Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Sample Collection
2.3. DNA Isolation
2.4. Bacterial Composition
2.5. Fungal Composition
2.6. Bioinformatics Analysis
2.7. Statistics
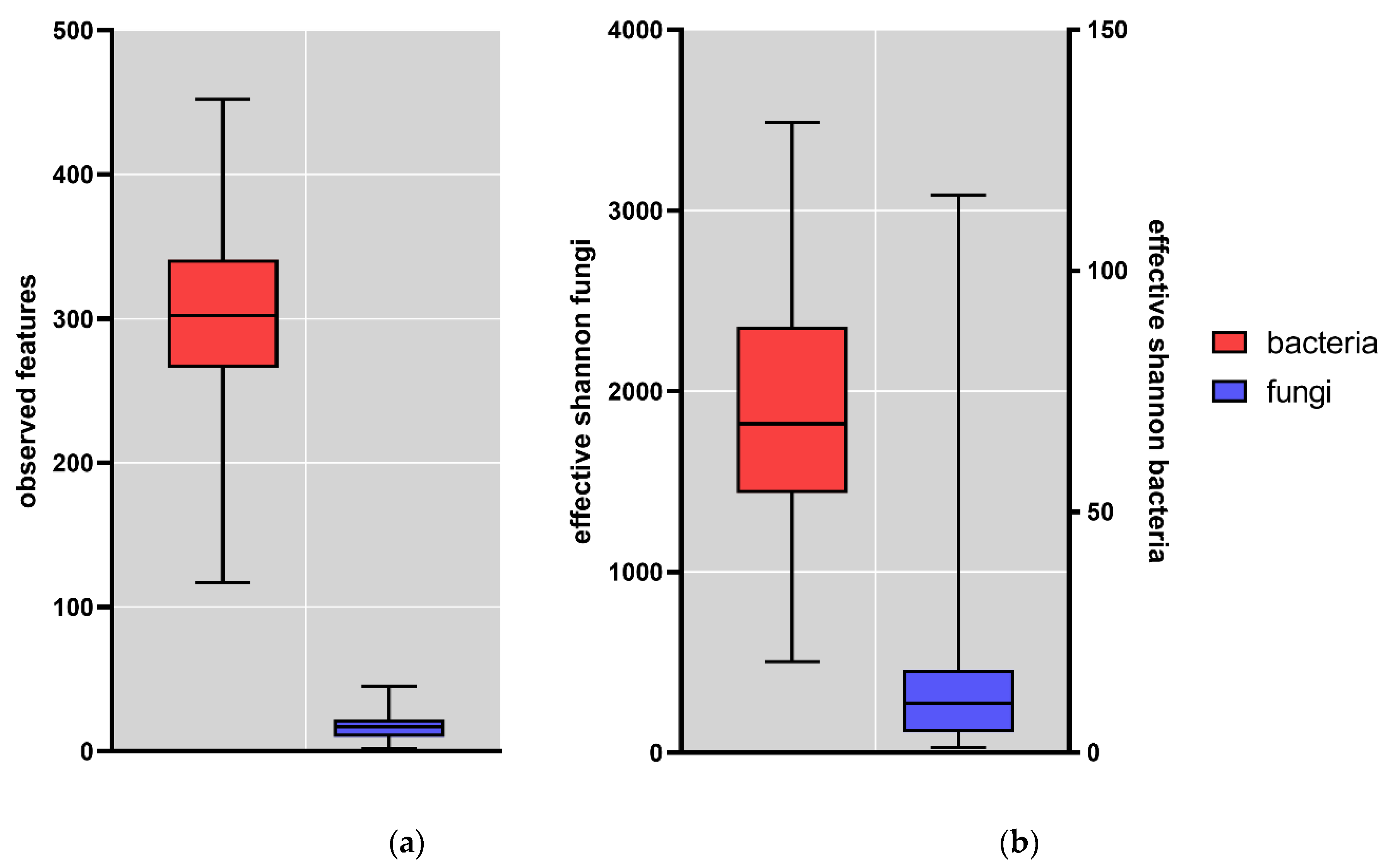
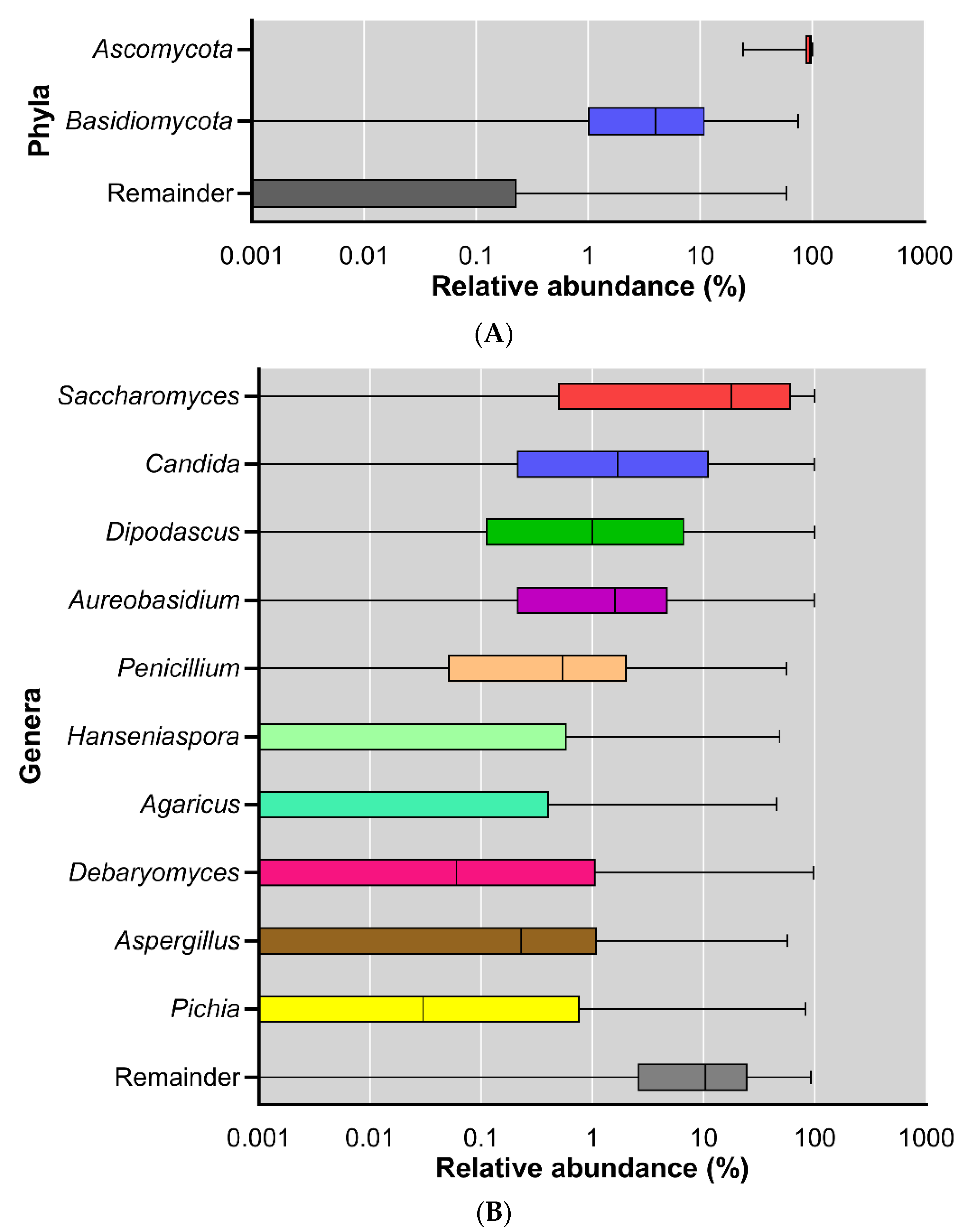
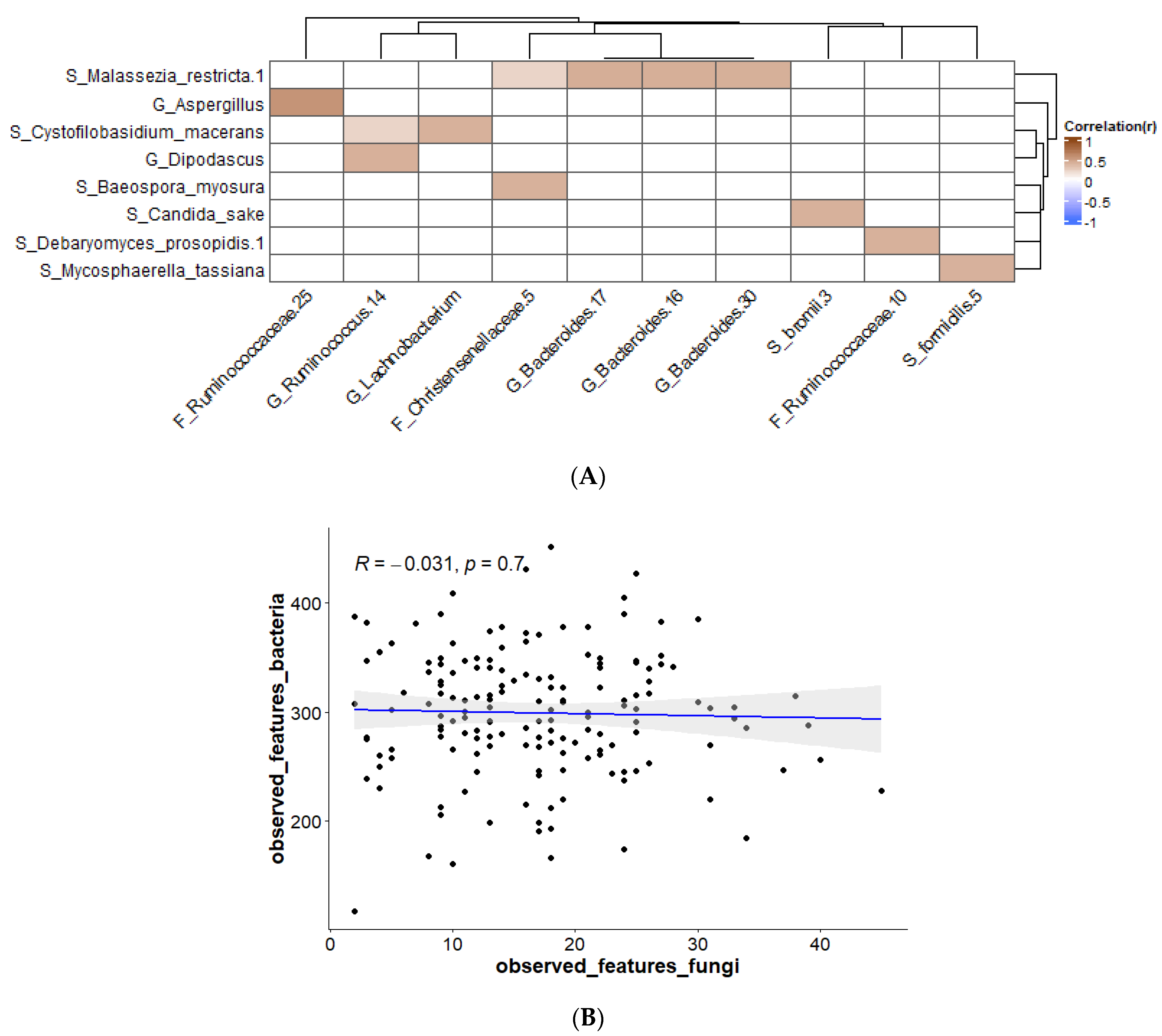
3. Results
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef] [Green Version]
- Breitbart, M.; Hewson, I.; Felts, B.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003, 185, 6220–6223. [Google Scholar] [CrossRef] [Green Version]
- Hamad, I.; Raoult, D.; Bittar, F. Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: Taxonomy and detection methods. Parasite Immunol. 2016, 38, 12–36. [Google Scholar] [CrossRef]
- Simon, G.L.; Gorbach, S.L. Intestinal flora in health and disease. Gastroenterology 1984, 86, 174–193. [Google Scholar] [CrossRef]
- Macfarlane, S.; Macfarlane, G.T. Bacterial diversity in the human gut. Gastroenterology 2004, 54, 261–290. [Google Scholar]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Finegold, S.M.; Attebery, H.R.; Sutter, V.L. Effect of diet on human fecal flora: Comparison of Japanese and American diets. Am. J. Clin. Nutr. 1974, 27, 1456–1469. [Google Scholar] [CrossRef] [Green Version]
- Hamad, I.; Sokhna, C.; Raoult, D.; Bittar, F. Molecular detection of eukaryotes in a single human stool sample from Senegal. PloS ONE 2012, 7, e40888. [Google Scholar] [CrossRef] [Green Version]
- Scanlan, P.D.; Marchesi, J.R. Micro-eukaryotic diversity of the human distal gut microbiota: Qualitative assessment using culture-dependent and-independent analysis of faeces. ISME J. 2008, 2, 1183–1193. [Google Scholar] [CrossRef]
- Heisel, T.; Podgorski, H.; Staley, C.M.; Knights, D.; Sadowsky, M.J.; Gale, C.A. Complementary amplicon-based genomic approaches for the study of fungal communities in humans. PloS One 2015, 10, e0116705. [Google Scholar] [CrossRef] [Green Version]
- Beheshti-Maal, A.; Shahrokh, S.; Ansari, S.; Mirsamadi, E.S.; Yadegar, A.; Mirjalali, H.; Zali, M.R. Gut mycobiome: The probable determinative role of fungi in IBD patients. Mycoses 2021, 64, 468–476. [Google Scholar] [CrossRef]
