Importance of Clinical Isolates in Cryptococcus neoformans Research
Abstract
1. Introduction
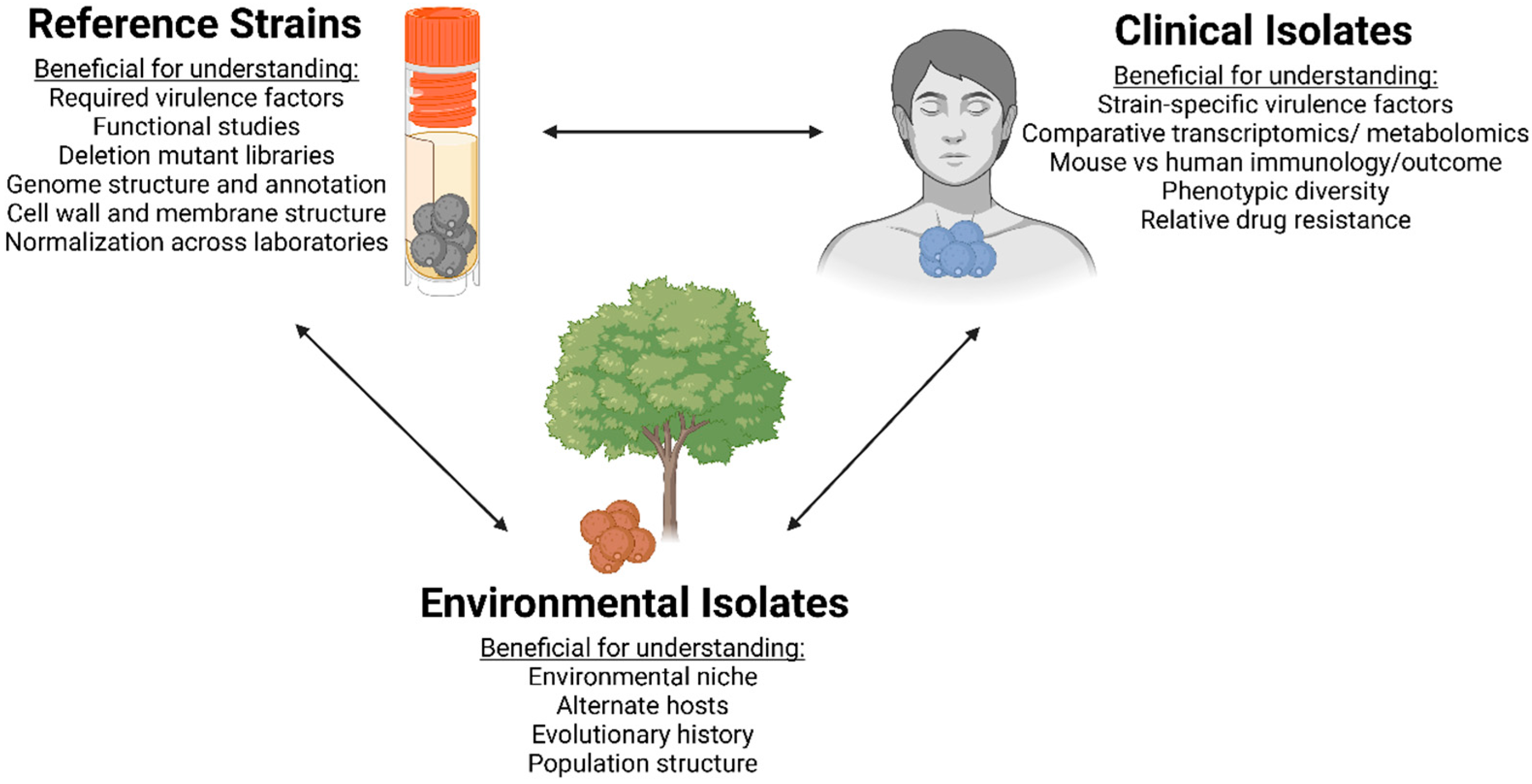
2. Advantages and Disadvantages of Reference Isolates
3. Clinical and Phenotypic Variation in Clinical Isolates
4. Genomic Plasticity in Clinical Isolates and Its Impact on Virulence
5. Genetic Polymorphisms in Clinical Isolates and Their Impact on Disease Outcomes
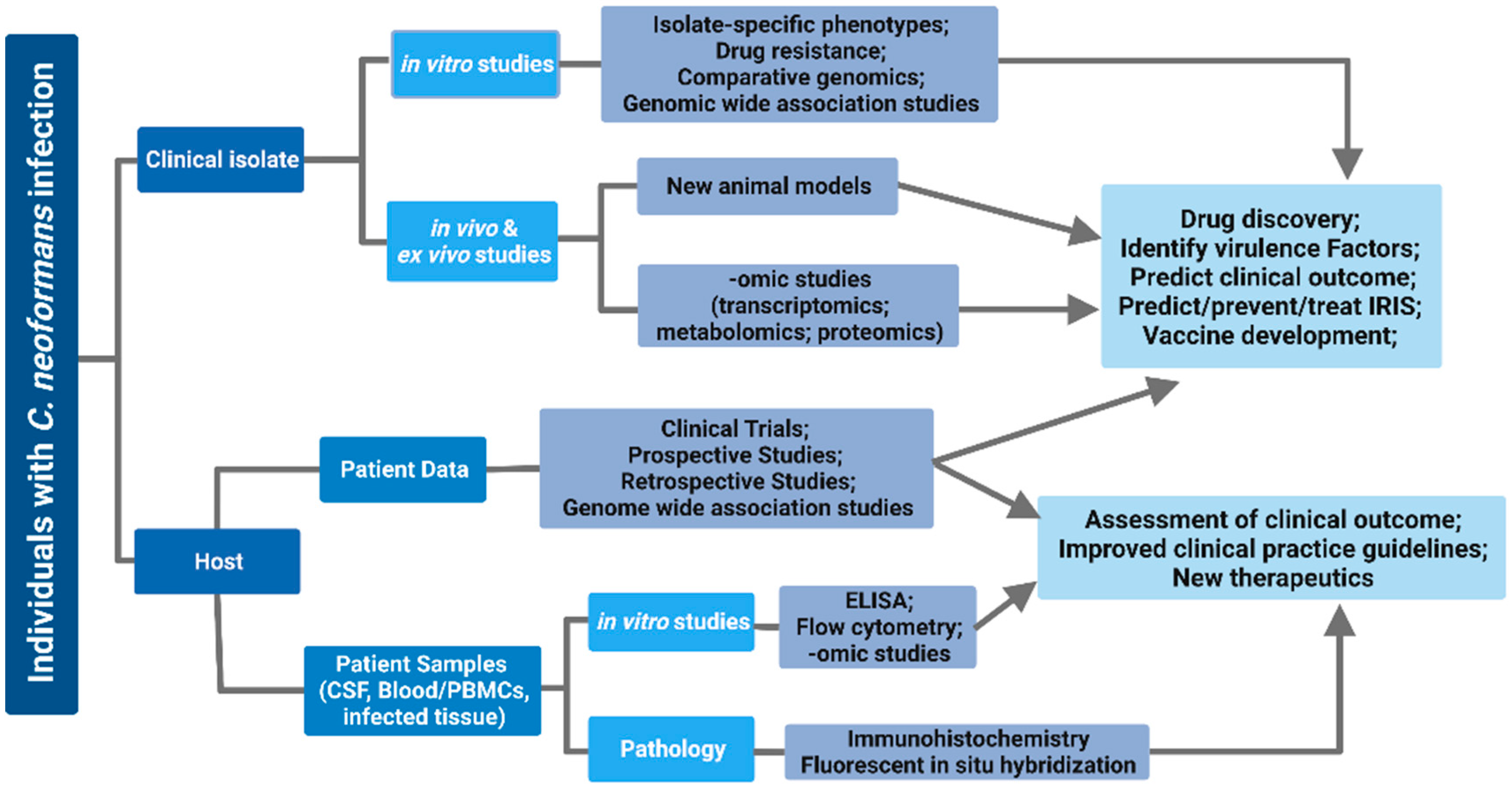
6. Use of Clinical Isolates to Study Host–Pathogen Interactions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Altamirano, S.; Jackson, K.M.; Nielsen, K. The interplay of phenotype and genotype in Cryptococcus neoformans disease. Biosci. Rep. 2020, 40, BSR20190337. [Google Scholar] [CrossRef]
- Rajasingham, R.; Smith, R.M.; Park, B.J.; Jarvis, J.N.; Govender, N.P.; Chiller, T.M.; Denning, D.W.; Loyse, A.; Boulware, D.R. Global burden of disease of HIV-associated cryptococcal meningitis: An updated analysis. Lancet Infect. Dis. 2017, 17, 873–881. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Gertz, A.M.; Lawrence, D.S.; Wills, N.K.; Guthrie, B.L.; Farquhar, C.; Jarvis, J.N. Mortality from HIV-associated meningitis in sub-Saharan Africa: A systematic review and meta-analysis. J. Int. AIDS Soc. 2020, 23, e25416. [Google Scholar] [CrossRef]
- Jarvis, J.N.; Lawrence, D.S.; Meya, D.B.; Kagimu, E.; Kasibante, J.; Mpoza, E.; Rutakingirwa, M.K.; Ssebambulidde, K.; Tugume, L.; Rhein, J.; et al. Single-dose liposomal amphotericin B treatment for cryptococcal meningitis. N. Engl. J. Med. 2022, 386, 1109–1120. [Google Scholar] [CrossRef]
- Boulware, D.R.; Meya, D.B.; Muzoora, C.; Rolfes, M.A.; Huppler Hullsiek, K.; Musubire, A.; Taseera, K.; Nabeta, H.W.; Schutz, C.; Williams, D.A.; et al. Timing of antiretroviral therapy after diagnosis of cryptococcal meningitis. N. Engl. J. Med. 2014, 370, 2487–2498. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Guidelines for the Diagnosis, Prevention and Management of Cryptococcal Disease in HIV-Infected Adults, Adolescents and Children; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Bicanic, T.; Bottomley, C.; Loyse, A.; Brouwer, A.E.; Muzoora, C.; Taseera, K.; Jackson, A.; Phulusa, J.; Hosseinipour, M.C.; van der Horst, C.; et al. Toxicity of amphotericin B deoxycholate-based induction therapy in patients with HIV-associated cryptococcal meningitis. Antimicrob. Agents Chemother. 2015, 59, 7224–7231. [Google Scholar] [CrossRef]
- Khayhan, K.; Hagen, F.; Pan, W.; Simwami, S.; Fisher, M.C.; Wahyuningsih, R.; Chakrabarti, A.; Chowdhary, A.; Ikeda, R.; Taj-Aldeen, S.J.; et al. Geographically structured populations of Cryptococcus neoformans variety grubii in Asia correlate with HIV status and show a clonal population structure. PLoS ONE 2013, 8, e72222. [Google Scholar] [CrossRef]
- Litvintseva, A.P.; Thakur, R.; Vilgalys, R.; Mitchell, T.G. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 2006, 172, 2223–2238. [Google Scholar] [CrossRef]
- Andrade-Silva, L.E.; Ferreira-Paim, K.; Ferreira, T.B.; Vilas-Boas, A.; Mora, D.J.; Manzato, V.M.; Fonseca, F.M.; Buosi, K.; Andrade-Silva, J.; Prudente, B.D.S.; et al. Genotypic analysis of clinical and environmental Cryptococcus neoformans isolates from Brazil reveals the presence of VNB isolates and a correlation with biological factors. PLoS ONE 2018, 13, e0193237. [Google Scholar] [CrossRef]
- Bovers, M.; Hagen, F.; Kuramae, E.E.; Boekhout, T. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet. Biol. 2008, 45, 400–421. [Google Scholar] [CrossRef]
- Nielsen, K.; Cox, G.M.; Wang, P.; Toffaletti, D.L.; Perfect, J.R.; Heitman, J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 2003, 71, 4831–4841. [Google Scholar] [CrossRef]
