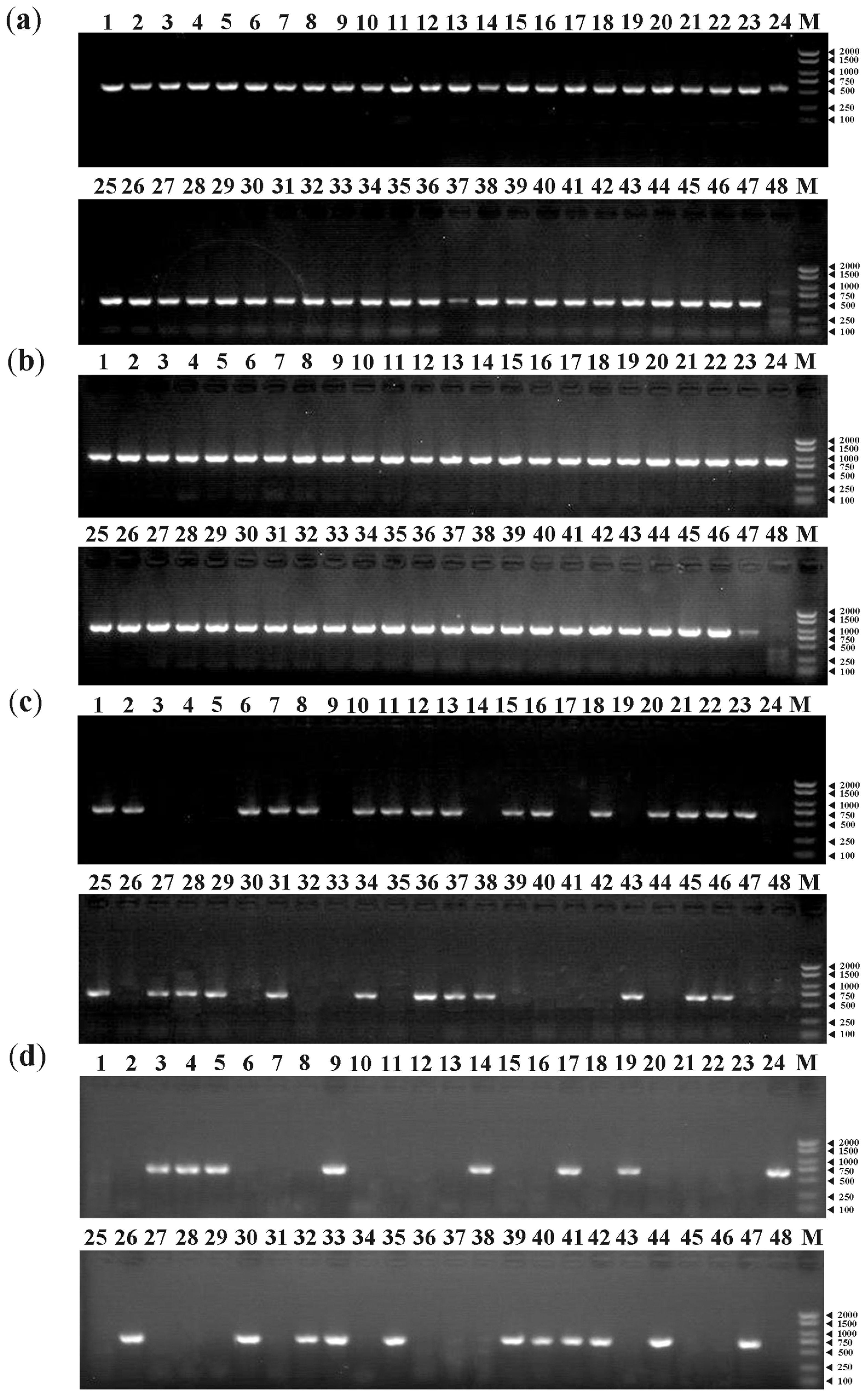
3.6. PCR Amplifications of MAT Genes
4. Discussion
4.1. MAT Loci and Mating-Type Genes
4.2. Structure of Mating-Type Loci between Verpa bohemica and Verpa conica
4.3. Homothallism in V. bohemica and Heterothallism in V. conica
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bunyard, B.A.; Nicholson, M.S.; Royse, D.J. Phylogenetic resolution of Morchella, Verpa, and Disciotis [Pezizales: Morchellaceae] based on restriction enzyme analysis of the 28S ribosomal RNA gene. Exp. Mycol. 1995, 19, 223–233. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Cigelnik, E.; Weber, N.S.; Trappe, J.M. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia 1997, 89, 48–65. [Google Scholar] [CrossRef]
- Aktas, S.; Karaselek, M.A. The taxonomicalch characterization of some helvella and its relatives by morphological and molecular data from turkey. Appl. Ecol. Environ. Res. 2019, 17, 7395–7405. [Google Scholar] [CrossRef]
- Anand, N.; Chowdhry, P.N. Taxonomic and molecular identification of Verpa bohemica: A newly explored fungi from Rajouri (J&K), India. Recent Res. Sci. Technol. 2013, 5, 9–12. [Google Scholar]
- Shameem, N.; Kamili, A.N.; Ahmad, M.; Masoodi, F.A.; Parray, J.A. Antimicrobial activity of crude fractions and morel compounds from wild edible mushrooms of North Western Himalaya. Microb. Pathog. 2017, 105, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Lagrange, E.; Vernoux, J.P. Warning on false or true morels and button mushrooms with potential toxicity linked to hydrazinic toxins: An update. Toxins 2020, 12, 482. [Google Scholar] [CrossRef]
- Misirikl, D.; Elmastas, M.; Turkekul, İ. Determination of antioxidant activities of some wild mushrooms species in Tokat region. Bütünleyici Ve Anadolu Tıbbı Derg. 2019, 1, 24–32. [Google Scholar]
- Sheikh, P.; Dar, G.H.H.; Dar, A.; Shah, S.; Bhat, K. Chemical composition and anti-oxidant activities of some edible mushrooms of western Himalayas of India. Int. J. Plant Res. 2015, 28, 124–133. [Google Scholar] [CrossRef]
- Lin, S.Y.; Chen, Y.K.; Yu, H.T.; Barseghyan, G.S.; Asatiani, M.D.; Wasser, S.P.; Mau, J.L. Comparative study of contents of several bioactive components in fruiting bodies and mycelia of culinary-medicinal mushrooms. Int. J. Med. Mushrooms 2013, 15, 315–323. [Google Scholar] [CrossRef]
- Elmastas, M.; Isildak, O.; Turkekul, I.; Temur, N. Determination of antioxidant activity and antioxidant compounds in wild edible mushrooms. J. Food Compos. Anal. 2007, 20, 337–345. [Google Scholar] [CrossRef]
- Kellner, H.; Luis, P.; Buscot, F. Diversity of laccase-like multicopper oxidase genes in morchellaceae: Identification of genes potentially involved in extracellular activities related to plant litter decay. FEMS Microbiol. Ecol. 2007, 61, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Coppin, E.; Debuchy, R.; Arnaise, S.; Picard, M. Mating types and sexual development in filamentous ascomycetes. Microbiol. Mol. Biol. Rev. 1997, 61, 411–428. [Google Scholar] [CrossRef] [PubMed]