- Salamon, D.; Sroka-Oleksiak, A.; Gurgul, A.; Arent, Z.; Szopa, M.; Bulanda, M.; Małecki, M.T.; Gosiewski, T. Analysis of the Gut Mycobiome in Adult Patients with Type 1 and Type 2 Diabetes Using Next-Generation Sequencing (NGS) with Increased Sensitivity—Pilot Study. Nutrients 2021, 13, 1066. [Google Scholar] [CrossRef]
- García-Gamboa, R.; Kirchmayr, M.R.; Gradilla-Hernández, M.S.; Pérez-Brocal, V.; Moya, A.; González-Avila, M. The intestinal mycobiota and its relationship with overweight, obesity and nutritional aspects. J. Hum. Nutr. Diet. 2021, 34, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Polvi, E.J.; Li, X.; O’Meara, T.R.; Leach, M.D.; Cowen, L.E. Opportunistic yeast pathogens: Reservoirs, virulence mechanisms, and therapeutic strategies. Cell Mol. Life Sci. 2015, 72, 2261–2287. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, R.B.; Atobe, J.H.; Souza, S.A.; de Castro Lima Santos, D.W. Epidemiology of invasive fungal infections in patients with acquired immunodeficiency syndrome at a reference hospital for infectious diseases in Brazil. Mycopathologia 2014, 178, 71–78. [Google Scholar] [CrossRef]
- Kazantseva, J.; Malv, E.; Kaleda, A.; Kallastu, A.; Meikas, A. Optimisation of sample storage and DNA extraction for human gut microbiota studies. BMC Microbiol. 2021, 21, 158. [Google Scholar] [CrossRef]
- Knudsen, B.E.; Bergmark, L.; Munk, P.; Lukjancenko, O.; Priemé, A.; Aarestrup, F.M.; Pamp, S.J.; Jansson, J.K. Impact of Sample Type and DNA Isolation Procedure on Genomic Inference of Microbiome Composition. mSystems 2016, 1, e00095-16. [Google Scholar] [CrossRef] [Green Version]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [Green Version]
- Schultz, J.; Maisel, S.; Gerlach, D.; Muller, T.; Wolf, M. A common core of secondary structure of the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef] [Green Version]
