Identification of Mycoparasitism-Related Genes against the Phytopathogen Botrytis cinerea via Transcriptome Analysis of Trichoderma harzianum T4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Source of Fungal Strains
2.2. Culture Conditions
2.3. Total RNA Extraction, cDNA Synthesis and Sequencing
2.4. Analysis of DEGs
2.5. RT-qPCR
2.6. Analysis of the Expression of Genes Related to Mycoparasitism in the Dual Culture
3. Results
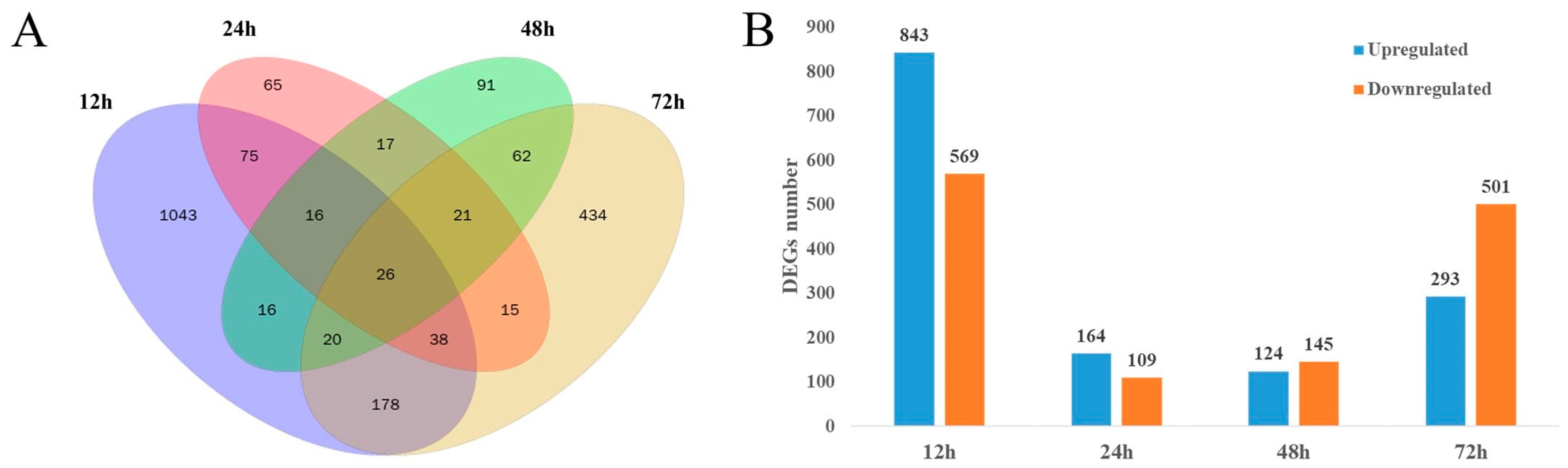
3.1. Data Analysis of the Transcriptome
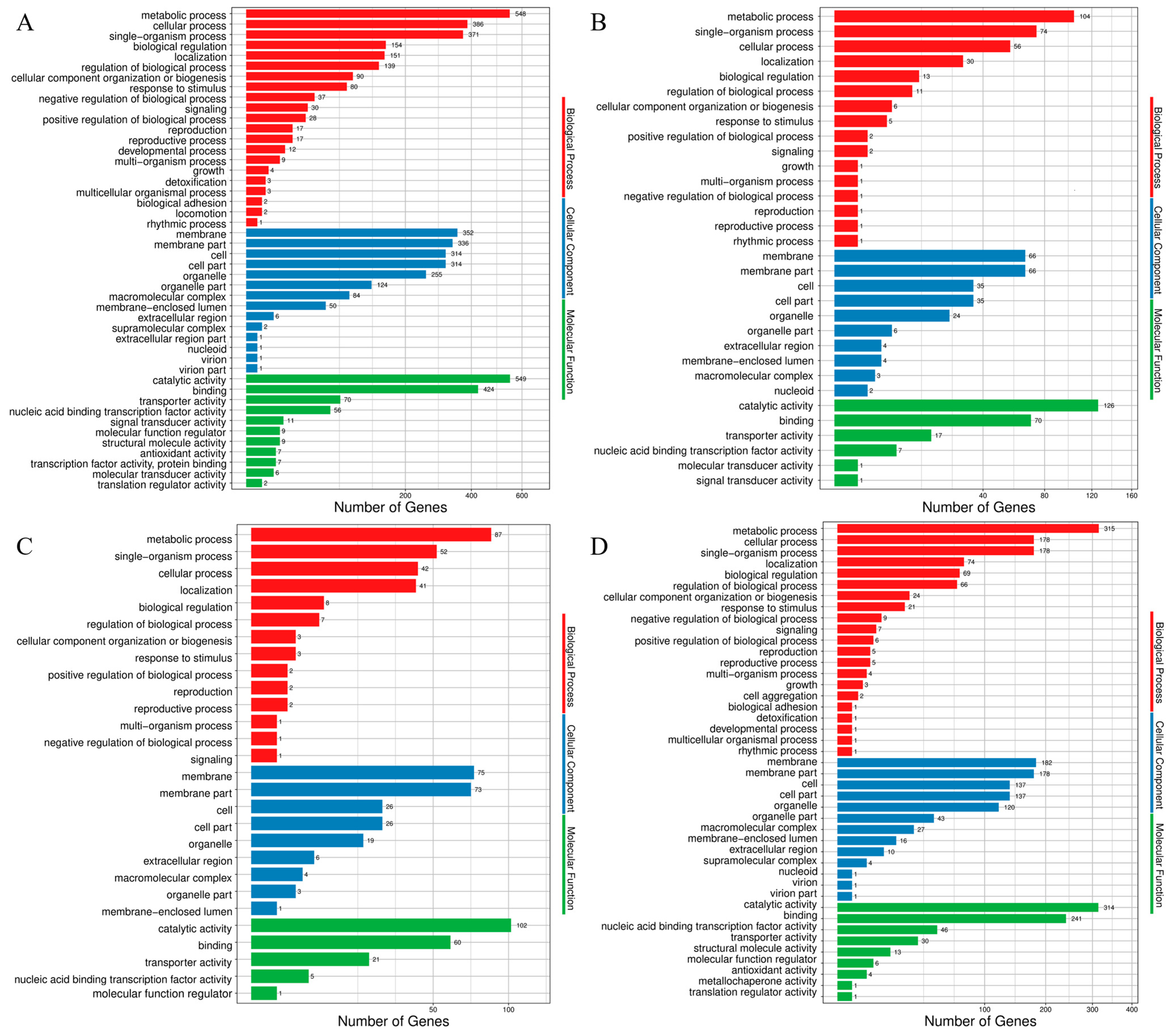
3.2. GO Enrichment Analysis of DEGs
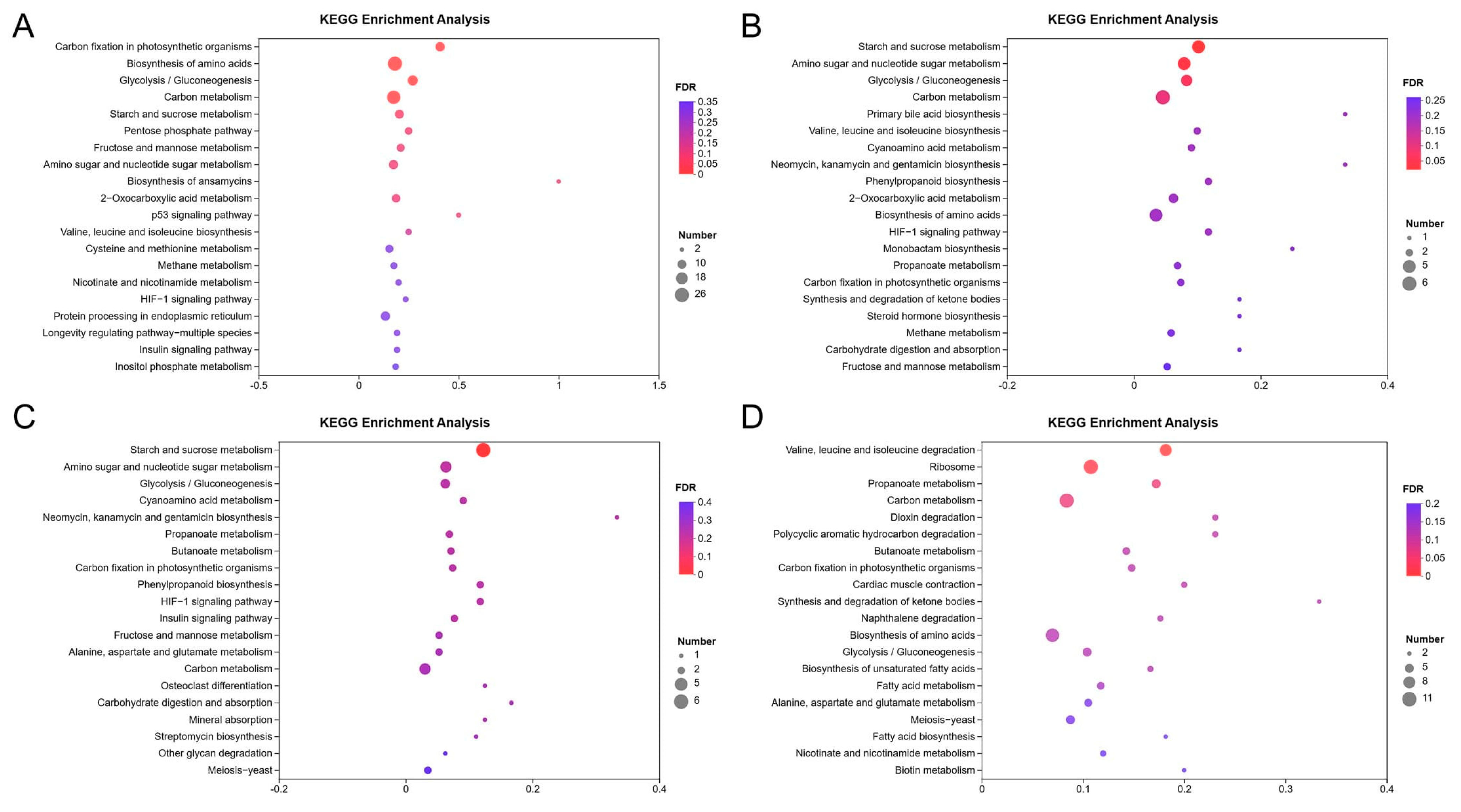
3.3. KEGG Enrichment Analysis of DEGs
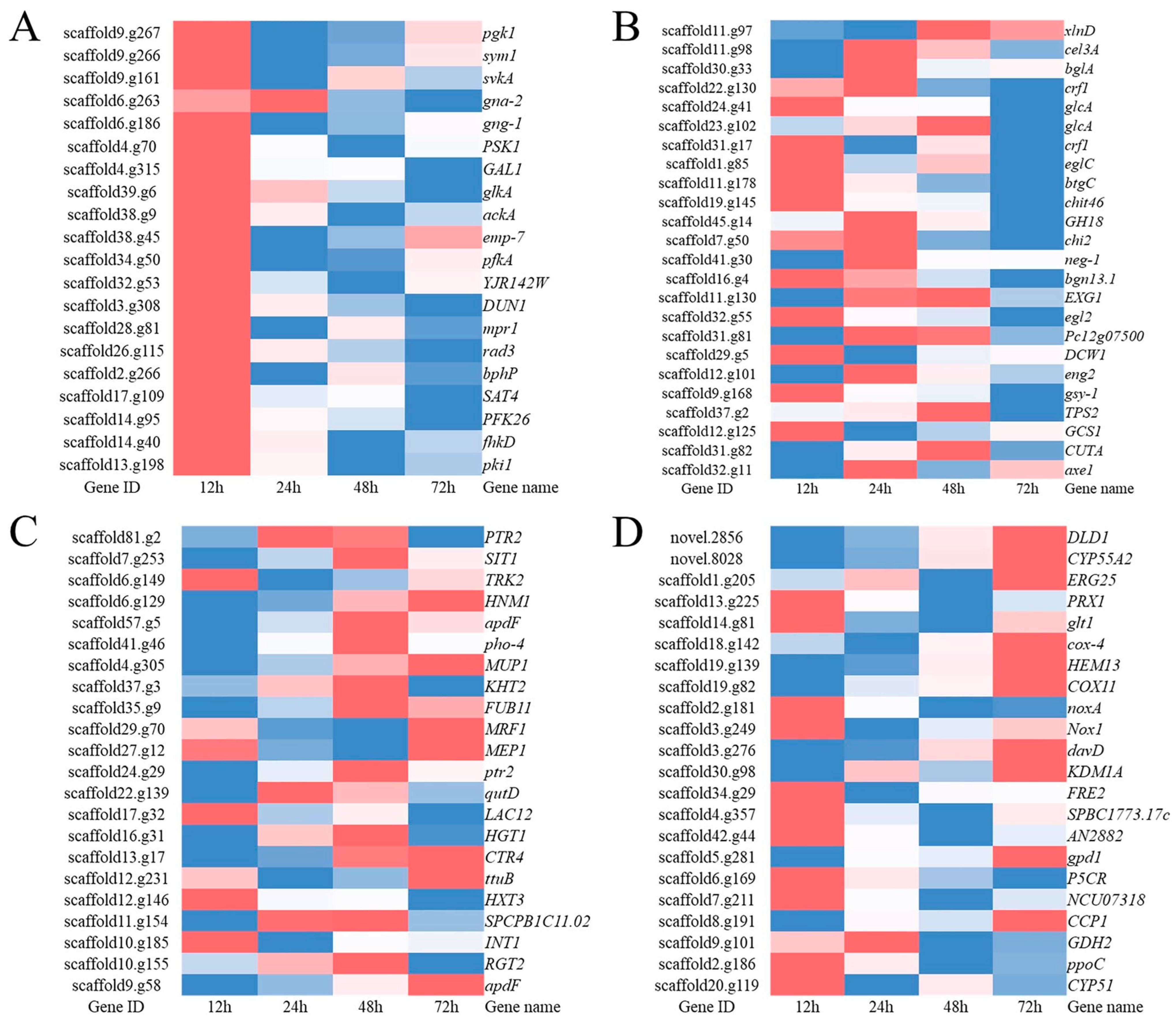
3.4. Analysis of Genes Related to T. harzianum Mycoparasitism Based on GO and KEGG Enrichment
3.4.1. Kinase Activity and Signal Transducer Activity
3.4.2. Carbohydrate Active Enzymes (CAZymes)
3.4.3. Transmembrane Transport
3.4.4. Antioxidant Enzymes
3.5. The Top 10 Significantly Upregulated Genes
3.6. Real-Time Quantitative PCR Verification (RT-qPCR)
3.7. Dual-Culture of T. harzianum T4 and B. cinerea to Verify the Genes Related to Mycoparasitism
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fendrihan, S.; Lixandru, M.; Dinu, S. Control methods of Botrytis cinerea. Int. J. Life Sci. Technol. 2018, 11, 31–36. [Google Scholar]
- Yang, Q.; Song, L.; Miao, Z.; Su, M.; Liang, W.; He, Y. Acetylation of BcHpt Lysine 161 Regulates Botrytis cinerea Sensitivity to Fungicides, Multistress Adaptation and Virulence. Front. Microbiol. 2020, 10, 2965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vela-Corcía, D.; Aditya Srivastava, D.; Dafa-Berger, A.; Rotem, N.; Barda, O.; Levy, M. MFS transporter from Botrytis cinerea provides tolerance to glucosinolate-breakdown products and is required for pathogenicity. Nat. Commun. 2019, 10, 2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Cheng, J.; Tang, J.; Fu, Y.; Jiang, D.; Baker, T.S.; Ghabrial, S.A.; Xie, J. A novel partitivirus that confers hypovirulence on plant pathogenic fungi. J. Virol. 2014, 88, 10120–10133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, T.; Lei, L.; He, L.; Yi, J.; Li, L.; Dai, L.; Hong, Y. Symbiotic Fungus Affected the Asian Citrus Psyllid (ACP) Resistance to Imidacloprid and Thiamethoxam. Front. Microbiol. 2020, 11, 522164. [Google Scholar] [CrossRef]
- Wang, X.; Gong, C.; Zhao, Y.; Shen, L. Transcriptome and Resistance-Related Genes Analysis of Botrytis cinerea B05.10 Strain to Different Selective Pressures of Cyprodinil and Fenhexamid. Front. Microbiol. 2018, 9, 2591. [Google Scholar] [CrossRef]
- Soulie, M.C.; Koka, S.M.; Floch, K.; Vancostenoble, B.; Barbe, D.; Daviere, A.; Soubigou-Taconnat, L.; Brunaud, V.; Poussereau, N.; Loisel, E.; et al. Plant nitrogen supply affects the Botrytis cinerea infection process and modulates known and novel virulence factors. Mol. Plant Pathol. 2020, 21, 1436–1450. [Google Scholar] [CrossRef]
- Calderón, C.E.; Rotem, N.; Harris, R.; Vela-Corcía, D.; Levy, M. Pseudozyma aphidis activates reactive oxygen species production, programmed cell death and morphological alterations in the necrotrophic fungus Botrytis cinerea. Mol. Plant Pathol. 2019, 20, 562–574. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.; Ser, H.L.; Khan, T.M.; Chuah, L.H.; Pusparajah, P.; Chan, K.G.; Goh, B.H.; Lee, L.H. The Potential of Streptomyces as Biocontrol Agents against the Rice Blast Fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 2017, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Kottb, M.; Gigolashvili, T.; Großkinsky, D.K.; Piechulla, B. Trichoderma volatiles effecting Arabidopsis: From inhibition to protection against phytopathogenic fungi. Front. Microbiol. 2015, 6, 995. [Google Scholar] [CrossRef] [Green Version]
- Wu, Q.; Ni, M.; Wang, G.; Liu, Q.; Yu, M.; Tang, J. Omics for understanding the tolerant mechanism of Trichoderma asperellum TJ01 to organophosphorus pesticide dichlorvos. BMC Genom. 2018, 19, 596. [Google Scholar] [CrossRef] [PubMed]