- Ashton, P.M.; Thanh, L.T.; Trieu, P.H.; Van Anh, D.; Trinh, N.M.; Beardsley, J.; Kibengo, F.; Chierakul, W.; Dance, D.A.B.; Rattanavong, S.; et al. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat. Commun. 2019, 10, 2035. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Edman, J.C.; Wickes, B.L. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 1992, 60, 602–605. [Google Scholar] [CrossRef]
- Casadevall, A.; Perfect, J.R. Cryptococcus neoformans; American Society of Microbiology: Washington, DC, USA, 1998. [Google Scholar] [CrossRef]
- Perfect, J.R.; Casadevall, A. Cryptococcosis. Infect. Dis. Clin. N. Am. 2002, 16, 837–874. [Google Scholar] [CrossRef]
- Janbon, G.; Ormerod, K.L.; Paulet, D.; Byrnes, E.J.; Yadav, V.; Chatterjee, G.; Mullapudi, N.; Hon, C.C.; Billmyre, R.B.; Brunel, F.; et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014, 10, e1004261. [Google Scholar] [CrossRef]
- Arras, S.D.M.; Ormerod, K.L.; Erpf, P.E.; Espinosa, M.I.; Carpenter, A.C.; Blundell, R.D.; Stowasser, S.R.; Schulz, B.L.; Tanurdzic, M.; Fraser, J.A. Convergent microevolution of Cryptococcus neoformans hypervirulence in the laboratory and the clinic. Sci. Rep. 2017, 7, 17918. [Google Scholar] [CrossRef]
- Janbon, G.; Maeng, S.; Yang, D.H.; Ko, Y.J.; Jung, K.W.; Moyrand, F.; Floyd, A.; Heitman, J.; Bahn, Y.S. Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet. Biol. 2010, 47, 1070–1080. [Google Scholar] [CrossRef]
- Chun, C.D.; Madhani, H.D. Applying genetics and molecular biology to the study of the human pathogen Cryptococcus neoformans. Methods Enzym. 2010, 470, 797–831. [Google Scholar] [CrossRef]
- Chun, C.D.; Madhani, H.D. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS ONE 2010, 5, e12503. [Google Scholar] [CrossRef]
- Kraus, P.R.; Boily, M.J.; Giles, S.S.; Stajich, J.E.; Allen, A.; Cox, G.M.; Dietrich, F.S.; Perfect, J.R.; Heitman, J. Identification of Cryptococcus neoformans temperature-regulated genes with a genomic-DNA microarray. Eukaryot. Cell 2004, 3, 1249–1260. [Google Scholar] [CrossRef]
- Chow, E.D.; Liu, O.W.; O’Brien, S.; Madhani, H.D. Exploration of whole-genome responses of the human AIDS-associated yeast pathogen Cryptococcus neoformans var grubii: Nitric oxide stress and body temperature. Curr. Genet. 2007, 52, 137–148. [Google Scholar] [CrossRef]
- Jung, W.H.; Saikia, S.; Hu, G.; Wang, J.; Fung, C.K.; D’Souza, C.; White, R.; Kronstad, J.W. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010, 6, e1001209. [Google Scholar] [CrossRef]
- Haynes, B.C.; Skowyra, M.L.; Spencer, S.J.; Gish, S.R.; Williams, M.; Held, E.P.; Brent, M.R.; Doering, T.L. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS Pathog. 2011, 7, e1002411. [Google Scholar] [CrossRef]
- Pukkila-Worley, R.; Gerrald, Q.D.; Kraus, P.R.; Boily, M.J.; Davis, M.J.; Giles, S.S.; Cox, G.M.; Heitman, J.; Alspaugh, J.A. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot. Cell 2005, 4, 190–201. [Google Scholar] [CrossRef]
- Fan, W.; Kraus, P.R.; Boily, M.J.; Heitman, J. Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot. Cell 2005, 4, 1420–1433. [Google Scholar] [CrossRef]
- Hu, G.; Cheng, P.Y.; Sham, A.; Perfect, J.R.; Kronstad, J.W. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol. Microbiol. 2008, 69, 1456–1475. [Google Scholar] [CrossRef]
- Liew, K.L.; Jee, J.M.; Yap, I.; Yong, P.V. In vitro analysis of metabolites secreted during infection of lung epithelial cells by Cryptococcus neoformans. PLoS ONE 2016, 11, e0153356. [Google Scholar] [CrossRef]
- Mukaremera, L.; Lee, K.K.; Wagener, J.; Wiesner, D.L.; Gow, N.A.R.; Nielsen, K. Titan cell production in Cryptococcus neoformans reshapes the cell wall and capsule composition during infection. Cell Surf. 2018, 1, 15–24. [Google Scholar] [CrossRef]
