Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Media and Culture Conditions
2.2. Identification of KA Gene Clusters in Aspergillus Species
2.3. Phylogenetic Study of Section Flavi Aspergilli
2.4. Disruption of the kojR Gene in A. flavus
2.5. Construction of kojR Expression Vectors with gpdA or gpiA Promoter
2.6. Determination of KA Production and Fungal Mycelial Dry Weight
2.7. Determination of kojR Copy Numbers of gpdA and gpiA Promoter-Driven Overexpression Transformants
2.8. Time-Course Quantitative Reverse Transcription PCR (qRT-PCR) Analysis
2.9. KojR DNA-Binding Motif Analysis
2.10. Generation of Defects in Putative KojR DNA-binding Sites by the Established CRISPR/Cas9 Genome-Editing Approach
2.11. Identification of Molecular Defects in kojA and kojT Transformants
3. Results
3.1. Phylogeny of Aspergillus Section Flavi Species Bases on Complete KA Gene Cluster Sequences
3.2. Partial KA Gene Clusters in Other Aspergilli and Penicillia
3.3. Expressing kojR Restored Expression of kojA and kojT in Overexpression Strains
3.4. Zn(II)2Cys6 Zinc Cluster Domains and Downstream Basic Regions of Aspergilli
3.5. Identification of Putative KojR-Binding Sites in kojA and kojR Promoters of Aspergilli
3.6. Involvement of the KojR-Binding Site in the A. flavus kojA Promoter in KA Production
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parrish, F.W.; Wiley, B.J.; Simmons, E.G.; Long, L., Jr. Production of aflatoxins and kojic acid by species of Aspergillus and Penicillium. Appl Microbiol 1966, 14, 139. [Google Scholar] [CrossRef] [PubMed]
- Katagiri, H.; Kitahara, K. The formation of kojic acid by Aspergillus oryzae: Part I. The formation of kojic acid from pentoses, sugar alcohols and gluconic acid. Bull. Agric. Chem. Soc. Jpn. 1929, 5, 38–47. [Google Scholar] [CrossRef]
- Bentley, R. From miso, sake and shoyu to cosmetics: A century of science for kojic acid. Nat. Prod. Rep. 2006, 23, 1046–1062. [Google Scholar] [CrossRef] [PubMed]
- Yabuta, T. LXXIII.—The constitution of kojic acid, a γ-pyrone derivative formed by Aspergillus oryzæ from carbohydrates. J. Chem. Soc. Trans. 1924, 125, 575–587. [Google Scholar] [CrossRef]
- Cabanes, J.; Chazarra, S.; Garcia-Carmona, F. Kojic acid, a cosmetic skin whitening agent, is a slow-binding inhibitor of catecholase activity of tyrosinase. J. Pharm. Pharm. 1994, 46, 982–985. [Google Scholar] [CrossRef]
- Brtko, J.; Rondahl, L.; Fickova, M.; Hudecova, D.; Eybl, V.; Uher, M. Kojic acid and its derivatives: History and present state of art. Cent. Eur. J. Public Health 2004, 12, S16–S18. [Google Scholar]
- Saeedi, M.; Eslamifar, M.; Khezri, K. Kojic acid applications in cosmetic and pharmaceutical preparations. Biomed. Pharm. 2019, 110, 582–593. [Google Scholar] [CrossRef]
- Mohamad, R.; Mohamed, M.S.; Suhaili, N.B.; Salleh, M.M.; Ariff, A.B. Kojic acid: Applications and development of fermentation process for production. Biotechnol. Mol. Biol. Rev. 2010, 5, 24–37. [Google Scholar]
- ResearchReportsWorld. Global Kojic Acid Market Growth (Status and Outlook) 2022–2028. 2022. Available online: https://www.researchreportsworld.com/global-kojic-acid-market-19888650 (accessed on 29 September 2022).