- Wenliang, P.; Wei, W. Preparation and Field Efficacy of Chlamydospore Water Dispersible Granule of Trichoderma harzianum. Chin. J. Biol. Control 2020, 36, 241–248. [Google Scholar]
- Xia, F.; Zhang, Y.; Re, X.U.; Wang, W. Effect of Trichoderma harzianum T4 on Bacterial Community in Watermelon (Citrullus lanatus) Rhizosphere Soil. Chin. J. Biol. Control 2013, 29, 232–241. [Google Scholar]
- Yu, X.X.; Zhao, Y.T.; Cheng, J.; Wang, W. Biocontrol effect of Trichoderma harzianum T4 on brassica clubroot and analysis of rhizosphere microbial communities based on T-RFLP. Biocontrol Sci. Technol. 2015, 25, 1493–1505. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, Y.; Wang, X.; Wang, W. Exogenous Regulators Enhance the Yield and Stress Resistance of Chlamydospores of the Biocontrol Agent Trichoderma harzianum T4. J. Fungi 2022, 8, 1017. [Google Scholar] [CrossRef]
- Moreno-Ruiz, D.; Lichius, A.; Turrà, D.; Di Pietro, A.; Zeilinger, S. Chemotropism Assays for Plant Symbiosis and Mycoparasitism Related Compound Screening in Trichoderma atroviride. Front. Microbiol. 2020, 11, 601251. [Google Scholar] [CrossRef]
- Harman, G.E. Overview of Mechanisms and Uses of Trichoderma spp. Phytopathology 2006, 96, 190–194. [Google Scholar] [CrossRef] [Green Version]
- Borin, G.P.; Sanchez, C.C.; de Santana, E.S.; Zanini, G.K.; dos Santos, R.A.C.; de Oliveira Pontes, A.; de Souza, A.T.; Dal’Mas, R.M.M.T.S.; Riaño-Pachón, D.M.; Goldman, G.H.; et al. Comparative transcriptome analysis reveals different strategies for degradation of steam-exploded sugarcane bagasse by Aspergillus niger and Trichoderma reesei. BMC Genom. 2017, 18, 501. [Google Scholar] [CrossRef] [Green Version]
- Steindorff, A.S.; Soller Ramada, M.H.; Guedes Coelho, A.S. Identification of mycoparasitism-related genes against the phytopathogen Sclerotinia sclerotiorum through transcriptome and expression profile analysis in Trichoderma harzianum. BMC Genom. 2014, 15, 204. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumar, K.; Wang, M.-H. Isolation and molecular identification of Trichoderma species from wetland soil and their antagonistic activity against phytopathogens. Physiol. Mol. Plant Pathol. 2020, 109, 101458. [Google Scholar] [CrossRef]
- Liu, N.; Bao, Z.; Li, J.; Ao, X.; Zhu, J.; Chen, Y. Identification of differentially expressed genes from Trichoderma atroviride strain SS003 in the presence of cell wall of Cronartium ribicola. Genes Genom. 2017, 39, 473–484. [Google Scholar] [CrossRef]
- Steindorff, A.S.; Silva, R.d.N.; Coelho, A.S.G.; Nagata, T.; Noronha, E.F.; Ulhoa, C.J. Trichoderma harzianum expressed sequence tags for identification of genes with putative roles in mycoparasitism against Fusarium solani. Biol. Control 2012, 61, 134–140. [Google Scholar] [CrossRef]
- Vieira, P.; Coelho, A.; Steindorff, A.; deSiqueira, S.; Silva, R.; Ulhoa, C. Identification of differentially expressed genes from Trichoderma harzianum during growth on cell wall of Fusarium solani as a tool for biotechnological application. BMC Genom. 2013, 14, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullnig, C.; Mach, R.L.; Lorito, M.; Kubicek, C.P. Enzyme diffusion from Trichoderma atroviride (=T. harzianum P1) to Rhizoctonia solani is a prerequisite for triggering of Trichoderma ech42 gene expression before mycoparasitic contact. Appl. Environ. Microbiol. 2000, 66, 2232–2234. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Yang, X.; Sun, D.; Song, J.; Chen, G.; Juba, O.; Yang, Q. Expressed sequence tags-based identification of genes in a biocontrol strain Trichoderma asperellum. Mol. Biol. Rep. 2010, 37, 3673–3681. [Google Scholar] [CrossRef]
- Troian, R.; Steindorff, A.; Ramada, M.; Arruda, W.; Ulhoa, C. Mycoparasitism studies of Trichoderma harzianum against Sclerotinia sclerotiorum: Evaluation of antagonism and expression of cell wall-degrading enzymes genes. Biotechnol. Lett. 2014, 36, 2095–2101. [Google Scholar] [CrossRef]