- Mukaremera, L. The Cryptococcus wall: A different wall for a unique lifestyle. PLoS Pathog. 2023, 19, e1011141. [Google Scholar] [CrossRef]
- Mukaremera, L.; Nielsen, K. Adaptive immunity to Cryptococcus neoformans infections. J. Fungi 2017, 3, 64. [Google Scholar] [CrossRef]
- Nielsen, K.; Cox, G.M.; Litvintseva, A.P.; Mylonakis, E.; Malliaris, S.D.; Benjamin, D.K.; Giles, S.S.; Mitchell, T.G.; Casadevall, A.; Perfect, J.R.; et al. Cryptococcus neoformans α strains preferentially disseminate to the central nervous system during coinfection. Infect. Immun. 2005, 73, 4922–4933. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Portillo, E.; Orts, D.; Llorca, E.; Fernández, C.; Antón, J.; Ferrer, C.; Gálvez, B.; Esteban, V.; Revelles, E.; Pérez-Martín, C.; et al. The domestic environment and the lung mycobiome. Microorganisms 2020, 8, 1717. [Google Scholar] [CrossRef]
- Ding, M.; Smith, K.D.; Wiesner, D.L.; Nielsen, J.N.; Jackson, K.M.; Nielsen, K. Use of clinical isolates to establish criteria for a mouse model of latent Cryptococcus neoformans infection. Front. Cell. Infect. Microbiol. 2022, 11, 804059. [Google Scholar] [CrossRef]
- Pirofski, L.A.; Casadevall, A. The state of latency in microbial pathogenesis. J. Clin. Investig. 2020, 130, 4525–4531. [Google Scholar] [CrossRef]
- Wang, P.; Cutler, J.; King, J.; Palmer, D. Mutation of the regulator of G protein signaling Crg1 increases virulence in Cryptococcus neoformans. Eukaryot. Cell 2004, 3, 1028–1035. [Google Scholar] [CrossRef]
- Dromer, F.; Casadevall, A.; Perfect, J.; Sorrell, T. Cryptococcus neoformans: Latency and disease. In Cryptococcus: From Human Pathogen to Model Yeast; ASM Press: Washington, DC, USA, 2010; pp. 429–439. [Google Scholar] [CrossRef]
- Goldman, D.; Lee, S.C.; Casadevall, A. Pathogenesis of pulmonary Cryptococcus neoformans infection in the rat. Infect. Immun. 1994, 62, 4755–4761. [Google Scholar] [CrossRef]
- Goldman, D.L.; Lee, S.C.; Mednick, A.J.; Montella, L.; Casadevall, A. Persistent Cryptococcus neoformans pulmonary infection in the rat is associated with intracellular parasitism, decreased inducible nitric oxide synthase expression, and altered antibody responsiveness to cryptococcal polysaccharide. Infect. Immun. 2000, 68, 832–838. [Google Scholar] [CrossRef]
- Wiesner, D.L.; Moskalenko, O.; Corcoran, J.M.; McDonald, T.; Rolfes, M.A.; Meya, D.B.; Kajumbula, H.; Kambugu, A.; Bohjanen, P.R.; Knight, J.F.; et al. Cryptococcal genotype influences immunologic response and human clinical outcome after meningitis. mBio 2012, 3, e00196-12. [Google Scholar] [CrossRef]
- Beale, M.A.; Sabiiti, W.; Robertson, E.J.; Fuentes-Cabrejo, K.M.; O’Hanlon, S.J.; Jarvis, J.N.; Loyse, A.; Meintjes, G.; Harrison, T.S.; May, R.C.; et al. Genotypic diversity is associated with clinical outcome and phenotype in cryptococcal meningitis across southern Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003847. [Google Scholar] [CrossRef]
- Gerstein, A.C.; Jackson, K.M.; McDonald, T.R.; Wang, Y.; Lueck, B.D.; Bohjanen, S.; Smith, K.D.; Akampurira, A.; Meya, D.B.; Xue, C.; et al. Identification of pathogen genomic differences that impact human immune response and disease during Cryptococcus neoformans infection. mBio 2019, 10, e01440-19. [Google Scholar] [CrossRef]
- Day, J.N.; Hoang, T.N.; Duong, A.V.; Hong, C.T.; Diep, P.T.; Campbell, J.I.; Sieu, T.P.; Hien, T.T.; Bui, T.; Boni, M.F.; et al. Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VN1. J. Clin. Microbiol. 2011, 49, 658–664. [Google Scholar] [CrossRef]