- Chaudhary, J.; Pathak, A.N.; Lakhawat, S. Production technology and applications of kojic acid. Annu. Res. Rev. Biol. 2014, 4, 3165–3196. [Google Scholar] [CrossRef]
- El-Kady, I.A.; Zohri, A.-N.A.; Hamed, S.R. Kojic Acid production from agro-industrial by-products using fungi. Biotechnol. Res. Int. 2014, 2014, 642385. [Google Scholar] [CrossRef] [Green Version]
- Feng, W.; Liang, J.-S.; Wang, B.; Chen, J. Improvement of kojic acid production in Aspergillus oryzae AR-47 mutant strain by combined mutagenesis. Bioprocess Biosyst. Eng. 2019, 42, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Futamura, T.; Okabe, M.; Tamura, T.; Toda, K.; Matsunobu, T.; Park, Y.S. Improvement of production of kojic acid by a mutant strain Aspergillus oryzae, MK107-39. J. Biosci. Bioeng. 2001, 91, 272–276. [Google Scholar] [CrossRef]
- Arnstein, H.R.; Bentley, R. Kojic acid biosynthesis from 1-C14-glucose. Nature 1950, 166, 948–949. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, P.; Agrawala, P.K.; Vishwanathan, L. Enzymes relevant to kojic acid biosynthesis in Aspergillus flavus. Microbiology 1981, 127, 131–136. [Google Scholar] [CrossRef] [Green Version]
- Terabayashi, Y.; Sano, M.; Yamane, N.; Marui, J.; Tamano, K.; Sagara, J.; Dohmoto, M.; Oda, K.; Ohshima, E.; Tachibana, K.; et al. Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 2010, 47, 953–961. [Google Scholar] [CrossRef] [PubMed]
- Marui, J.; Yamane, N.; Ohashi-Kunihiro, S.; Ando, T.; Terabayashi, Y.; Sano, M.; Ohashi, S.; Ohshima, E.; Tachibana, K.; Higa, Y.; et al. Kojic acid biosynthesis in Aspergillus oryzae is regulated by a Zn(II)2Cys6 transcriptional activator and induced by kojic acid at the transcriptional level. J. Biosci. Bioeng. 2011, 112, 40–43. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.-K.; Chang, T.D.; Katoh, K. Deciphering the origin of Aspergillus flavus NRRL21882, the active biocontrol agent of Afla-Guard®. Lett. Appl. Microbiol. 2021, 72, 509–516. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Chang, P.-K.; Scharfenstein, L.L.; Wei, Q.; Bhatnagar, D. Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 2010, 81, 240–246. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Mack, B.; Ehrlich, K.C. Deletion of the Aspergillus flavus orthologue of A. nidulans fluG reduces conidiation and promotes production of sclerotia but does not abolish aflatoxin biosynthesis. Appl. Env. Microbiol. 2012, 78, 7557–7563. [Google Scholar] [CrossRef] [Green Version]
