A Polyphasic Approach including Whole Genome Sequencing Reveals Paecilomyces paravariotii sp. nov. as a Cryptic Sister Species to P. variotii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Whole Genome Sequencing and Assembly
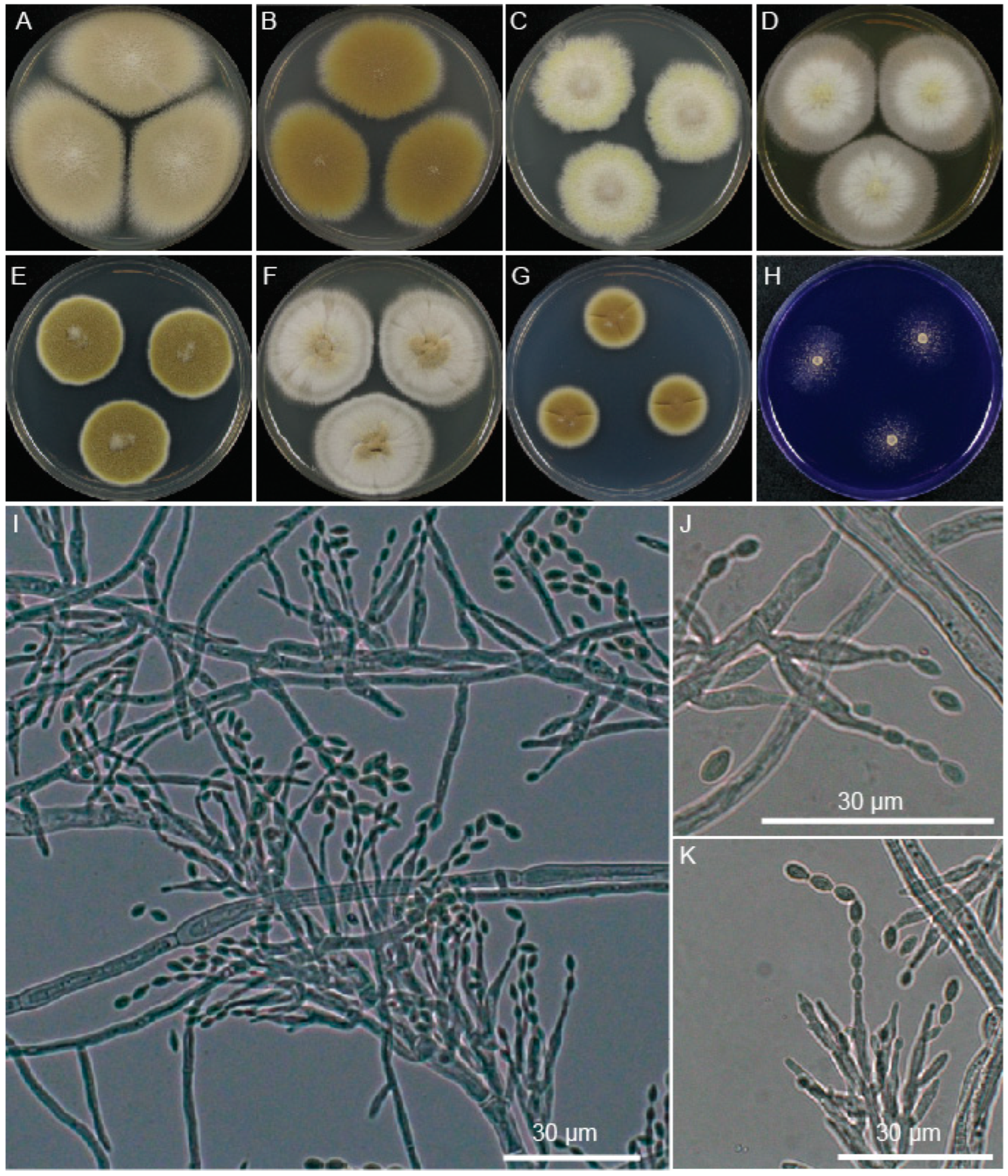
2.2. Properties of P. paravariotii in Culture
2.3. Phylogenetic Analyses
2.4. Metabolite Analysis Using High-Performance Liquid Chromatography (HPLC) and Mass Spectrometry
2.5. Sexual Crosses
2.6. Transformation
2.7. Microscopy
3. Results and Discussion
3.1. ‘Paecilomyces variotii’ Strain FRR 5287 Is Phylogenetically Separated from Previously Described Paecilomyces Species
3.2. Genome-Wide BLAST Comparisons Confirm Divergence between P. variotii and P. paravariotii and Reveal a Previously Unrecognised Horizontal Gene Transfer (HGT) Event
3.3. FRR 5287 Is Unable to Form Ascospores with a Fertile P. variotii Strain of Opposite Mating Type
3.4. Strain FRR 5287 Is Chemically Distinguished from P. variotii Due to Genetic Differences in Viriditoxin Biosynthesis
3.5. Features and Taxonomic Description of Paecilomyces Paravariotii
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samson, R.A.; Houbraken, J.; Varga, J.; Frisvad, J.C. Polyphasic taxonomy of the heat resistant ascomycete genus Byssochlamys and its Paecilomyces anamorphs. Persoonia 2009, 22, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Houbraken, J.; Kocsubé, S.; Visagie, C.M.; Yilmaz, N.; Wang, X.C.; Meijer, M.; Kraak, B.; Hubka, V.; Bensch, K.; Samson, R.A.; et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020, 95, 5–169. [Google Scholar] [CrossRef]
- Spetik, M.; Berraf-Tebbal, A.; Gramaje, D.; Mahamedi, A.E.; Stuskkova, K.; Burgova, J.; Eichmeier, A. Paecilomyces clematidis (Eurotiales, Thermoascaceae): A new species from Clematis root. Phytotaxa 2022, 559, 238–246. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.M.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet description sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Richardson, D.M.; Le Roux, J.J.; Strasberg, D.; Edwards, J.; Roets, F.; Hubka, V.; Taylor, P.W.J.; Heykoop, M.; et al. Fungal Planet description sheets: 400–468. Persoonia 2016, 36, 316–458. [Google Scholar] [CrossRef] [Green Version]
- Inglis, P.W.; Tigano, M.S. Identification and taxonomy of some entomopathogenic Paecilomyces spp. (Ascomycota) isolates using rDNA-ITS sequences. Genet. Mol. Biol. 2006, 29, 132–136. [Google Scholar] [CrossRef] [Green Version]
- Houbraken, J.; Verweij, P.E.; Rijs, A.; Borman, A.M.; Samson, R.A. Identification of Paecilomyces variotii in clinical samples and settings. J. Clin. Microbiol. 2010, 48, 2754–2761. [Google Scholar] [CrossRef] [Green Version]
- Sprute, R.; Salmanton-García, J.; Sal, E.; Malaj, X.; Falces-Romero, I.; Hatvani, L.; Heinemann, M.; Klimko, N.; López-Soria, L.; Meletiadis, J.; et al. Characterization and outcome of invasive infections due to Paecilomyces variotii: Analysis of patients from the FungiScope (R) registry and literature reports. J. Antimicrob. Chemother. 2021, 76, 765–774. [Google Scholar] [CrossRef]