- Metzenberg, R.L.; Glass, N.L. Mating type and mating strategies in Neurospora. BioEssays 1990, 12, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, B.G.; Yoder, O.C. Proposed nomenclature for mating type genes of filamentous ascomycetes. Fungal Genet. Biol. 2000, 31, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Debuchy, R.; Turgeon, B.G. Mating-type structure, evolution, and function in euascomycetes. In Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2006; pp. 293–323. [Google Scholar]
- Nelson, M.A. Mating systems in ascomycetes: A romp in the Sac. Trends Genet. 1996, 12, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; de Beer, Z.W.; Wingfield, B.D. Which MAT gene? Pezizomycotina (ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol. Rev. 2017, 31, 199–211. [Google Scholar] [CrossRef]
- Wilson, A.M.; Wilken, P.M.; Wingfield, M.J.; Wingfield, B.D. Genetic networks that govern sexual reproduction in the pezizomycotina. Microbiol. Mol. Biol. Rev. 2021, 85, e00020-21. [Google Scholar] [CrossRef]
- Liu, W.; Chen, L.; Cai, Y.; Zhang, Q.; Bian, Y. Opposite polarity monospore genome de novo sequencing and comparative analysis reveal the possible heterothallic life cycle of Morchella importuna. Int. J. Mol. Sci. 2018, 19, 2525. [Google Scholar] [CrossRef]
- Chai, H.; Chen, W.; Zhang, X.; Su, K.; Zhao, Y. Structural variation and phylogenetic analysis of the mating-type locus in the genus Morchella. Mycologia 2019, 111, 551–562. [Google Scholar] [CrossRef]
- Chai, H.; Liu, P.; Ma, Y.; Chen, W.; Tao, N.; Zhao, Y. Organization and unconventional integration of the mating-type loci in Morchella species. J. Fungi 2022, 8, 746. [Google Scholar] [CrossRef]
- Brock, T.D.; Madigan, M.T.; Martinko, J.M. Brock Biology of Microorganisms, 13th ed.; Benjamin Cummings: San Francisco, CA, USA, 2012. [Google Scholar]
- Inglis, P.W.; Pappas, M.D.C.R.; Resende, L.V.; Grattapaglia, D. Fast and inexpensive protocols for consistent extraction of high quality DNA and RNA from challenging plant and fungal samples for high-throughput SNP genotyping and sequencing applications. PLoS ONE 2018, 13, e0206085. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; D’Hert, S.; Schultz, D.T.; Cruts, M.; Van Broeckhoven, C. NanoPack: Visualizing and processing long-read sequencing data. Bioinformatics 2018, 34, 2666–2669. [Google Scholar] [CrossRef] [PubMed]
- Allaire, J. RStudio: Integrated Development Environment for R; Boston, MA, USA, 2012; Volume 770, pp. 165–171. Available online: http://www.rstudio.com/ (accessed on 1 December 2023).