- Sun, Z.; Sun, M.; Li, S. Identification of mycoparasitism-related genes in Clonostachys rosea 67-1 active against Sclerotinia sclerotiorum. Sci. Rep. 2015, 14, 18169. [Google Scholar] [CrossRef]
- Suárez, M.B.; Sanz, L.; Chamorro, M.I.; Rey, M.; González, F.J.; Llobell, A.; Monte, E. Proteomic analysis of secreted proteins from Trichoderma harzianum. Identification of a fungal cell wall-induced aspartic protease. Fungal Genet. Biol. 2005, 42, 924–934. [Google Scholar] [CrossRef]
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Moreno-Ruiz, D.; Salzmann, L.; Fricker, M.D.; Zeilinger, S.; Lichius, A. Stress-Activated Protein Kinase Signalling Regulates Mycoparasitic Hyphal-Hyphal Interactions in Trichoderma atroviride. J. Fungi 2021, 7, 365. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, M.; Li, L.; Dong, Y.; Jiang, Y.; Liu, K.; Zhang, R.; Jiang, B.; Niu, K.; Fang, X. Role of Trichoderma reesei mitogen-activated protein kinases (MAPKs) in cellulase formation. Biotechnol. Biofuels 2017, 10, 99. [Google Scholar] [CrossRef]
- Chen, F.; Chen, X.-Z.; Su, X.-Y.; Qin, L.-N.; Huang, Z.-B.; Tao, Y.; Dong, Z.-Y. An Ime2-like mitogen-activated protein kinase is involved in cellulase expression in the filamentous fungus Trichoderma reesei. Biotechnol. Lett. 2015, 37, 2055–2062. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Hinterdobler, W.; Monroy, A.A.; Bazafkan, H.; Schmoll, M. The Kinase USK1 Regulates Cellulase Gene Expression and Secondary Metabolite Biosynthesis in Trichoderma reesei. Front. Microbiol. 2020, 11, 974. [Google Scholar] [CrossRef]
- Mohanan, V.C.; Chandarana, P.M.; Chattoo, B.B.; Patkar, R.N.; Manjrekar, J. Fungal Histidine Phosphotransferase Plays a Crucial Role in Photomorphogenesis and Pathogenesis in Magnaporthe oryzae. Front. Chem. 2017, 5, 31. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Copeland, D.M.; Soares, A.S.; West, A.H. Crystal Structure of a Complex between the Phosphorelay Protein YPD1 and the Response Regulator Domain of SLN1 Bound to a Phosphoryl Analog. J. Mol. Biol. 2008, 375, 1141–1151. [Google Scholar] [CrossRef] [Green Version]
- Carapia-Minero, N.; Castelán-Vega, J.A.; Pérez, N.O.; Rodríguez-Tovar, A.V. The phosphorelay signal transduction system in Candida glabrata: An in silico analysis. J. Mol. Model. 2018, 24, 13. [Google Scholar] [CrossRef]
- Rodrigues, C.M.; Takita, M.A.; Silva, N.V.; Ribeiro-Alves, M.; Machado, M.A. Comparative genome analysis of Phyllosticta citricarpa and Phyllosticta capitalensis, two fungi species that share the same host. BMC Genom. 2019, 20, 554. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Sailsbery, J.K.; Eyre, A.W.; Dean, R.A. Comparative genome analysis and genome evolution of members of the magnaporthaceae family of fungi. BMC Genom. 2016, 17, 135. [Google Scholar] [CrossRef] [Green Version]
- Geremía, R.A.; Goldman, G.H.; Jacobs, D.; Ardrtes, W.; Vila, S.B.; van Montagu, M.C.; Herrera-Estrella, A. Molecular characterization of the proteinase-encoding gene, prb1, related to mycoparasitism by Trichoderma harzianum. Mol. Microbiol. 1993, 8, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Lorito, M.; Woo, S.L.; Garcia, I.; Colucci, G.; Harman, G.E.; Pintor-Toro, J.A.; Filippone, E.; Muccifora, S.; Lawrence, C.B.; Zoina, A.; et al. Genes from mycoparasitic fungi as a source for improving plant resistance to fungal pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 7860–7865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jatav, P.; Ahirwar, S.S.; Gupta, A.; Kushwaha, K.; Jatav, S. Antagonistic activity of cellulase enzyme produced by Trichoderma Viride against Xanthomonas Citri. Indian J. Agric. Res. 2018, 52, 497–504. [Google Scholar]
- Saravanakumar, K.; Dou, K.; Lu, Z.; Wang, X.; Li, Y.; Chen, J. Enhanced biocontrol activity of cellulase from Trichoderma harzianum against Fusarium graminearum through activation of defense-related genes in maize. Physiol. Mol. Plant Pathol. 2018, 103, 130–136. [Google Scholar] [CrossRef]