- Heidarian, R.; Fotouhifar, K.B.; Debets, A.J.M.; Aanen, D.K. Phylogeny of Paecilomyces, the causal agent of pistachio and some other trees dieback disease in Iran. PLoS ONE 2018, 13, e0200794. [Google Scholar] [CrossRef] [Green Version]
- Biango-Daniels, M.N.; Ayer, K.M.; Cox, K.D.; Hodge, K.T. Paecilomyces niveus: Pathogenicity in the orchard and sensitivity to three fungicides. Plant Dis. 2019, 103, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.C.; Liu, H.F.; Jiang, D.P.; Wang, L.L.; Jiang, Y.K.; Tang, S.Y.; Hou, X.W.; Han, X.Y.; Liu, Z.G.; Zhang, M.; et al. Paecilomyces variotii extracts (ZNC) enhance plant immunity and promote plant growth. Plant Soil 2019, 441, 383–397. [Google Scholar] [CrossRef]
- Suárez-Estrella, F.; Arcos-Nievas, M.A.; López, M.J.; Vargas-García, M.C.; Moreno, J. Biological control of plant pathogens by microorganisms isolated from agro-industrial composts. Biol. Control 2013, 67, 509–515. [Google Scholar] [CrossRef]
- Moreno-Gavíra, A.; Diánez, F.; Sánchez-Montesinos, B.; Santos, M. Biocontrol effects of Paecilomyces variotii against fungal plant diseases. J. Fungi 2021, 7, 415. [Google Scholar] [CrossRef]
- van den Brule, T.; Punt, M.; Teertstra, W.; Houbraken, J.; Wösten, H.; Dijksterhuis, J. The most heat-resistant conidia observed to date are formed by distinct strains of Paecilomyces variotii. Environ. Microbiol. 2020, 22, 986–999. [Google Scholar] [CrossRef] [Green Version]
- Houbraken, J.; Varga, J.; Rico-Munoz, E.; Johnson, S.; Samson, R.A. Sexual reproduction as the cause of heat resistance in the food spoilage fungus Byssochlamys spectabilis (anamorph Paecilomyces variotii). Appl. Environ. Microbiol. 2008, 74, 1613–1619. [Google Scholar] [CrossRef] [Green Version]
- Urquhart, A.; Mondo, S.; Mäkelä, M.; Hane, J.; Wiebenga, A.; He, G.; Mihaltcheva, S.; Pangilinan, J.; Lipzen, A.; Barry, K.; et al. Genomic and genetic insights into a cosmopolitan fungus, Paecilomyces variotii (Eurotiales). Front. Microbiol. 2018, 9, 3058. [Google Scholar] [CrossRef] [Green Version]
- Seekles, S.J.; Teunisse, P.P.P.; Punt, M.; van den Brule, T.; Dijksterhuis, J.; Houbraken, H.; Wösten, H.A.B.; Ram, A.F.J. Preservation stress resistance of melanin deficient conidia from Paecilomyces variotii and Penicillium roqueforti mutants generated via CRISPR/Cas9 genome editing. Fungal Biol. Biotechnol. 2021, 8, 4. [Google Scholar] [CrossRef]
- Lim, S.; Bijlani, S.; Blachowicz, A.; Chiang, Y.M.; Lee, M.S.; Torok, T.; Venkateswaran, K.; Wang, C.C.C. Identification of the pigment and its role in UV resistance in Paecilomyces variotii, a Chernobyl isolate, using genetic manipulation strategies. Fungal Genet. Biol. 2021, 152, 103567. [Google Scholar] [CrossRef]
- Urquhart, A.S.; Hu, J.; Chooi, Y.H.; Idnurm, A. The fungal gene cluster for biosynthesis of the antibacterial agent viriditoxin. Fungal Biol. Biotechnol. 2019, 6, 10. [Google Scholar] [CrossRef] [Green Version]
- Urquhart, A.S.; Chong, N.F.; Yang, Y.; Idnurm, A. A large transposable element mediates metal resistance in the fungus Paecilomyces variotii. Curr. Biol. 2022, 32, 937–950.e935. [Google Scholar] [CrossRef]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar] [CrossRef] [Green Version]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Basturkmen, M.; Spevak, C.C.; Clutterbuck, J.; et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar] [CrossRef] [Green Version]