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Du, X.H.; Yang, Z.L. Mating systems in true morels (Morchella). Microbiol. Mol. Biol. Rev. 2021, 85, e00220-20. [Google Scholar] [CrossRef]
- Ferris, P.J.; Pavlovic, C.; Fabry, S.; Goodenough, U.W. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 1997, 94, 8634–8639. [Google Scholar] [CrossRef]
- Martin, F.; Kohler, A.; Murat, C.; Balestrini, R.; Coutinho, P.M.; Jaillon, O.; Montanini, B.; Morin, E.; Noel, B.; Percudani, R.; et al. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 2010, 464, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Corby Kistler, H.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Grotelueschen, J.; Metzenberg, R.L. Neurospora crassa a mating-type region. Proc. Natl. Acad. Sci. USA 1990, 87, 4912–4916. [Google Scholar] [CrossRef] [PubMed]
- Kanematsu, S.; Adachi, Y.; Ito, T. Mating-type loci of heterothallic Diaporthe spp.: Homologous genes are present in opposite mating-types. Curr. Genet. 2007, 52, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Waalwijk, C.; Mendes, O.; Verstappen, E.C.P.; De Waard, M.A.; Kema, G.H.J. Isolation and characterization of the mating-type idiomorphs from the wheat septoria leaf blotch fungus Mycosphaerella graminicola. Fungal Genet. Biol. 2002, 35, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Wilken, P.M.; Steenkamp, E.T.; Wingfield, M.J.; de Beer, Z.W.; Wingfield, B.D. DNA loss at the Ceratocystis fimbriata mating locus results in self-sterility. PLoS ONE 2014, 9, e92180. [Google Scholar] [CrossRef]
- Teixeira, M.D.M.; Rodrigues, A.M.; Tsui, C.K.M.; De Almeida, L.G.P.; Van Diepeningen, A.D.; Van Den Ende, B.G.; Fernandes, G.F.; Kano, R.; Hamelin, R.C.; Lopes Bezerra, L.M.; et al. Asexual propagation of a virulent clone complex in a human and feline outbreak of Sporotrichosis. Eukaryot. Cell 2015, 14, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Ait Benkhali, J.; Coppin, E.; Brun, S.; Peraza Reyes, L.; Martin, T.; Dixelius, C.; Lazar, N.; Van Tilbeurgh, H.; Debuchy, R. A network of HMG-box transcription factors regulates sexual cycle in the fungus Podospora anserina. PLoS Genet. 2013, 9, e1003642. [Google Scholar] [CrossRef]
- Debuchy, R.; Berteaux Lecellier, V.; Silar, P. Mating systems and sexual morphogenesis in ascomycetes. In Cellular and Molecular Biology of Filamentous Fungi; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2010; pp. 499–535. ISBN 978-1-68367-129-9. [Google Scholar]
- Jacobi, V.; Dufour, J.; Bouvet, G.F.; Aoun, M.; Bernier, L. Identification of transcripts up-regulated in asexual and sexual fruiting bodies of the dutch elm disease pathogen Ophiostoma novo-ulmi. Can. J. Microbiol. 2010, 56, 697–705. [Google Scholar] [CrossRef]
- Arnaise, S.; Zickler, D.; Le Bilcot, S.; Poisier, C.; Debuchy, R. Mutations in mating-type genes of the heterothallic fungus Podospora anserina lead to self-fertility. Genetics 2001, 159, 545–556. [Google Scholar] [CrossRef]