- Ghasemi, S.; Safaie, N.; Shahbazi, S.; Shams-Bakhsh, M.; Askari, H. The Role of Cell Wall Degrading Enzymes in Antagonistic Traits of Trichoderma virens against Rhizoctonia solani. Iran. J. Biotechnol. 2020, 18, e2333. [Google Scholar]
- Monteiro, V.N.; Ulhoa, C.J. Biochemical Characterization of a β-1,3-Glucanase from Trichoderma koningii Induced by Cell Wall of Rhizoctonia solani. Curr. Microbiol. 2006, 52, 92–96. [Google Scholar] [CrossRef]
- Martin, K.; McDougall, B.M.; McIlroy, S.; Jayus Chen, J.; Seviour, R.J. Biochemistry and molecular biology of exocellular fungal β-(1,3)- and β-(1,6)-glucanases. FEMS Microbiol. Rev. 2007, 31, 168–192. [Google Scholar] [CrossRef]
- Lingner, U.; Münch, S.; Deising, H.B.; Sauer, N. Hexose transporters of a hemibiotrophic plant pathogen: Functional variations and regulatory differences at different stages of infection. J. Biol. Chem. 2011, 286, 20913–20922. [Google Scholar] [CrossRef] [Green Version]
- Wei, H.; Vienken, K.; Weber, R.; Bunting, S.; Requena, N.; Fischer, R. A putative high affinity hexose transporter, hxtA, of Aspergillus nidulans is induced in vegetative hyphae upon starvation and in ascogenous hyphae during cleistothecium formation. Fungal Genet. Biol. B 2004, 41, 148–156. [Google Scholar] [CrossRef]
- Chikamori, M.; Fukushima, K. A new hexose transporter from Cryptococcus neoformans: Molecular cloning and structural and functional characterization. Fungal Genet. Biol. 2005, 42, 646–655. [Google Scholar] [CrossRef]
- Droce, A.; Holm, K.; Olsson, S.; Frandsen, R.; Sondergaard, T.; Sorensen, J.; Giese, H. Expression profiling and functional analyses of BghPTR2, a peptide transporter from Blumeria graminis f. sp. hordei. Fungal Biol. 2015, 119, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Marini, A.M.; Vissers, S.; Urrestarazu, A.; André, B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994, 13, 3456–3463. [Google Scholar] [CrossRef] [PubMed]
- Tangen, K.L.; Jung, W.H.; Sham, A.P.; Lian, T.; Kronstad, J.W. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 2007, 153, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguiar, M.; Orasch, T.; Misslinger, M.; Dietl, A.-M.; Gsaller, F.; Haas, H. The Siderophore Transporters Sit1 and Sit2 Are Essential for Utilization of Ferrichrome-, Ferrioxamine- and Coprogen-Type Siderophores in Aspergillus fumigatus. J. Fungi 2021, 7, 768. [Google Scholar] [CrossRef]
- Dietl, A.-M.; Misslinger, M.; Aguiar, M.M.; Ivashov, V.; Teis, D.; Pfister, J.; Decristoforo, C.; Hermann, M.; Sullivan, S.M.; Smith, L.R.; et al. The Siderophore Transporter Sit1 Determines Susceptibility to the Antifungal VL-2397. Antimicrob. Agents Chemother. 2019, 63, e00807–e00819. [Google Scholar] [CrossRef]
- Chand Arya, G.; Aditya Srivastava, D.; Manasherova, E.; Prusky, D.B.; Elad, Y.; Frenkel, O.; Harel, A. BcHnm1, a predicted choline transporter, modulates conidial germination and virulence in Botrytis cinerea. Fungal Genet. Biol. 2022, 158, 103653. [Google Scholar] [CrossRef]
- Chauhan, N.; Latge, J.-P.; Calderone, R. Signalling and oxidant adaptation in Candida albicans and Aspergillus fumigatus. Nat. Rev. Microbiol. 2006, 4, 435–444. [Google Scholar] [CrossRef]
- Zhu, X.; Sayari, M.; Islam, M.R.; Daayf, F. NOXA Is Important for Verticillium dahliae’s Penetration Ability and Virulence. J. Fungi 2021, 7, 814. [Google Scholar] [CrossRef]
- Sun, C.C.; Dong, W.R.; Shao, T.; Li, J.Y.; Zhao, J.; Nie, L.; Xiang, L.X.; Zhu, G.; Shao, J.Z. Peroxiredoxin 1 (Prx1) is a dual-function enzyme by possessing Cys-independent catalase-like activity. Biochem. J. 2017, 474, 1373–1394. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Kim, K.-J. Crystal structure of γ-aminobutyrate aminotransferase in complex with a PLP-GABA adduct from Corynebacterium glutamicum. Biochem. Biophys. Res. Commun. 2019, 514, 601–606. [Google Scholar] [CrossRef]