- Op De Beeck, M.; Lievens, B.; Busschaert, P.; Declerck, S.; Vangronsveld, J.; Colpaert, J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PloS ONE 2014, 9, e97629. [Google Scholar] [CrossRef] [PubMed]
- Estaki, M.; Jiang, L.; Bokulich, N.A.; McDonald, D.; González, A.; Kosciolek, T.; Martino, C.; Zhu, Q.; Birmingham, A.; Vázquez-Baeza, Y.; et al. QIIME 2 Enables Comprehensive End-to-End Analysis of Diverse Microbiome Data and Comparative Studies with Publicly Available Data. Curr. Protoc. Bioinform. 2020, 70, e100. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S.D. ITSxpress: Software to rapidly trim internally transcribed spacer sequences with quality scores for marker gene analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef]
- Nash, A.K.; Auchtung, T.A.; Wong, M.C.; Smith, D.P.; Gesell, J.R.; Ross, M.C.; Stewart, C.J.; Metcalf, G.A.; Muzny, D.M.; Gibbs, R.A.; et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 2017, 5, 153. [Google Scholar] [CrossRef]
- Shuai, M.; Fu, Y.; Zhong, H.-l.; Gou, W.; Jiang, Z.; Liang, Y.; Miao, Z.; Xu, J.-J.; Huynh, T.; Wahlqvist, M.L. Mapping the human gut mycobiome in middle-aged and elderly adults: Multiomics insights and implications for host metabolic health. Gut 2022, 71, 1812–1820. [Google Scholar] [CrossRef]
- Mar Rodríguez, M.; Pérez, D.; Javier Chaves, F.; Esteve, E.; Marin-Garcia, P.; Xifra, G.; Vendrell, J.; Jové, M.; Pamplona, R.; Ricart, W. Obesity changes the human gut mycobiome. Sci. Rep. 2015, 5, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Santus, W.; Devlin, J.R.; Behnsen, J. Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect. Immun. 2021, 89, e00648-20. [Google Scholar] [CrossRef]
- Raimondi, S.; Amaretti, A.; Gozzoli, C.; Simone, M.; Righini, L.; Candeliere, F.; Brun, P.; Ardizzoni, A.; Colombari, B.; Paulone, S. Longitudinal survey of fungi in the human gut: ITS profiling, phenotyping, and colonization. Front. Microbiol. 2019, 10, 1575. [Google Scholar] [CrossRef] [PubMed]