- Klix, V.; Nowrousian, M.; Ringelberg, C.; Loros, J.J.; Dunlap, J.C.; Pöggeler, S. Functional characterization of MAT1-1-specific mating-type genes in the homothallic ascomycete Sordaria macrospora provides new insights into essential and nonessential sexual regulators. Eukaryot. Cell 2010, 9, 894–905. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Hou, R.; Zhang, J.; Ma, J.; Wu, Z.; Wang, G.; Wang, C.; Xu, J.R. The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS ONE 2013, 8, e66980. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Kenny, C.; Fagan, A.; Kurischko, C.; Gaillardin, C.; Wolfe, K.H. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. USA 2004, 101, 1632–1637. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J. Evolution of sexual reproduction: A view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biol. Rev. 2015, 29, 108–117. [Google Scholar] [CrossRef]
- Heitman, J.; Sun, S.; James, T.Y. Evolution of fungal sexual reproduction. Mycologia 2013, 105, 1–27. [Google Scholar] [CrossRef]
- Casselton, L.; Challen, M. The mating type genes of the basidiomycetes. In Growth, Differentiation and Sexuality; Springer: Berlin/Heidelberg, Germany, 2006; pp. 357–374. [Google Scholar]



| Ascocarp | Species | Origin | Genbank |
|---|---|---|---|
| No. 20020 | Verpa bohemica | Shennongjia Hubei | JAUFSG000000000 |
| No. 20124 | Verpa bohemica | Linxia Gansu | JAUFSF000000000 |
| No. 21108 | Verpa bohemica | Arba Sichuan | JAUFSE000000000 |
| No. 21110 | Verpa conica | Pengshui Chongqing | JAUFSD000000000 |
| No. 21117 | Verpa conica | Tiaiyuan Shanxi | JAUFSC000000000 |
| No. 21120 | Verpa conica | Xinyuan Xinjiang | JAUFSB000000000 |
| Gene | Code | Sequence (5′–3′) | Annealing Temperature (°C) | Product Size (bp) |
|---|---|---|---|---|
| MAT1-1-1 gene of V. bohemica | b1f | TCTCGCAGAAAGCCACGATT | 61 | 481 |
| b1r | ATCCTCATTGCGAGCGTAGG | |||
| MAT1-2-1 gene of V. bohemica | b2f | CTCCATCAAACGTGCGGTTC | 61 | 654 |
| b2r | GGGGGTTTCTTGTCCGAAGT | |||
| MAT1-1-10 gene of V. bohemica | b10f | GGGCCTATCAATCAACGCTT | 60 | 396 |
| b10r | AGGTAGCGTCTCAATGGGGT | |||
| MAT1-1-1 gene of V. conica | c1f | ATCTCGCAGAAGTCCACGATT | 61 | 580 |
| c1r | TTGATACCACAGCCGAGAAGG | |||
| MAT1-2-1 gene of V. conica | c2f | GTCGTGACTAAAACCGGCCT | 61 | 500 |
| c2r | TCATTGCTGCGGTGGATCTT | |||
| MAT1-1-10 gene of V. conica | c10f | CGCGTCGTTAGTAGTGCGAT | 60 | 819 |
| c10r | GAAGCTCAGTCCGGTGACAT | |||
| MAT1-1-11 gene of V. conica | c11f | TGGACGAAACGCTGGACTTT | 61 | 648 |
| c11r | CCTTCTGGAACCCTGCAAGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Liu, W.; Cai, Y.; Shi, X.; Wu, L.; Zhang, J.; Er, L.; Huang, Q.; Yin, Q.; Zhao, Z.; et al. Structure of the Mating-Type Genes and Mating Systems of Verpa bohemica and Verpa conica (Ascomycota, Pezizomycotina). J. Fungi 2023, 9, 1202. https://doi.org/10.3390/jof9121202
Sun W, Liu W, Cai Y, Shi X, Wu L, Zhang J, Er L, Huang Q, Yin Q, Zhao Z, et al. Structure of the Mating-Type Genes and Mating Systems of Verpa bohemica and Verpa conica (Ascomycota, Pezizomycotina). Journal of Fungi. 2023; 9(12):1202. https://doi.org/10.3390/jof9121202
Chicago/Turabian StyleSun, Wenhua, Wei Liu, Yingli Cai, Xiaofei Shi, Liyuan Wu, Jin Zhang, Lingfang Er, Qiuchen Huang, Qi Yin, Zhiqiang Zhao, and et al. 2023. "Structure of the Mating-Type Genes and Mating Systems of Verpa bohemica and Verpa conica (Ascomycota, Pezizomycotina)" Journal of Fungi 9, no. 12: 1202. https://doi.org/10.3390/jof9121202
APA StyleSun, W., Liu, W., Cai, Y., Shi, X., Wu, L., Zhang, J., Er, L., Huang, Q., Yin, Q., Zhao, Z., He, P., & Yu, F. (2023). Structure of the Mating-Type Genes and Mating Systems of Verpa bohemica and Verpa conica (Ascomycota, Pezizomycotina). Journal of Fungi, 9(12), 1202. https://doi.org/10.3390/jof9121202