- Forlani, G.; Nocek, B.; Chakravarthy, S.; Joachimiak, A. Functional Characterization of Four Putative δ1-Pyrroline-5-Carboxylate Reductases from Bacillus subtilis. Front. Microbiol. 2017, 8, 1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, R.; Meena, M.; Prasad, V.; Zehra, A.; Gupta, V. Mannitol metabolism during pathogenic fungal–host interactions under stressed conditions. Front. Microbiol. 2015, 6, 1019. [Google Scholar]
- Hur, G.H.; Vickery, C.R.; Burkart, M.D. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat. Prod. Rep. 2012, 29, 1074–1098. [Google Scholar] [CrossRef] [Green Version]
- Niu, X.; Thaochan, N.; Hu, Q. Diversity of Linear Non-Ribosomal Peptide in Biocontrol Fungi. J. Fungi 2020, 6, 61. [Google Scholar] [CrossRef]
- Petrov, A.; Meskauskas, A.; Dinman, J.D. Ribosomal protein L3: Influence on ribosome structure and function. RNA Biol. 2004, 1, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Cohen, E.; Gamliel, A.; Katan, J. Glutathione and glutathione-S-transferase in fungi: Effect of pentachloronitrobenzene and 1-choro-2,4-dinitrobenzene; purification and characterization of the transferase from Fusarium. Pestic. Biochem. Physiol. 1986, 26, 1–9. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Kirly, L.; Schrder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munyampundu, J.; Xu, Y.; Cai, X. Phi Class of Glutathione S-transferase Gene Superfamily Widely Exists in Nonplant Taxonomic Groups. Evol. Bioinform. Online 2016, 12, 59–71. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Chi, Y.; Li, S.; Gu, X.; Ye, Y. Cloning, homology modeling, heterologous expression and bioinformatic analysis of Ure2pA glutathione S-transferase gene from white rot fungus Trametes gibbosa. Biotechnol. Biotechnol. Equip. 2021, 35, 1560–1573. [Google Scholar] [CrossRef]
- Calmes, B.; Morel-Rouhier, M.; Bataillé-Simoneau, N.; Gelhaye, E.; Guillemette, T.; Simoneau, P.; Simoneau, P. Characterization of glutathione transferases involved in the pathogenicity of Alternaria brassicicola. BMC Microbiol. 2015, 15, 123. [Google Scholar] [CrossRef] [Green Version]
- Imai, S.-I.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Vall-llaura, N.; Mir, N.; Garrido, L.; Vived, C.; Cabiscol, E. Redox control of yeast Sir2 activity is involved in acetic acid resistance and longevity. Redox Biol. 2019, 24, 101229. [Google Scholar] [CrossRef] [PubMed]
- Lin, W. A trehalose-6-phosphate synthase gene from Phellinus igniarius. J. Chem. Pharm. Res. 2015, 7, 343–347. [Google Scholar]
- Vicente, R.L.; Spina, L.; Gómez, J.P.L.; Dejean, S.; Parrou, J.-L.; François, J.M. Trehalose-6-phosphate promotes fermentation and glucose repression in Saccharomyces cerevisiae. Microb. Cell 2018, 5, 444–459. [Google Scholar] [CrossRef]
- Maddi, A.; Dettman, A.; Fu, C.; Seiler, S.; Free, S.J.; Zhou, R. WSC-1 and HAM-7 Are MAK-1 MAP Kinase Pathway Sensors Required for Cell Wall Integrity and Hyphal Fusion in Neurospora crassa. PLoS ONE 2012, 7, e42374. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.; Chen, Y.; Zhu, J.; Ying, S.; Feng, M. Subcellular localization of five singular WSC domain-containing proteins and their roles in Beauveria bassiana responses to stress cues and metal ions. Environ. Microbiol. Rep. 2016, 8, 295–304. [Google Scholar] [CrossRef]
- Dobrowolski, A.; Mirończuk, A.M. The influence of transketolase on lipid biosynthesis in the yeast Yarrowia lipolytica. Microb. Cell Factories 2020, 19, 138. [Google Scholar] [CrossRef]
- Iwata, H.; Kobayashi, Y.; Mizushima, D.; Watanabe, T.; Ogihara, J.; Kasumi, T. Complementary function of two transketolase isoforms from Moniliella megachiliensis in relation to stress response. AMB Express 2017, 7, 45. [Google Scholar] [CrossRef] [Green Version]




| Putative Function | Gene Name | Primers for qPCR (5′ to 3′) | |
|---|---|---|---|
| Forward | Reverse | ||