- Lofgren, S.; Hullsiek, K.H.; Morawski, B.M.; Nabeta, H.W.; Kiggundu, R.; Taseera, K.; Musubire, A.; Schutz, C.; Abassi, M.; Bahr, N.C.; et al. Differences in immunologic factors among patients presenting with altered mental status during cryptococcal meningitis. J. Infect. Dis. 2017, 215, 693–697. [Google Scholar] [CrossRef]
- Meya, D.B.; Okurut, S.; Zziwa, G.; Cose, S.; Bohjanen, P.R.; Mayanja-Kizza, H.; Joloba, M.; Boulware, D.R.; Yukari Manabe, C.; Wahl, S.; et al. Monocyte ohenotype and IFN-γ-inducible cytokine responses are associated with cryptococcal immune reconstitution inflammatory syndrome. J. Fungi 2017, 3, 28. [Google Scholar] [CrossRef]
- Meya, D.B.; Okurut, S.; Zziwa, G.; Rolfes, M.A.; Kelsey, M.; Cose, S.; Joloba, M.; Naluyima, P.; Palmer, B.E.; Kambugu, A.; et al. Cellular immune activation in cerebrospinal fluid from Ugandans with cryptococcal meningitis and immune reconstitution inflammatory syndrome. J. Infect. Dis. 2015, 211, 1597–1606. [Google Scholar] [CrossRef]
- Musubire, A.K.; Meya, D.B.; Rhein, J.; Meintjes, G.; Bohjanen, P.R.; Nuwagira, E.; Muzoora, C.; Boulware, D.R.; Hullsiek, K.H.; COAT and ASTRO Trial Teams. Blood neutrophil counts in HIV-infected patients with cryptococcal meningitis: Association with mortality. PLoS ONE 2018, 13, e0209337. [Google Scholar] [CrossRef]
- Boulware, D.R.; von Hohenberg, M.; Rolfes, M.A.; Bahr, N.C.; Rhein, J.; Akampurira, A.; Williams, D.A.; Taseera, K.; Schutz, C.; McDonald, T.; et al. human immune response varies by the degree of relative cryptococcal antigen shedding. Open. Forum Infect. Dis. 2016, 3, ofv194. [Google Scholar] [CrossRef]
- Scriven, J.; Rhein, J.; Hullsiek, K.; von Hohenberg, M.; Linder, G.; Rolfes, M.; Williams, D.; Taseera, K.; Meya, D.; Meintjes, G.; et al. Early ART after cryptococcal meningitis is associated with cerebrospinal fluid pleocytosis and macrophage activation in a multisite randomized trial. J. Infect. Dis. 2015, 212, 769–778. [Google Scholar] [CrossRef]
- Mukaremera, L.; McDonald, T.R.; Nielsen, J.N.; Molenaar, C.J.; Akampurira, A.; Schutz, C.; Taseera, K.; Muzoora, C.; Meintjes, G.; Meya, D.B.; et al. The mouse inhalation model of Cryptococcus neoformans infection recapitulates strain virulence in humans and shows closely related strains can possess differential virulence. Infect. Immun. 2019, 87, e00046-19. [Google Scholar] [CrossRef] [PubMed]
- Chau, T.T.; Mai, N.H.; Phu, N.H.; Nghia, H.D.; Chuong, L.V.; Sinh, D.X.; Duong, V.A.; Diep, P.T.; Campbell, J.I.; Baker, S.; et al. A prospective descriptive study of cryptococcal meningitis in HIV uninfected patients in Vietnam—High prevalence of Cryptococcus neoformans var grubii in the absence of underlying disease. BMC Infect. Dis. 2010, 10, 199. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Varma, A.; Diaz, M.R.; Litvintseva, A.P.; Wollenberg, K.K.; Kwon-Chung, K.J. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg. Infect. Dis. 2008, 14, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Thanh, L.T.; Toffaletti, D.L.; Tenor, J.L.; Giamberardino, C.; Sempowski, G.D.; Asfaw, Y.; Phan, H.T.; Van Duong, A.; Trinh, N.M.; Thwaites, G.E.; et al. Assessing the virulence of Cryptococcus neoformans causing meningitis in HIV infected and uninfected patients in Vietnam. Med. Mycol. 2020, 58, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Qifeng, X.; Tohyama, M.; Qureshi, M.H.; Saito, A. Contribution of tumour necrosis factor-alpha (TNF-alpha) in host defence mechanism against Cryptococcus neoformans. Clin. Exp. Immunol. 1996, 106, 468–474. [Google Scholar] [CrossRef]
- Herring, A.C.; Lee, J.; McDonald, R.A.; Toews, G.B.; Huffnagle, G.B. Induction of interleukin-12 and gamma interferon requires tumor necrosis factor alpha for protective T1-cell-mediated immunity to pulmonary Cryptococcus neoformans infection. Infect. Immun. 2002, 70, 2959–2964. [Google Scholar] [CrossRef] [PubMed]