- Punt, P.J.; Dingemanse, M.A.; Jacobs-Meijsing, B.J.; Pouwels, P.H.; van den Hondel, C.A. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene 1988, 69, 49–57. [Google Scholar] [CrossRef]
- Chang, P.-K.; Zhang, Q.; Scharfenstein, L.; Mack, B.; Yoshimi, A.; Miyazawa, K.; Abe, K. Aspergillus flavus GPI-anchored protein-encoding ecm33 has a role in growth, development, aflatoxin biosynthesis, and maize infection. Appl. Microbiol. Biotechnol. 2018, 102, 5209–5220. [Google Scholar] [CrossRef] [PubMed]
- Umemura, M.; Kuriiwa, K.; Dao, L.V.; Okuda, T.; Terai, G. Promoter tools for further development of Aspergillus oryzae as a platform for fungal secondary metabolite production. Fungal Biol. Biotechnol. 2020, 7, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Jin, F.J.; Wang, B.T.; Wang, Z.D.; Jin, L.; Han, P. CRISPR/Cas9-based genome editing and its application in Aspergillus species. J. Fungi 2022, 8, 467. [Google Scholar] [CrossRef]
- Chang, P.-K.; Scharfenstein, L.L.; Abbas, H.K.; Bellaloui, N.; Accinelli, C.; Ebelhar, M.W. Prevalence of NRRL21882-like (Afla-Guard®) Aspergillus flavus on sesame seeds grown in research fields in the Mississippi Delta. Biocontrol Sci. Technol. 2020, 30, 1090–1099. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Novakova, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenar, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Gilchrist, C.L.M.; Lacey, H.J.; Vuong, D.; Pitt, J.I.; Lange, L.; Lacey, E.; Pilgaard, B.; Chooi, Y.H.; Piggott, A.M. Comprehensive chemotaxonomic and genomic profiling of a biosynthetically talented Australian fungus, Aspergillus burnettii sp. nov. Fungal Genet. Biol. 2020, 143, 103435. [Google Scholar] [CrossRef]
- Kjaerbolling, I.; Vesth, T.; Frisvad, J.C.; Nybo, J.L.; Theobald, S.; Kildgaard, S.; Petersen, T.I.; Kuo, A.; Sato, A.; Lyhne, E.K.; et al. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 2020, 11, 1106. [Google Scholar] [CrossRef] [Green Version]
- Arias, R.S.; Orner, V.A.; Martinez-Castillo, J.; Sobolev, V.S. Aspergillus Section Flavi, Need for a robust taxonomy. Microbiol. Resour. Announc. 2021, 10, e0078421. [Google Scholar] [CrossRef] [PubMed]