- Fiers, W.D.; Gao, I.H.; Iliev, I.D. Gut mycobiota under scrutiny: Fungal symbionts or environmental transients? Curr. Opin. Microbiol. 2019, 50, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Graves, R.; Hesseltine, C. Fungi in flour and refrigerated dough products. Mycopathol. Et Mycol. Appl. 1966, 29, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Suhr, M.J.; Hallen-Adams, H.E. The human gut mycobiome: Pitfalls and potentials—A mycologist’s perspective. Mycologia 2015, 107, 1057–1073. [Google Scholar] [CrossRef] [Green Version]
- Iliev, I.D.; Funari, V.A.; Taylor, K.D.; Nguyen, Q.; Reyes, C.N.; Strom, S.P.; Brown, J.; Becker, C.A.; Fleshner, P.R.; Dubinsky, M. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 2012, 336, 1314–1317. [Google Scholar] [CrossRef] [Green Version]
- Scupham, A.J.; Presley, L.L.; Wei, B.; Bent, E.; Griffith, N.; McPherson, M.; Zhu, F.; Oluwadara, O.; Rao, N.; Braun, J. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006, 72, 793–801. [Google Scholar] [CrossRef] [Green Version]
- Ponomarova, O.; Gabrielli, N.; Sévin, D.C.; Mülleder, M.; Zirngibl, K.; Bulyha, K.; Andrejev, S.; Kafkia, E.; Typas, A.; Sauer, U. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017, 5, 345–357.e346. [Google Scholar] [CrossRef] [Green Version]
- van Leeuwen, P.T.; van der Peet, J.M.; Bikker, F.J.; Hoogenkamp, M.A.; Oliveira Paiva, A.M.; Kostidis, S.; Mayboroda, O.A.; Smits, W.K.; Krom, B.P. Interspecies interactions between Clostridium difficile and Candida albicans. Msphere 2016, 1, e00187-16. [Google Scholar] [CrossRef] [Green Version]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 2018, 6, 1–16. [Google Scholar] [CrossRef]
- Hoarau, G.; Mukherjee, P.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.-F. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. MBio 2016, 7, e01250-16. [Google Scholar] [CrossRef] [Green Version]
- Lambooij, J.M.; Hoogenkamp, M.A.; Brandt, B.W.; Janus, M.M.; Krom, B.P. Fungal mitochondrial oxygen consumption induces the growth of strict anaerobic bacteria. Fungal Genet. Biol. 2017, 109, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kong, E.F.; Tsui, C.; Kucharíková, S.; Van Dijck, P.; Jabra-Rizk, M.A. Modulation of Staphylococcus aureus response to antimicrobials by the Candida albicans quorum sensing molecule farnesol. Antimicrob. Agents Chemother. 