- Koguchi, Y.; Kawakami, K. Cryptococcal infection and Th1-Th2 cytokine balance. Int. Rev. Immunol. 2002, 21, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Olszewski, M.A.; Tsang, T.M.; McDonald, R.A.; Toews, G.B.; Huffnagle, G.B. Effect of cytokine interplay on macrophage polarization during chronic pulmonary infection with Cryptococcus neoformans. Infect. Immun. 2011, 79, 1915–1926. [Google Scholar] [CrossRef]
- Garelnabi, M.; May, R.C. Variability in innate host immune responses to cryptococcosis. Mem. Inst. Oswaldo Cruz 2018, 113, e180060. [Google Scholar] [CrossRef]
- Casadevall, A.; Pirofski, L.A. Host-pathogen interactions: Redefining the basic concepts of virulence and pathogenicity. Infect. Immun. 1999, 67, 3703–3713. [Google Scholar] [CrossRef]
- Pirofski, L.A.; Casadevall, A. Immune-mediated damage completes the parabola: Cryptococcus neoformans pathogenesis can reflect the outcome of a weak or strong immune response. mBio 2017, 8, e02063-17. [Google Scholar] [CrossRef]
- Yu, C.H.; Chen, Y.; Desjardins, C.A.; Tenor, J.L.; Toffaletti, D.L.; Giamberardino, C.; Litvintseva, A.; Perfect, J.R.; Cuomo, C.A. Landscape of gene expression variation of natural isolates of Cryptococcus neoformans in response to biologically relevant stresses. Microb. Genom. 2020, 6, e000319. [Google Scholar] [CrossRef]
- Litvintseva, A.P.; Mitchell, T.G. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect. Immun. 2009, 77, 3188–3195. [Google Scholar] [CrossRef]
- Casadevall, A.; Coelho, C.; Alanio, A. Mechanisms of Cryptococcus neoformans-mediated host damage. Front. Immunol. 2018, 9, 855. [Google Scholar] [CrossRef]
- Coelho, C.; Bocca, A.L.; Casadevall, A. The tools for virulence of Cryptococcus neoformans. Adv. Appl. Microbiol. 2014, 87, 1–41. [Google Scholar] [CrossRef] [PubMed]
- Kwon-Chung, K.J.; Rhodes, J.C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect. Immun. 1986, 51, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Bulmer, G.S.; Sans, M.D.; Gunn, C.M. Cryptococcus neoformans. I. Nonencapsulated mutants. J. Bacteriol. 1967, 94, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Litvintseva, A.P.; Kestenbaum, L.; Vilgalys, R.; Mitchell, T.G. Comparative analysis of environmental and clinical populations of Cryptococcus neoformans. J. Clin. Microbiol. 2005, 43, 556–564. [Google Scholar] [CrossRef]
- Da Silva, E.G.; Baroni, F.A.; Viani, F.C.; Ruiz, L.D.S.; Gandra, R.F.; Auler, M.E.; Dias, A.L.T.; Gambale, W.; Paula, C.R. Virulence profile of strains of Cryptococcus neoformans var. grubii evaluated by experimental infection in BALB/c mice and correlation with exoenzyme activity. J. Med. Microbiol. 2006, 55, 139–142. [Google Scholar] [CrossRef]
- Desjardins, C.A.; Giamberardino, C.; Sykes, S.M.; Yu, C.H.; Tenor, J.L.; Chen, Y.; Yang, T.; Jones, A.M.; Sun, S.; Haverkamp, M.R.; et al. Population genomics and the evolution of virulence in the fungal pathogen. Genome Res. 2017, 27, 1207–1219. [Google Scholar] [CrossRef]
- Yu, C.H.; Sephton-Clark, P.; Tenor, J.L.; Toffaletti, D.L.; Giamberardino, C.; Haverkamp, M.; Cuomo, C.A.; Perfect, J.R. Gene expression of diverse Cryptococcus isolates during infection of the human central nervous system. mBio 2021, 12, e0231321. [Google Scholar] [CrossRef]
- Chen, Y.; Toffaletti, D.L.; Tenor, J.L.; Litvintseva, A.P.; Fang, C.; Mitchell, T.G.; McDonald, T.R.; Nielsen, K.; Boulware, D.R.; Bicanic, T.; et al. The Cryptococcus neoformans transcriptome at the site of human meningitis. mBio 2014, 5, e01087-13. [Google Scholar] [CrossRef]
- Montoya, M.C.; Magwene, P.M.; Perfect, J.R. Associations between Cryptococcus genotypes, phenotypes, and clinical parameters of human disease. J. Fungi 2021, 7, 260. [Google Scholar] [CrossRef]