- Frisvad, J.C.; Larsen, T.O.; Dalsgaard, P.W.; Seifert, K.A.; Louis-Seize, G.; Lyhne, E.K.; Jarvis, B.B.; Fettinger, J.C.; Overy, D.P. Four psychrotolerant species with high chemical diversity consistently producing cycloaspeptide A, Penicillium jamesonlandense sp. nov., Penicillium ribium sp. nov., Penicillium soppii and Penicillium lanosum. Int. J. Syst. Evol. Microbiol. 2006, 56, 1427–1437. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-K.; Ehrlich, K.C. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Aspergillus flavus. Appl. Microbiol. Biotechnol. 2013, 97, 4289–4300. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S.; Moreno, I.; Nieto, A.; Gomez, M.; Sentandreu, R.; Valentin, E. In silico analysis for transcription factors with Zn(II)2Cys6 binuclear cluster DNA-binding domains in Candida albicans. Comp. Funct. Genom. 2005, 6, 345–356. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Huang, H.; Deng, W.; Li, T. Genome-wide analysis of the Zn(II)2Cys6 zinc cluster-encoding gene family in Tolypocladium guangdongense and its light-induced expression. Genes 2019, 10, 179. [Google Scholar] [CrossRef] [Green Version]
- MacPherson, S.; Larochelle, M.; Turcotte, B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol. Mol. Biol. Rev. 2006, 70, 583–604. [Google Scholar] [CrossRef] [Green Version]
- Crews, S.T.; Pearson, J.C. Transcriptional autoregulation in development. Curr. Biol. 2009, 19, R241–R246. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, B.; Valerius, O.; Andermann, M.; Braus, G.H. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 2001, 12, 2846–2857. [Google Scholar] [CrossRef] [Green Version]
- Gerosa, L.; Sauer, U. Regulation and control of metabolic fluxes in microbes. Curr. Opin. Biotechnol. 2011, 22, 566–575. [Google Scholar] [CrossRef]
- Park, P.J. ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009, 10, 669–680. [Google Scholar] [CrossRef] [Green Version]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Chang, P.-K.; Li, C.; Hu, Z.; Zheng, M.; Sun, Q.; Shan, S. Identification of AflR binding sites in the genome of Aspergillus flavus by ChIP-Seq. J. Fungi 2020, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Montalbano, B.G.; Cary, J.W. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.; Keller, N.P.; Adams, T.H. Sequence-specific binding by Aspergillus nidulans AflR, a C6 zinc cluster protein regulating mycotoxin biosynthesis. Mol. Microbiol. 1998, 28, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