| Glucan endo-1,3-beta-glucosidase | bgn13.1 | CAGCAGTAGCAATGAATGTAAG | GAAGTCCTCAGCGATGTG |
| Cytochrome P450 55A1 | CYP55A2 | CATTGCCAGTTCCATCAT | CTTCCGTTGGTTCGTATG |
| D-lactate dehydrogenase | DLD1 | GGACAGCATCAACCAGAC | CCGACCAGAGCGTATGAG |
| 4-aminobutyrate aminotransferase | gatA | TGACTTCTCCTCCTGACAT | TGTTGAACTGGCGGTAAG |
| Pyruvate decarboxylase | pdcA | ACAACGACATCTCTAACTG | ATTCCTCCTTGGTCTTGA |
| Glyceraldehyde-3-phosphate dehydrogenase | gpd1 | GTCTTGGTTGTCGCTGAG | CCCTTGTCCTTCCTCTGG |
| Aspartyl-tRNA synthetase | DPS1 | TGCTGTTTCTTCGGATGA | CTTTCCAGATTCCCACAATAG |
| Sedoheptulose-1,7-bisphosphatase | E3.1.3.37 | AGACACATGACCGAGTTC | AATAGCACGCCTTCCAATA |
| Mutanase precursor | Pc12g07500 | CCGCCAATGTTGCTATTCGG | ATCATAGTCTGCCGAGCTGC |
| Glucan endo-1,3-beta-glucosidase | eglC | CGGCACTCTGGTCAACTACA | AAGGGTATGTGTCCATGCCG |
| Peroxidase | PRX1 | CGACAACTGGGTCGTCTTCT | ATCCAGCCATTGTGGGAGTC |
| Acid phosphatase | aphA | GCGAATCCTCCGTTCTGGTT | GCAAAGTCCGTTTCCGTGTC |
| HAL protein kinase | SAT4 | GCTGCAGATGCTCAACCCTA | TTGCCGCCGTAATCCTTCTT |
| Guanine nucleotide-binding protein gamma subunit 1 | gng-1 | ACTACGCGAGGATTTGGACC | CTTTGGCACGGTTCCCCATA |
| Nitrilase | blr3397 | AAGTGAGGCATGGACGAAGG | TTCGTGATTGCTCTTCCCCC |
| Glutathione S-transferase | GST, gst | GCATCAGTAAGTTTGGCGGC | CGGTCAATGTACTCTCCCGC |
| Murein transglycosylase | btgC | GAGAGCGAACCTCTCGATGG | GAGGTTGATGCTGGTCGTGA |
| Exo-beta-1,3-glucanase | EXG1 | CGCGGTCTGTTGGTTGAAAG | CTGGAATTGGGTTGCGTTGG |
| Glutamate/leucine/phenylalanine/valine dehydrogenase | GDH2 | GGGATACCGTGTGCAGTTCA | TCTCGTTGTCAGACTTGCCC |
| Endochitinase 42 | chit46 | CAGACGGCACAGTTGTCTCT | CAGAGGGGAAGTTGGTGGAC |
| Beta-glucosidase | bglA | GACACTGCGATCCAGAACCA | GTAACCCTCACCACTGTCCG |
| Hexokinase | glkA | CGCTAGATCGAGACAGCGTT | GACCAGTCATTGGTGCTGGA |
| Aspyridones efflux protein | apdF | CACAACGAGTGTCTCGACCA | GACATGTTGGCAATCGTCGG |
| Leucine-2 | LEUA | AAGCTGTCCGAGTACACTGC | GCATGCCGGTTGAATCACTC |
| Lysine-specific histone demethylase 1 | VPS33 | TCGGAGAAAGCGGATGCAAT | AAGAAGCGTCCCCGATTTGT |
| Osomolarity two-component system, phosphorelay intermediate protein YPD1 | mpr1 | TAAAGGTCCGAGACGGTTGC | AGACTTGACTGCTGTGAGCG |
| Trehalose 6-phosphate synthase | TPS2 | ATGGCATGAACACGACCAGT | GGATTGCATCGCGGAGACTA |
| Delta-aminolevulinic acid dehydratase | HEM2 | TGTCTGTGCCGCAGTACTAAT | GTCAGAGTAGATGCCGGGAG |
| Glutamate synthase precursor | glt1 | AATGGGCTGCTTGCCAAATG | GAGGCGCAAATGATTCGGTC |
| 6-phosphofructo-2-kinase | PFK26 | ATTGAGCGCATCACTGACCA | AATGCCATAAGGCTTGGGCT |
| Pyruvate decarboxylase | pdcA | CAAGTACCTCCGAGCTGCAA | CTCTTCGTTGACAGCACCCT |
| Mannitol-1-phosphate dehydrogenase | mtlD | GCAACCCTCACCTGGAAGAC | CTGGAAGCGGAACGTCATCT |
| 18s rRNA | 18s rRNA | CAACCATAAACGATGCCGA | AGCCTTGCGACCATACTCC |
| α-tubulin | α-tubulin | TATCTGCTACCAGGCTCCCGAGAA | TGGTGTTGGACAGCATGCAGACAG |
| Culture Time | Regulated | Pathway Hierarchy1 | Pathway Hierarchy2 | KEGG Pathway | Pathway ID | Gene Number | Background Number | q-Value |
|---|---|---|---|---|---|---|---|---|
| 12 h | Up | Metabolism | Energy metabolism | Carbon fixation in photosynthetic organisms | ko00710 | 11 | 27 | 1.46 × 10−4 |
| Metabolism | Overview | Biosynthesis of amino acids | ko01230 | 26 | 143 | 4.15 × 10−4 | ||
| Metabolism | Carbohydrate metabolism | Glycolysis/Gluconeogenesis | ko00010 | 13 | 48 | 9.70 × 10−4 | ||
| Metabolism | Overview | Carbon metabolism | ko01200 | 23 | 131 | 1.36 × 10−3 | ||
| Down | Cellular processes | Transport and catabolism | Peroxisome | ko04146 | 20 | 53 | 1.66 × 10−11 | |
| Metabolism | Amino acid metabolism | Valine, leucine and isoleucine degradation | ko00280 | 14 | 44 | 6.14 × 10−7 | ||
| Metabolism | Lipid metabolism | Fatty acid degradation | ko00071 | 8 | 28 | 1.65 × 10−3 | ||
| Metabolism | Lipid metabolism | Primary bile acid biosynthesis | ko00120 | 3 | 3 | 2.86 × 10−3 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Benzoate degradation | ko00362 | 5 | 11 | 2.86 × 10−3 | ||