- Fernandes, K.E.; Brockway, A.; Haverkamp, M.; Cuomo, C.A.; van Ogtrop, F.; Perfect, J.R.; Carter, D.A. Phenotypic variability correlates with clinical outcome in Cryptococcus isolates obtained from Botswanan HIV/AIDS patients. mBio 2018, 9, e02016-18. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, K.E.; Fraser, J.A.; Carter, D.A. Lineages derived from Cryptococcus neoformans type strain H99 support a link between the capacity to be pleomorphic and virulence. mBio 2022, 13, e0028322. [Google Scholar] [CrossRef] [PubMed]
- Zaragoza, O.; García-Rodas, R.; Nosanchuk, J.D.; Cuenca-Estrella, M.; Rodríguez-Tudela, J.L.; Casadevall, A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010, 6, e1000945. [Google Scholar] [CrossRef]
- Okagaki, L.H.; Strain, A.K.; Nielsen, J.N.; Charlier, C.; Baltes, N.J.; Chrétien, F.; Heitman, J.; Dromer, F.; Nielsen, K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Cruickshank, J.G.; Cavill, R.; Jelbert, M. Cryptococcus neoformans of unusual morphology. Appl. Microbiol. 1973, 25, 309–312. [Google Scholar] [CrossRef]
- Love, G.L.; Boyd, G.D.; Greer, D.L. Large Cryptococcus neoformans isolated from brain abscess. J. Clin. Microbiol. 1985, 22, 1068–1070. [Google Scholar] [CrossRef]
- Wang, J.M.; Zhou, Q.; Cai, H.R.; Zhuang, Y.; Zhang, Y.F.; Xin, X.Y.; Meng, F.Q.; Wang, Y.P. Clinicopathological features of pulmonary cryptococcosis with cryptococcal titan cells: A comparative analysis of 27 cases. Int. J. Clin. Exp. Pathol. 2014, 7, 4837–4846. [Google Scholar]
- Okagaki, L.H.; Nielsen, K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot. Cell 2012, 11, 820–826. [Google Scholar] [CrossRef]
- Wiesner, D.L.; Specht, C.A.; Lee, C.K.; Smith, K.D.; Mukaremera, L.; Lee, S.T.; Lee, C.G.; Elias, J.A.; Nielsen, J.N.; Boulware, D.R.; et al. Chitin recognition via chitotriosidase promotes pathologic type-2 helper T cell responses to cryptococcal infection. PLoS Pathog. 2015, 11, e1004701. [Google Scholar] [CrossRef]
- Gerstein, A.C.; Fu, M.S.; Mukaremera, L.; Li, Z.; Ormerod, K.L.; Fraser, J.A.; Berman, J.; Nielsen, K. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 2015, 6, e01340-15. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; de Oliveira, H.C.; García-Rodas, R.; Rossi, S.A.; Llorente, I.; Zaballos, Á.; Janbon, G.; Ariño, J.; Zaragoza, Ó. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 2018, 14, e1007007. [Google Scholar] [CrossRef]
- Dambuza, I.M.; Drake, T.; Chapuis, A.; Zhou, X.; Correia, J.; Taylor-Smith, L.; LeGrave, N.; Rasmussen, T.; Fisher, M.C.; Bicanic, T.; et al. The Cryptococcus neoformans Titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 2018, 14, e1006978. [Google Scholar] [CrossRef]
- Hu, G.; Liu, I.; Sham, A.; Stajich, J.E.; Dietrich, F.S.; Kronstad, J.W. Comparative hybridization reveals extensive genome variation in the AIDS-associated pathogen Cryptococcus neoformans. Genome Biol. 2008, 9, R41. [Google Scholar] [CrossRef]
- Sionov, E.; Lee, H.; Chang, Y.C.; Kwon-Chung, K.J. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 2010, 6, e1000848. [Google Scholar] [CrossRef]
- Stone, N.R.; Rhodes, J.; Fisher, M.C.; Mfinanga, S.; Kivuyo, S.; Rugemalila, J.; Segal, E.S.; Needleman, L.; Molloy, S.F.; Kwon-Chung, J.; et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Investig. 2019, 129, 999–1014. [Google Scholar] [CrossRef]
- Semighini, C.P.; Averette, A.F.; Perfect, J.R.; Heitman, J. Deletion of Cryptococcus neoformans AIF ortholog promotes chromosome aneuploidy and fluconazole-resistance in a metacaspase-independent manner. PLoS Pathog. 2011, 7, e1002364. [Google Scholar] [CrossRef] [PubMed]