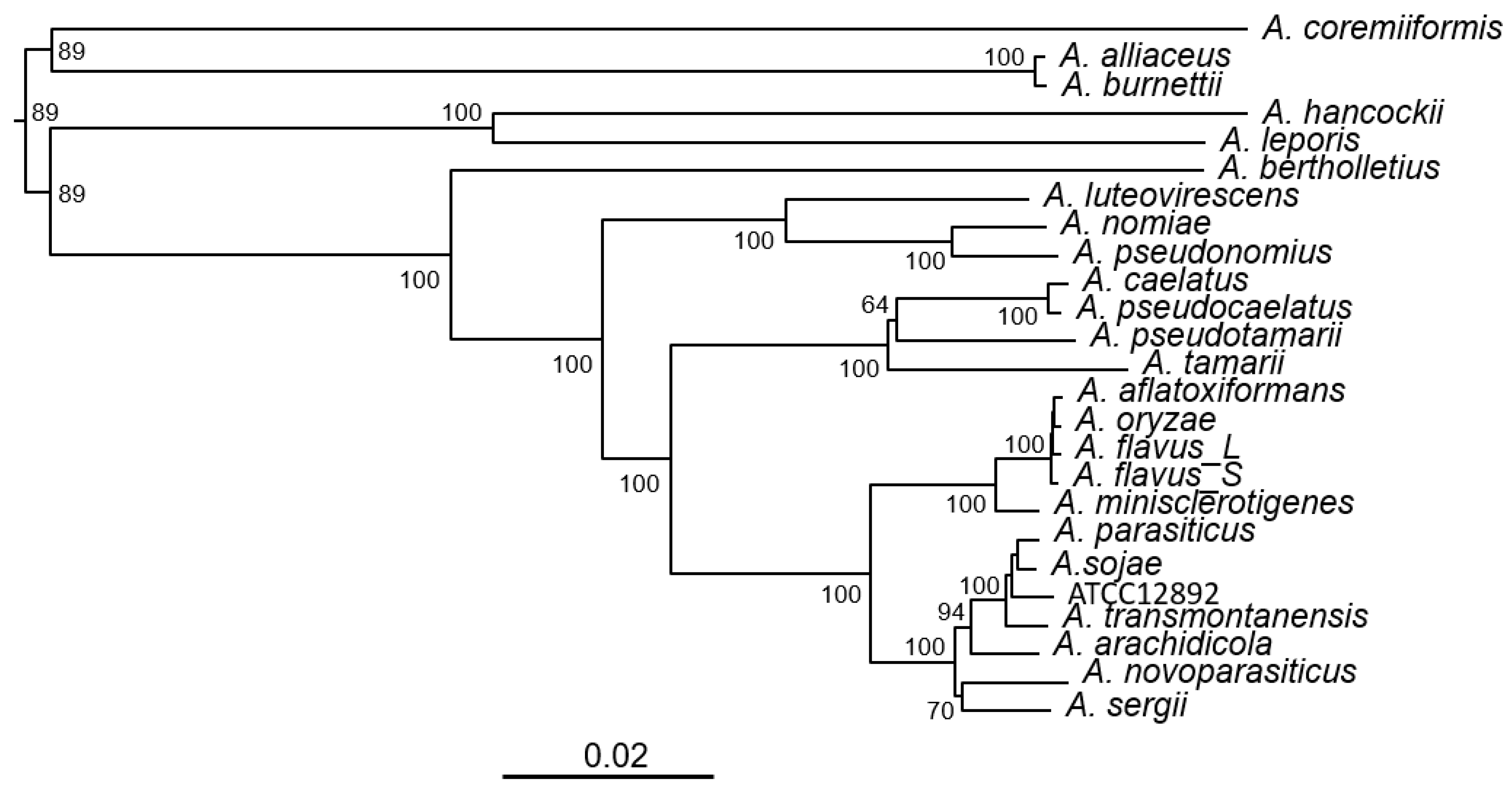
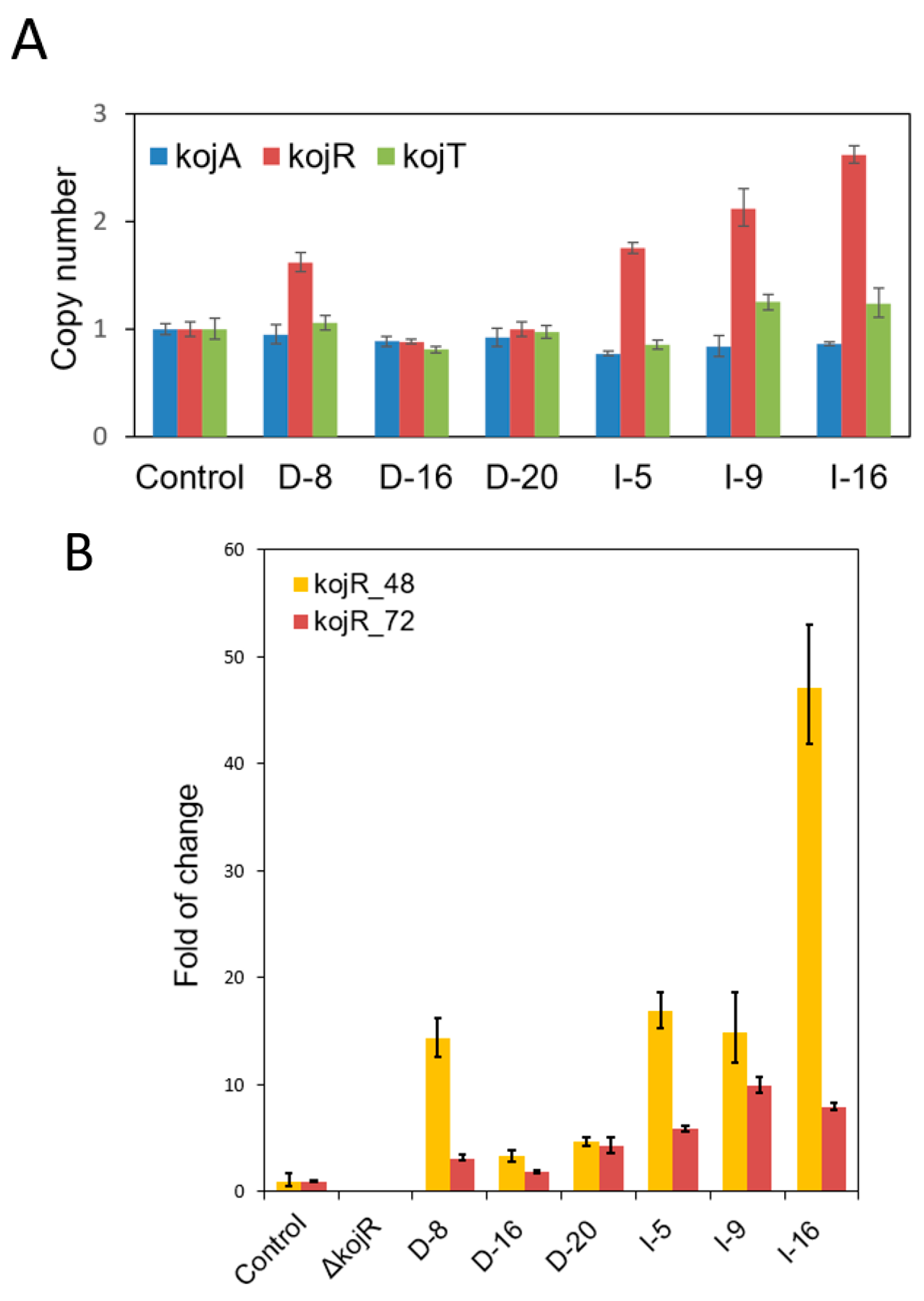
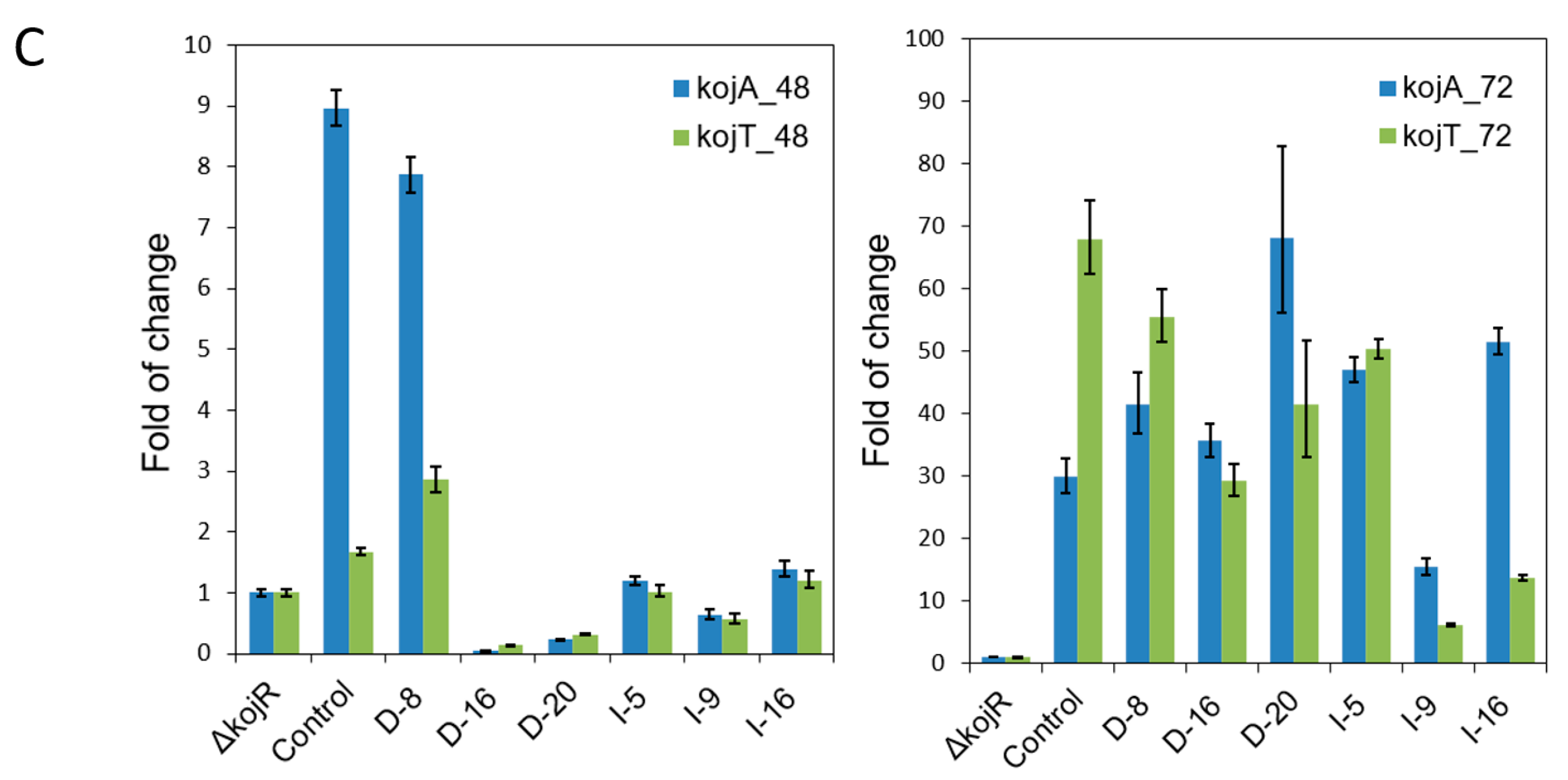


| Species | Strain | Genome (Mb) | GenBank (WGS) | Identity (%) a | Note |
|---|---|---|---|---|---|
| A. aflatoxiformans | CBS 121.62 | 37.6 | SWAT00000000.1 | 99.8 | =A. parvisclerotigenus |
| A. alliaceus | CBS 536.65 | 40.2 | SWAS00000000.1 | 81.6 | =A. albertensis |
| A. arachidicola | CBS 117610 | 38.9 | NEXV00000000.1 | 96.5 | |
| A. avenaceus | IBT 18842 | 33.8 | STFI00000000.1 | Not significant | |
| A. bertholletius | IBT 29228 | 37.0 | STFP00000000.1 | 87.2 | |
| A. burnettii | FRR 5400 | 41.0 | SPNV00000000.1 | 81.6 | |
| A. caelatus | CBS 763.97 | 40.0 | STFO00000000.1 | 92.5 | |
| A. coremiiformis | CBS 553.77 | 30.1 | STFN00000000.1 | 78.3 | 80.09% to A. leporis 81.53% to A. alliiaceus 81.53% to A. burnettii |
| A. flavus | NRRL 3357 | 36.9 | AAIH00000000.3 | 100.0 | L-morphotype |
| A. flavus | AF12 | 38.0 | NLCN00000000.1 | 99.8 | S-morphotype |
| A. hancockii | FRR 3425 | 39.9 | MBFL00000000.1 | 80.4 | |
| A. leporis | CBS 151.66 | 39.4 | SWBU00000000.1 | 80.9 | |
| A. luteovirescens | NRRL 26010 | 37.5 | LYCR00000000.1 | 91.6 | =A. bombycis |
| A. minisclerotigenes | CBS 117635 | 37.1 | SWDZ00000000.1 | 98.9 | |
| A. nomiae | NRRL 13137 | 36.1 | JNOM00000000.1 | 91.2 | =A. nomius |
| A. novoparasiticus | CBS 126849 | 40.9 | SWDA00000000.1 | 96.3 | |
| A. oryzae | RIB40 | 37.1 | JZJM00000000.1 | 99.8 | |
| A. parasiticus | SU-1 | 39.5 | JZEE00000000.1 | 96.6 | |
| A. pseudocaelatus | CBS 117616 | 39.7 | STFS00000000.1 | 92.6 | |
| A. pseudonomius | CBS 119388 | 37.8 | STFR00000000.1 | 91.3 | |
| A. pseudotamarii | CBS 117625 | 38.2 | STFH00000000.1 | 92.5 | |
| A. sergii | CBS 130017 | 38.3 | STFL00000000.1 | 96.3 | |
| A. sojae | NBRC 4239 | 39.8 | BACA00000000.2 | 96.5 | |
| A. tamarii | CBS 117626 | 38.5 | STFJ00000000.1 | 92.1 | |
| A. transmontanensis | CBS 130015 | 39.3 | STFK00000000.1 | 96.5 | |
| A. oryzaeb | ATCC 12892 | 41.2 | NVQI00000000.1 | 96.5 |
| Species | KojR(%) a | #AA | Protein ID b | KojT(%) a | #AA | Protein ID b |
|---|---|---|---|---|---|---|
| A. niger CBS 513.88 | 100.0 | 561 | XP_001393818.1 c | 100.0 | 572 | XP_001393819.2 c |
| A. welwitschiae CBS 139.54b | 99.5 | 561 | XP_026625778.1 | 99.0 | 585 | XP_026625777.1 |