- Machida, M.; Asai, K.; Sano, M.; Tanaka, T.; Kumagai, T.; Terai, G.; Kusumoto, K.I.; Arima, T.; Akita, O.; Kashiwagi, Y.; et al. Genome sequencing and analysis of Aspergillus oryzae. Nature 2005, 438, 1157–1161. [Google Scholar] [CrossRef] [Green Version]
- Urquhart, A.S.; Vogan, A.A.; Gardiner, D.M.; Idnurm, A. Starships are active eukaryotic transposable elements mobilized by a new family of tyrosine recombinases. bioRxiv 2022. [Google Scholar] [CrossRef]
- Singh, N.K.; Blachowicz, A.; Romsdahl, J.; Wang, C.; Torok, T.; Venkateswaran, K. Draft genome sequences of several fungal strains selected for exposure to microgravity at the international space station. Genome Announc. 2017, 5, e01602–e01616. [Google Scholar] [CrossRef] [Green Version]
- Urbaniak, C.; Dadwal, S.; Bagramyan, K.; Venkateswaran, K. Draft genome sequence of a clinical isolate of Fusarium fujikuroi isolated from a male patient with acute myeloid leukemia. Genome Announc. 2018, 6, e00476-18. [Google Scholar] [CrossRef] [Green Version]
- Oka, T.; Ekino, K.; Fukuda, K.; Nomura, Y. Draft genome sequence of the formaldehyde-resistant fungus Byssochlamys spectabilis No. 5 (anamorph Paecilomyces variotii No. 5) (NBRC109023). Genome Announc. 2014, 2, e01162-13. [Google Scholar] [CrossRef] [Green Version]
- Stamps, B.W.; Andrade, O.C.; Lyon, W.J.; Floyd, J.G.; Nunn, H.S.; Bojanowski, C.L.; Crookes-Goodson, W.J.; Stevenson, B.S. Genome sequence of a Byssochlamys sp. strain isolated from fouled B20 biodiesel. Microbiol. Resour. Ann. 2018, 6, e00085-18. [Google Scholar] [CrossRef] [Green Version]
- Biango-Daniels, M.N.; Wang, T.W.; Hodge, K.T. Draft genome sequence of the patulin-producing fungus Paecilomyces niveus strain CO7. Genome Announc. 2018, 6, e00556-18. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, Y.; Zhang, Q.; Du, G.; Yang, F.; Li, J. Whole-genome sequencing and analysis of Paecilomyces variotii MTDF-01 from Moutai-flavor liquor brewing process. Food Sci. 2019, 40, 185–192. [Google Scholar]
- Pitkin, J.W.; Panaccione, D.G.; Walton, J.D. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 1996, 142, 1557–1565. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [Green Version]
- Frisvad, J.C. Creatine-sucrose agar, a differential medium for mycotoxin producing terverticillate Penicillium species. Lett. Appl. Microbiol. 1985, 1, 109–113. [Google Scholar]
- Filtenborg, O.; Frisvad, J.C.; Thrane, U. The significance of yeast extract composition on metabolite production in Penicillium. In Modern Concepts in Penicillium and Aspergillus Classification; Samson, R.A., Pitt, J.I., Eds.; Springer: Boston, MA, USA, 1990; pp. 433–441. [Google Scholar]