2017, 61, e01573-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruger, W.; Vielreicher, S.; Kapitan, M.; Jacobsen, I.D.; Niemiec, M.J. Fungal-Bacterial Interactions in Health and Disease. Pathogens 2019, 8, 70. [Google Scholar] [CrossRef] [Green Version]
- Prohic, A.; Jovovic Sadikovic, T.; Krupalija-Fazlic, M.; Kuskunovic-Vlahovljak, S. Malassezia species in healthy skin and in dermatological conditions. Int. J. Dermatol. 2016, 55, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Hallen-Adams, H.E.; Suhr, M.J. Fungi in the healthy human gastrointestinal tract. Virulence 2017, 8, 352–358. [Google Scholar] [CrossRef]
- Limon, J.J.; Tang, J.; Li, D.; Wolf, A.J.; Michelsen, K.S.; Funari, V.; Gargus, M.; Nguyen, C.; Sharma, P.; Maymi, V.I. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host. Microbe 2019, 25, 377–388.e376. [Google Scholar] [CrossRef] [Green Version]
- Bloom, S.M.; Bijanki, V.N.; Nava, G.M.; Sun, L.; Malvin, N.P.; Donermeyer, D.L.; Dunne Jr, W.M.; Allen, P.M.; Stappenbeck, T.S. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host. Microbe 2011, 9, 390–403. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhao, J.; Zhang, H.; Lee, Y.-K.; Zhai, Q.; Chen, W. Roles of intestinal bacteroides in human health and diseases. Crit. Rev. Food Sci. Nutr. 2021, 61, 3518–3536. [Google Scholar] [CrossRef]
- Hallen-Adams, H.E.; Kachman, S.D.; Kim, J.; Legge, R.M.; Martínez, I. Fungi inhabiting the healthy human gastrointestinal tract: A diverse and dynamic community. Fungal Ecol. 2015, 15, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Ze, X.; Duncan, S.H.; Louis, P.; Flint, H.J. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 2012, 6, 1535–1543. [Google Scholar] [CrossRef] [Green Version]
- Rangarajan, A.A.; Chia, H.E.; Azaldegui, C.A.; Olszewski, M.H.; Koropatkin, N.M.; Biteen, J.S. Ruminococcus bromii enables the growth of proximal Bacteroides thetaiotaomicron by releasing glucose during starch degradation. bioRxiv 2022, 168, 001180. [Google Scholar] [CrossRef] [PubMed]