- Sephton-Clark, P.; Tenor, J.L.; Toffaletti, D.L.; Meyers, N.; Giamberardino, C.; Molloy, S.F.; Palmucci, J.R.; Chan, A.; Chikaonda, T.; Heyderman, R.; et al. Genomic Variation across a clinical Cryptococcus population linked to disease outcome. mBio 2022, 13, e0262622. [Google Scholar] [CrossRef]
- Hu, G.; Wang, J.; Choi, J.; Jung, W.H.; Liu, I.; Litvintseva, A.P.; Bicanic, T.; Aurora, R.; Mitchell, T.G.; Perfect, J.R.; et al. Variation in chromosome copy number influences the virulence of Cryptococcus neoformans and occurs in isolates from AIDS patients. BMC Genom. 2011, 12, 526. [Google Scholar] [CrossRef]
- Ormerod, K.L.; Morrow, C.A.; Chow, E.W.; Lee, I.R.; Arras, S.D.; Schirra, H.J.; Cox, G.M.; Fries, B.C.; Fraser, J.A. Comparative genomics of serial isolates of Cryptococcus neoformans reveals gene associated with carbon utilization and virulence. G3 2013, 3, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.; Beale, M.A.; Vanhove, M.; Jarvis, J.N.; Kannambath, S.; Simpson, J.A.; Ryan, A.; Meintjes, G.; Harrison, T.S.; Fisher, M.C.; et al. A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3 2017, 7, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.; Prolla, T.A.; Liskay, R.M.; Petes, T.D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 1993, 365, 274–276. [Google Scholar] [CrossRef]
- De Sousa, H.R.; Frazão, S.D.; Oliveira Júnior, G.P.D.; Albuquerque, P.; Nicola, A.M. Cryptococcal virulence in humans: Learning from translational studies with clinical isolates. Front. Cell. Infect. Microbiol. 2021, 11, 657502. [Google Scholar] [CrossRef] [PubMed]
- Luberto, C.; Martinez-Mariño, B.; Taraskiewicz, D.; Bolaños, B.; Chitano, P.; Toffaletti, D.L.; Cox, G.M.; Perfect, J.R.; Hannun, Y.A.; Balish, E.; et al. Identification of App1 as a regulator of phagocytosis and virulence of Cryptococcus neoformans. J. Clin. Investig. 2003, 112, 1080–1094. [Google Scholar] [CrossRef] [PubMed]
- Day, J.N.; Qihui, S.; Thanh, L.T.; Trieu, P.H.; Van, A.D.; Thu, N.H.; Chau, T.T.H.; Lan, N.P.H.; Chau, N.V.V.; Ashton, P.M.; et al. Comparative genomics of Cryptococcus neoformans var. grubii associated with meningitis in HIV infected and uninfected patients in Vietnam. PLoS Negl. Trop. Dis. 2017, 11, e0005628. [Google Scholar] [CrossRef]
- Abadi, J.; Pirofski, L. Antibodies reactive with the cryptococcal capsular polysaccharide glucuronoxylomannan are present in sera from children with and without human immunodeficiency virus infection. J. Infect. Dis. 1999, 180, 915–919. [Google Scholar] [CrossRef]
- Goldman, D.L.; Khine, H.; Abadi, J.; Lindenberg, D.J.; Pirofski, L.A.; Niang, R.; Casadevall, A. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 2001, 107, E66. [Google Scholar] [CrossRef]
- Baker, R.D.; Haugen, R.K. Tissue changes and tissue diagnosis in cryptococcosis; a study of 26 cases. Am. J. Clin. Pathol. 1955, 25, 14–24. [Google Scholar] [CrossRef]
- Baker, R.D. The primary pulmonary lymph node complex of cryptococcosis. Am. J. Clin. Pathol. 1976, 65, 83–92. [Google Scholar] [CrossRef]
- Shibuya, K.; Hirata, A.; Omuta, J.; Sugamata, M.; Katori, S.; Saito, N.; Murata, N.; Morita, A.; Takahashi, K.; Hasegawa, C.; et al. Granuloma and cryptococcosis. J. Infect. Chemother. 2005, 11, 115–122. [Google Scholar] [CrossRef]
- Giles, S.S.; Dagenais, T.R.; Botts, M.R.; Keller, N.P.; Hull, C.M. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 2009, 77, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Iseki, M.; Anzo, M.; Yamashita, N.; Matsuo, N. Hyper-IgM immunodeficiency with disseminated cryptococcosis. Acta Paediatr. 1994, 83, 780–782. [Google Scholar] [CrossRef] [PubMed]