| Metabolism | Carbohydrate metabolism | Propanoate metabolism | ko00640 | 7 | 29 | 8.06 × 10−3 | ||
| Metabolism | Amino acid metabolism | Phenylalanine metabolism | ko00360 | 7 | 38 | 3.82 × 10−2 | ||
| 24 h | Up | Metabolism | Carbohydrate metabolism | Starch and sucrose metabolism | ko00500 | 5 | 49 | 2.12 × 10−2 |
| Metabolism | Carbohydrate metabolism | Amino sugar and nucleotide sugar metabolism | ko00520 | 5 | 63 | 3.41 × 10−2 | ||
| Down | Metabolism | Amino acid metabolism | Valine, leucine and isoleucine degradation | ko00280 | 11 | 44 | 8.28 × 10−11 | |
| Metabolism | Carbohydrate metabolism | Propanoate metabolism | ko00640 | 5 | 29 | 6.10 × 10−4 | ||
| Metabolism | Metabolism of other amino acids | Taurine and hypotaurine metabolism | ko00430 | 3 | 10 | 3.77 × 10−3 | ||
| Metabolism | Lipid metabolism | Fatty acid degradation | ko00071 | 4 | 28 | 3.78 × 10−3 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Benzoate degradation | ko00362 | 3 | 11 | 3.78 × 10−3 | ||
| Metabolism | Amino acid metabolism | Tryptophan metabolism | ko00380 | 5 | 59 | 5.59 × 10−3 | ||
| Metabolism | Amino acid metabolism | Tyrosine metabolism | ko00350 | 4 | 33 | 5.59 × 10−3 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Aminobenzoate degradation | ko00627 | 4 | 38 | 7.79 × 10−3 | ||
| Metabolism | Lipid metabolism | Synthesis and degradation of ketone bodies | ko00072 | 2 | 6 | 1.38 × 10−2 | ||
| Metabolism | Amino acid metabolism | Arginine biosynthesis | ko00220 | 3 | 25 | 1.90 × 10−2 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Styrene degradation | ko00643 | 3 | 27 | 2.16 × 10−2 | ||
| 48 h | Up | Metabolism | Carbohydrate metabolism | Starch and sucrose metabolism | ko00500 | 6 | 49 | 1.75 × 10−3 |
| Down | Metabolism | Biosynthesis of other secondary metabolites | Isoquinoline alkaloid biosynthesis | ko00950 | 3 | 11 | 2.63 × 10−3 | |
| Metabolism | Amino acid metabolism | Phenylalanine metabolism | ko00360 | 4 | 38 | 3.32 × 10−3 | ||
| Metabolism | Amino acid metabolism | Glycine, serine and threonine metabolism | ko00260 | 4 | 46 | 4.71 × 10−3 | ||
| Metabolism | Amino acid metabolism | Tryptophan metabolism | ko00380 | 4 | 59 | 9.24 × 10−3 | ||
| Metabolism | Amino acid metabolism | Tyrosine metabolism | ko00350 | 3 | 33 | 1.31 × 10−2 | ||
| Metabolism | Biosynthesis of other secondary metabolites | Tropane, piperidine and pyridine alkaloid biosynthesis | ko00960 | 2 | 8 | 1.31 × 10−2 | ||
| 72 h | Up | Metabolism | Amino acid metabolism | Valine, leucine and isoleucine degradation | ko00280 | 8 | 44 | 5.56 × 10−3 |
| Genetic information processing | Translation | Ribosome | ko03010 | 11 | 102 | 1.63 × 10−2 | ||
| Down | Metabolism | Lipid metabolism | Fatty acid degradation | ko00071 | 8 | 28 | 2.43 × 10−5 | |
| Cellular processes | Transport and catabolism | Peroxisome | ko04146 | 9 | 53 | 2.44 × 10−4 | ||
| Metabolism | Amino acid metabolism | Tryptophan metabolism | ko00380 | 8 | 59 | 3.03 × 10−3 | ||
| Metabolism | Overview | Fatty acid metabolism | ko01212 | 6 | 34 | 4.03 × 10−3 | ||
| Metabolism | Metabolism of terpenoids and polyketides | Geraniol degradation | ko00281 | 2 | 4 | 4.42 × 10−2 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Dioxin degradation | ko00621 | 3 | 13 | 4.42 × 10−2 | ||
| Metabolism | Xenobiotics biodegradation and metabolism | Polycyclic aromatic hydrocarbon degradation | ko00624 | 3 | 13 | 4.42 × 10−2 | ||
| Organismal Systems | Endocrine system | Adipocytokine signaling pathway | ko04920 | 3 | 13 | 4.42 × 10−2 | ||
| Metabolism | Amino acid metabolism | Valine, leucine and isoleucine degradation | ko00280 | 5 | 44 | 4.52 × 10−2 | ||
| Metabolism | Carbohydrate metabolism | Amino sugar and nucleotide sugar metabolism | ko00520 | 6 | 63 | 4.52 × 10−2 |
| Times of Induction | Gene ID | Gene Name | Putative Function | Log2FC |
|---|---|---|---|---|