- Kovatcheva-Datchary, P.; Egert, M.; Maathuis, A.; Rajilić-Stojanović, M.; De Graaf, A.A.; Smidt, H.; De Vos, W.M.; Venema, K. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ. Microbiol. 2009, 11, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Rick, E.M.; Woolnough, K.F.; Seear, P.J.; Fairs, A.; Satchwell, J.; Richardson, M.; Monteiro, W.R.; Craner, M.; Bourne, M.; Wardlaw, A.J. The airway fungal microbiome in asthma. Clin. Exp. Allergy 2020, 50, 1325–1341. [Google Scholar] [CrossRef] [PubMed]
- Petrie, G.; Vanterpool, T. Mycosphaerella tassiana on Cruciferae in Western Canada. Can. Plant Dis. Surv. 1978, 58, 77–79. [Google Scholar]
- O’Gorman, C.M.; Fuller, H.T. Prevalence of culturable airborne spores of selected allergenic and pathogenic fungi in outdoor air. Atmos. Environ. 2008, 42, 4355–4368. [Google Scholar] [CrossRef]
- Mortensen, K.L.; Mellado, E.; Lass-Flörl, C.; Rodriguez-Tudela, J.L.; Johansen, H.K.; Arendrup, M.C. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrob. Agents Chemother. 2010, 54, 4545–4549. [Google Scholar] [CrossRef] [Green Version]
- Richardson, M.; Bowyer, P.; Sabino, R. The human lung and Aspergillus: You are what you breathe in? Med. Mycol. 2019, 57, S145–S154. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, A.A.; Aruni, A.; Rastogi, P.; Rana, S.; Gupta, R. Invasive aspergillosis causing gastric necrosis and perforation: A case report. JGH Open 2020, 4, 90–93. [Google Scholar] [CrossRef]
- Arfken, A.M.; Frey, J.F.; Ramsay, T.G.; Summers, K.L. Yeasts of burden: Exploring the Mycobiome–Bacteriome of the piglet GI tract. Front. Microbiol. 2019, 10, 2286. [Google Scholar] [CrossRef]
- Kabwe, M.H.; Vikram, S.; Mulaudzi, K.; Jansson, J.K.; Makhalanyane, T.P. The gut mycobiota of rural and urban individuals is shaped by geography. BMC Microbiol. 2020, 20, 257. [Google Scholar] [CrossRef]