- Barratt, R.W.; Johnson, G.B.; Ogata, W.N. Wild-type and mutant stocks of Aspergillus Nidulans. Genetics 1965, 52, 233. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Samson, R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium—A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud. Mycol. 2004, 49, 1–173. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Frisvad, J.C.; Hubka, V.; Ezekiel, C.N.; Hong, S.B.; Novakova, A.; Chen, A.J.; Arzanlou, M.; Larsen, T.O.; Sklenar, F.; Mahakarnchanakul, W.; et al. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Stud. Mycol. 2019, 93, 1–63. [Google Scholar] [CrossRef]
- Matute, D.R.; Sepulveda, V.E. Fungal species boundaries in the genomics era. Fungal Genet. Biol. 2019, 131, 103249. [Google Scholar] [CrossRef]





| GenBank Accession | GenBank Name | Corrected Name | Citation |
|---|---|---|---|
| JAPVCG000000000 | P. fulvus FRR 3794 | - | This study |
| JAPVCF000000000 | P. dactylethromorphus FRR 0699 | - | This study |
| JAPVCE000000000 | P. dactylethromorphus FRR 5262 | - | This study |
| JAPVCD000000000 | P. lecythidis FRR 3797 | - | This study |
| JAPVCC000000000 | P. lecythidis FRR 4481 | - | This study |
| JAPVCB000000000 | P. formosus FRR 3793 | - | This study |
| JAPVCA000000000 | P. maximus FRR 4742 | - | This study |
| JAPVBZ000000000 | P. maximus FRR 2337 | - | This study |
| JAPVBY000000000 | P. maximus FRR 4140 | - | This study |
| JANCMQ000000000 | P. variotii FRR 1658 | - | [24] |
| JANCMP000000000 | P. variotii FRR 2889 | - | [24] |
| JANCMO000000000 | P. variotii FRR 3823 | - | [24] |
| JANCMN000000000 | P. variotii FRR 5287 | P. paravariotii FRR 5287 | [16] |
| RHLL00000000.1 | P. variotii CBS 144490 | - | [16] |
| RCNU00000000.1 | P. variotii CBS 101075 | - | [16] |
| MSJH00000000.2 | Byssochlamys sp. IMV 00236 | P. variotii IMV 00236 | [25] (Cladosporium cladosporioides IMV 00236 in publication) |
| QBDR00000000.1 | Thermoascaceae sp. COH1141 | P. lecythidis COH1141 | [26] |
| JAGJCD000000000.1 | Aspergillus sp. MCCF 102 | P. lecythidis MCCF 102 | |
| BAUL01000000.1 | Byssochlamys spectabilis No. 5 | P. formosus no. 5 | [27] |
| JACXGS000000000.1 | Paecilomyces variotii WS011 | P. formosus WS011 | |
| PNEM00000000.1 | Byssochlamys sp. AF001 | P. dactylethromorphus AF001 | [28] |
| QEIL00000000.1 | Paecilomyces niveus Cornell no. 7 | - | [29] |
| RCHW01000000.1 | Paecilomyces variotii MTDF-01 | - | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Urquhart, A.S.; Idnurm, A. A Polyphasic Approach including Whole Genome Sequencing Reveals Paecilomyces paravariotii sp. nov. as a Cryptic Sister Species to P. variotii. J. Fungi 2023, 9, 285. https://doi.org/10.3390/jof9030285
Urquhart AS, Idnurm A. A Polyphasic Approach including Whole Genome Sequencing Reveals Paecilomyces paravariotii sp. nov. as a Cryptic Sister Species to P. variotii. Journal of Fungi. 2023; 9(3):285. https://doi.org/10.3390/jof9030285
Chicago/Turabian StyleUrquhart, Andrew S., and Alexander Idnurm. 2023. "A Polyphasic Approach including Whole Genome Sequencing Reveals Paecilomyces paravariotii sp. nov. as a Cryptic Sister Species to P. variotii" Journal of Fungi 9, no. 3: 285. https://doi.org/10.3390/jof9030285
APA StyleUrquhart, A. S., & Idnurm, A. (2023). A Polyphasic Approach including Whole Genome Sequencing Reveals Paecilomyces paravariotii sp. nov. as a Cryptic Sister Species to P. variotii. Journal of Fungi, 9(3), 285. https://doi.org/10.3390/jof9030285