| A. phoenicis ATCC 13157 | 99.1 | 561 | RDK41302.1 | 99.3 | 585 | RDK41301.1 |
| A. brasiliensis CBS 101740 | 90.2 | 561 | OJJ69434.1 | 94.1 | 572 | OJJ69433.1 |
| A. tubingensis CBS 134.48 | 88.8 | 560 | OJI88168.1 | 91.5 | 585 | OJI88169.1 |
| A. neoniger CBS 115656 | 87.9 | 560 | XP_025482490.1 | 93.2 | 567 | XP_025482489.1 |
| A. luchuensis CBS 106.47 | 87.7 | 560 | OJZ82270.1 | 92.8 | 563 | OJZ82269.1 |
| A. piperis CBS 112811 | 87.5 | 560 | XP_025520632.1 | 92.7 | 563 | XP_025520631.1 |
| A. vadensis CBS 113365 | 87.3 | 560 | XP_025563047.1 | 93.2 | 567 | XP_025563048.1 |
| A. eucalypticola CBS 122712 | 87.0 | 560 | XP_025388882.1 | 92.3 | 563 | XP_025388881.1 |
| A. costaricaensis CBS 115574 | 86.8 | 560 | XP_025545016.1 | 93.4 | 572 | XP_025545017.1 |
| A. sclerotioniger CBS 115572 | 76.7 | 560 | XP_025467076.1 | 85.7 | 574 | XP_025467075.1 |
| A. carbonarius ITEM 5010 | 75.9 | 559 | OOF94303.1 | 85.7 | 659 | OOF94304.1 |
| A. sclerotiicarbonarius CBS 121057 | 74.9 | 558 | PYI07967.1 | 85.3 | 571 | PYI07968.1 |
| A. ibericus CBS 121593 | 74.5 | 559 | XP_025576212.1 | 86.9 | 573 | XP_025576213.1 |
| A. transmontanensis CBS 130015 | 62.9 | 555 | KAE8316542.1 | 77.2 | 564 | KAE8316543.1 |
| A. avenaceus IBT 18842 | 61.7 | 553 | KAE8154428.1 | 75.0 | 563 | KAE8154429.1 |
| A. tanneri NIH1004 | 55.8 | 545 | XP_033424679.1 | 73.3 | 541 | XP_033424678.1 |
| A. melleus CBS 546.65 | 51.2 | 465 | XP_045944549.1 | 73.2 | 545 | XP_045944548.1 |
| A. steynii IBT 23096 | 49.4 | 546 | XP_024706771.1 | 73.7 | 546 | XP_024706772.1 |
| P. nordicum DAOMC 185683 | 100.0 | 544 | KOS46106.1 | 100.0 | 548 | KOS46105.1 |
| P. freii DAOM 242723 | 90.8 c | 488 | KUM59906.1 | 93.1 c | 548 | KUM59902.1 |
| P. polonicum IBT 4502 | 94.2 c | 292 | OQD63438.1 | 92.5 c | 557 | OQD63358.1 |
| Strain | mg KA/g Dry Mycelia b | Relative Amount |
|---|---|---|
| Control/(KuPG) a | 306.1 ± 187.0 | 1.00 |
| D-8 | 304.0 ± 24.1 | 0.99 |
| D-16 | 438.0 ± 57.8 | 1.43 |
| D-20 | 337.8 ± 15.5 | 1.10 |
| I-5 | 263.1 ± 35.2 | 0.86 |
| I-9 | 342.0 ± 19.1 | 1.12 |
| I-16 | 275.8 ± 39.2 | 0.90 |
| Species | Zinc-Finger Domain and Basic Region | KojR a | kojAb | kojTb |
|---|---|---|---|---|
| A. aflatoxiformans | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 100.0 | 99.2 |
| A. alliaceus | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRRPAVPKN | 91.7 | 100.0 | 67.4 |