- Schnorr, S.L.; Candela, M.; Rampelli, S.; Centanni, M.; Consolandi, C.; Basaglia, G.; Turroni, S.; Biagi, E.; Peano, C.; Severgnini, M. Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Libkind, D.; Gadanho, M.; van Broock, M.; Sampaio, J.P. Cystofilobasidium lacus-mascardii sp. nov., a basidiomycetous yeast species isolated from aquatic environments of the Patagonian Andes, and Cystofilobasidium macerans sp. nov., the sexual stage of Cryptococcus macerans. Int. J. Syst. Evol. Microbiol. 2009, 59, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Singer, R. Notes sur quelques Basidiomycetes. IV. Rev. Mycol. 1938, 3, 187–199. [Google Scholar]
- Phaff, H.J.; Vaughan-Martini, A.; Starmer, W.T. Debaryomyces prosopidis sp. nov., a yeast from exudates of mesquite trees. Int. J. Syst. Evol. Microbiol. 1998, 48, 1419–1424. [Google Scholar] [CrossRef]

| Sex | 38 male (23.3%); 125 female (76.7%) |
| Age a | 35.5 ± 12.1 years |
| Weight a | 67.9 ± 11.3 kg |
| Length a | 173.5 ± 8.7 cm |
| BMI a | 22.5 ± 2.7 |
| Overweight b | 23 |
| Most Abundant Genera | Reference |
|---|---|
| Penicillum, Candida, Saccharomyces, Mucor, Aspergillus | [29] |
| Saccharomyces, Candida, Aspergillus, Malassezia | [28] |
| Saccharomyces, Malassezia, Candida, Cyberlindnera, Penicillium, Cladosporium, Aspergillus, Agaricus, Fusarium, Pichia, Debaryomyces, Galactomyces, Alternaria, Clavispora | [27] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maas, E.; Penders, J.; Venema, K. Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals. J. Fungi 2023, 9, 139. https://doi.org/10.3390/jof9020139
Maas E, Penders J, Venema K. Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals. Journal of Fungi. 2023; 9(2):139. https://doi.org/10.3390/jof9020139
Chicago/Turabian StyleMaas, Evy, John Penders, and Koen Venema. 2023. "Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals" Journal of Fungi 9, no. 2: 139. https://doi.org/10.3390/jof9020139
APA StyleMaas, E., Penders, J., & Venema, K. (2023). Fungal-Bacterial Interactions in the Human Gut of Healthy Individuals. Journal of Fungi, 9(2), 139. https://doi.org/10.3390/jof9020139