| 12 h | Scaffold2.g181 | noxA | Superoxide-generating NADPH oxidase heavy chain subunit A | 9.14901 |
| Scaffold26.g60 | pha2 | Prephenate dehydratase | 8.255327 | |
| Scaffold3.g212 | QPCT | Glutaminyl-peptide cyclotransferase | 7.622853 | |
| Scaffold12.g184 | SPAC513.07 | NAD-dependent epimerase/dehydratase | 7.52363 | |
| Novel.3896 | ALDH6A1 | Methylmalonate-semialdehyde dehydrogenase, mitochondrial precursor | 6.657672 | |
| Novel.5121 | bgn13.1 | Endo-1,3(4)-beta-glucanase | 5.17254 | |
| Scaffold9.g19 | AIFM2 | Apoptosis-inducing factor | 4.56581 | |
| Scaffold4.g409 | FUB8 | Noncanonical nonribosomal peptide synthetase | 4.510308 | |
| Scaffold1.g129 | abf1 | Alpha-L-arabinofuranosidase B | 4.219728 | |
| Scaffold24.g41 | glcA | Glucan endo-1,3-beta-glucosidase | 4.149159 | |
| 24 h | Scaffold23.g56 | WSC domain-containing protein | 6.121065 | |
| Scaffold41.g30 | neg-1 | Endo-1,6-beta-D-glucanase | 4.952524 | |
| Novel.5121 | bgn13.1 | Endo-1,3(4)-beta-glucanase | 4.795401 | |
| Novel.5464 | ARB_02077 | Glucan endo-1,3-beta-glucosidase | 3.911272 | |
| Scaffold11.g98 | cel3A | Beta-glucosidase | 3.879044 | |
| Scaffold12.g101 | eng2 | Endo-1,3(4)-beta-glucanase | 3.805877 | |
| Scaffold12.g245 | prb1 | Subtilase | 3.391825 | |
| Scaffold18.g111 | mfnA | L-aspartate decarboxylase | 3.217549 | |
| Scaffold7.g216 | YEL023C | Uncharacterized protein | 3.112885 | |
| Scaffold45.g14 | chi2 | Endochitinase | 2.711266 | |
| 48 h | Scaffold23.g56 | WSC domain-containing protein | 5.267517 | |
| Novel.5121 | bgn13.1 | Endo-1,3(4)-beta-glucanase | 4.572012 | |
| Scaffold12.g245 | prb1 | Subtilase | 3.663304 | |
| Scaffold41.g30 | neg-1 | Endo-1,6-beta-D-glucanase | 3.622379 | |
| Scaffold31.g82 | CUTA | Cutinase | 3.521937 | |
| Scaffold3.g450 | arsH | NADPH-dependent FMN reductase | 3.228058 | |
| Scaffold11.g98 | cel3A | Beta-glucosidase | 3.177845 | |
| Scaffold8.g89 | gatA | 4-aminobutyrate aminotransferase | 2.890697 | |
| Scaffold10.g25 | YLL056C | Uncharacterized protein | 2.857343 | |
| Scaffold9.g58 | apdF | Aspyridones efflux protein | 2.820267 | |
| 72 h | Scaffold4.g229 | rpl-3 | 60S ribosomal protein L3 | 12.9414 |
| Novel.8070 | FRE7 | Ferric/cupric reductase transmembrane component 7 | 8.43363 | |
| Scaffold23.g56 | WSC domain-containing protein | 4.991655 | ||
| Scaffold65.g8 | hpcH | 4-hydroxy-2-oxo-heptane-1,7-dioate aldolase | 4.481846 | |
| Novel.5121 | bgn13.1 | Endo-1,3(4)-beta-glucanase | 4.327306 | |
| Scaffold8.g89 | gatA | 4-aminobutyrate aminotransferase | 4.17367 | |
| Scaffold5.g281 | gpd1 | Glyceraldehyde-3-phosphate dehydrogenase | 4.130136 | |
| Scaffold9.g58 | apdF | Aspyridones efflux protein | 4.031324 | |
| Novel.7601 | SPAC5H10.01 | Hydro-lyase C5H10.01 | 3.735899 | |
| Scaffold6.g107 | liuE | 3-hydroxy-3-isohexenylglutaryl-CoA/hydroxy-methylglutaryl-CoA lyase | 3.61478 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhu, X.; Wang, J.; Shen, C.; Wang, W. Identification of Mycoparasitism-Related Genes against the Phytopathogen Botrytis cinerea via Transcriptome Analysis of Trichoderma harzianum T4. J. Fungi 2023, 9, 324. https://doi.org/10.3390/jof9030324
Wang Y, Zhu X, Wang J, Shen C, Wang W. Identification of Mycoparasitism-Related Genes against the Phytopathogen Botrytis cinerea via Transcriptome Analysis of Trichoderma harzianum T4. Journal of Fungi. 2023; 9(3):324. https://doi.org/10.3390/jof9030324
Chicago/Turabian StyleWang, Yaping, Xiaochong Zhu, Jian Wang, Chao Shen, and Wei Wang. 2023. "Identification of Mycoparasitism-Related Genes against the Phytopathogen Botrytis cinerea via Transcriptome Analysis of Trichoderma harzianum T4" Journal of Fungi 9, no. 3: 324. https://doi.org/10.3390/jof9030324
APA StyleWang, Y., Zhu, X., Wang, J., Shen, C., & Wang, W. (2023). Identification of Mycoparasitism-Related Genes against the Phytopathogen Botrytis cinerea via Transcriptome Analysis of Trichoderma harzianum T4. Journal of Fungi, 9(3), 324. https://doi.org/10.3390/jof9030324