| A. arachidicola | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 94.9 | 90.1 |
| A. bertholletius | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPATSKS | 96.7 | 82.5 | 77.7 |
| A. burnettii | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRRPAVPKN | 91.7 | 82.9 | 68.0 |
| A. caelatus | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 99.2 | 91.9 | 86.3 |
| A. coremiiformis | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFNPHPRRKPAPTKS | 91.7 | 63.9 | 60.5 |
| A. flavus_L | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 100.0 | 100.0 |
| A. flavus_S | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 100.0 | 100.0 |
| A. hancockii | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPATSRS | 95.8 | 77.3 | 65.1 |
| A. leporis | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAGSKS | 95.8 | 75.1 | 66.3 |
| A. luteovirescens | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASRS | 98.3 | 87.4 | 85.6 |
| A. minisclerotigenes | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 98.9 | 97.4 |
| A. nomiae | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASRS | 95.8 | 88.4 | 83.6 |
| A. novoparasiticus | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPTASKS | 99.2 | 94.4 | 91.9 |
| A. oryzae | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 100.0 | 99.5 |
| A. parasiticus | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 95.4 | 91.9 |
| A. pseudocaelatus | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 99.2 | 92.3 | 86.3 |
| A. pseudonomius | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASRS | 95.8 | 88.6 | 84.7 |
| A. pseudotamarii | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 99.3 | 90.2 | 87.8 |
| A. sergii | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 94.3 | 92.7 |
| A. sojae | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 95.2 | 91.7 |
| A. tamarii | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPTASKS | 99.2 | 89.3 | 87.8 |
| A. transmontanensis | RAKRACETCKLRKRKCDGHEPCTYCLRYEYQCTFKPHPRRKPAASKS | 100.0 | 95.7 | 91.9 |
| Species | kojA Promoter | kojT Promoter | ||
|---|---|---|---|---|
| Start/Strand | Motif Sequence | Start/Strand | Motif Sequence | |
| A. aflatoxiformans | -277/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. alliaceus | -275/+ | CGACTTTGCCG | -205/+ | CGGCTATGTCG |
| A. arachidicola | -277/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. bertholletius | -278/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. burnettii | -276/+ | CGACTTTGCCG | -205/+ | CGGCTATGTCG |
| A. caelatus | -277/+ | CGACTTTGCCG | -206/+ | CGGCTAAGTCG |
| A. coremiiformis | -282/+ | CGACTTTGCCG | -212/+ | CGGGTAAGTCG |
| A. flavus | -277/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. hancockii | -298/+ | CGACTTTGCCG | -202/+ | CGGTTAAGTCG |
| A. leporis | -289/+ | CGACTTTGCCG | -207/+ | CGGCTAAGTCG |
| A. luteovirescens | -276/+ | CGACTTTGCCG | -204/+ | CGGTTAAGTCG |
| A. minisclerotigenes | -278/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. nomiae | -276/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. novoparasiticus | -278/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. oryzae | -277/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. parasiticus | -277/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. pseudocaelatus | -278/+ | CGACTTTGCCG | -206/+ | CGGCTAAGTCG |
| A. pseudonomius | -277/+ | CGACTTTGCCG | -207/+ | CGGCTAAGTCG |
| A. pseudotamarii | -278/+ | CGACTTTGCCG | -175/+ | CGGCTAAGTCG |
| A. sergii | -277/+ | CGACTTTGCCG | -205/+ | CGGCTAAGTCG |
| A. sojae | -277/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. tamarii | -278/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
| A. transmontanensis | -277/+ | CGACTTTGCCG | -204/+ | CGGCTAAGTCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, P.-K.; Scharfenstein, L.L.; Mahoney, N.; Kong, Q. Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes. J. Fungi 2023, 9, 259. https://doi.org/10.3390/jof9020259
Chang P-K, Scharfenstein LL, Mahoney N, Kong Q. Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes. Journal of Fungi. 2023; 9(2):259. https://doi.org/10.3390/jof9020259
Chicago/Turabian StyleChang, Perng-Kuang, Leslie L. Scharfenstein, Noreen Mahoney, and Qing Kong. 2023. "Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes" Journal of Fungi 9, no. 2: 259. https://doi.org/10.3390/jof9020259
APA StyleChang, P.-K., Scharfenstein, L. L., Mahoney, N., & Kong, Q. (2023). Kojic Acid Gene Clusters and the Transcriptional Activation Mechanism of Aspergillus flavus KojR on Expression of Clustered Genes. Journal of Fungi, 9(2), 259. https://doi.org/10.3390/jof9020259