- Winkelstein, J.A.; Marino, M.C.; Ochs, H.; Fuleihan, R.; Scholl, P.R.; Geha, R.; Stiehm, E.R.; Conley, M.E. The X-linked hyper-IgM syndrome: Clinical and immunologic features of 79 patients. Medicine 2003, 82, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Zonios, D.I.; Falloon, J.; Huang, C.Y.; Chaitt, D.; Bennett, J.E. Cryptococcosis and idiopathic CD4 lymphocytopenia. Medicine 2007, 86, 78–92. [Google Scholar] [CrossRef]
- Scott-Algara, D.; Balabanian, K.; Chakrabarti, L.A.; Mouthon, L.; Dromer, F.; Didier, C.; Arenzana-Seisdedos, F.; Lortholary, O. Idiopathic CD4+ T-cell lymphocytopenia is associated with impaired membrane expression of the chemokine receptor CXCR4. Blood 2010, 115, 3708–3717. [Google Scholar] [CrossRef] [PubMed]
- Vinh, D.C.; Patel, S.Y.; Uzel, G.; Anderson, V.L.; Freeman, A.F.; Olivier, K.N.; Spalding, C.; Hughes, S.; Pittaluga, S.; Raffeld, M.; et al. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood 2010, 115, 1519–1529. [Google Scholar] [CrossRef]
- Browne, S.K.; Burbelo, P.D.; Chetchotisakd, P.; Suputtamongkol, Y.; Kiertiburanakul, S.; Shaw, P.A.; Kirk, J.L.; Jutivorakool, K.; Zaman, R.; Ding, L.; et al. Adult-Onset Immunodeficiency in Thailand and Taiwan. N. Engl. J. Med. 2012, 367, 725–734. [Google Scholar] [CrossRef]
- Rosen, L.B.; Freeman, A.F.; Yang, L.M.; Jutivorakool, K.; Olivier, K.N.; Angkasekwinai, N.; Suputtamongkol, Y.; Bennett, J.E.; Pyrgos, V.; Williamson, P.R.; et al. Anti-GM-CSF autoantibodies in patients with cryptococcal meningitis. J. Immunol. 2013, 190, 3959–3966. [Google Scholar] [CrossRef]
- Jarvis, J.N.; Casazza, J.P.; Stone, H.H.; Meintjes, G.; Lawn, S.D.; Levitz, S.M.; Harrison, T.S.; Koup, R.A. The phenotype of the Cryptococcus-specific CD4+ memory T-cell response is associated with disease severity and outcome in HIV-associated cryptococcal meningitis. J. Infect. Dis. 2013, 207, 1817–1828. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Scriven, J.E.; Harrison, T.S.; Jarvis, J.N. Immune correlates of HIV-associated cryptococcal meningitis. PLOS Pathog. 2017, 13, e1006207. [Google Scholar] [CrossRef]
- Agrawal, C.; Sood, V.; Kumar, A.; Raghavan, V. Cryptococcal infection in transplant kidney manifesting as chronic allograft dysfunction. Indian J. Nephrol. 2017, 27, 392–394. [Google Scholar] [CrossRef]
- Marchand, T.; Revest, M.; Tattevin, P.; Chevrier, S.; Poullot, E.; Lamy, T.; Houot, R. Early cryptococcal meningitis following treatment with rituximab, fludarabine and cyclophosphamide in a patient with chronic lymphocytic leukemia. Leuk. Lymphoma 2013, 54, 643–645. [Google Scholar] [CrossRef]
- Wormley, F.L., Jr.; Perfect, J.R.; Steele, C.; Cox, G.M. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect. Immun. 2007, 75, 1453–1462. [Google Scholar] [CrossRef]
- Jarvis, J.N.; Meintjes, G.; Rebe, K.; Williams, G.N.; Bicanic, T.; Williams, A.; Schutz, C.; Bekker, L.G.; Wood, R.; Harrison, T.S. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: A randomized controlled trial. AIDS 2012, 26, 1105–1113. [Google Scholar] [CrossRef]
- Fu, M.S.; Drummond, R.A. The diverse roles of monocytes in Cryptococcosis. J. Fungi 2020, 6, 111. [Google Scholar] [CrossRef]
- Heung, L.J. Innate immune responses to Cryptococcus. J. Fungi 2017, 3, 35. [Google Scholar] [CrossRef]
- Nelson, B.N.; Hawkins, A.N.; Wozniak, K.L. Pulmonary macrophage and dendritic cell responses to Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 2020, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, S.; Pirofski, L.A. Host immunity to Cryptococcus neoformans. Future Microbiol. 2015, 10, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Smith, L. Cryptococcus–epithelial interactions. J. Fungi 2017, 3, 53. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.F.; Guillot, L.; Qureshi, S.T. Mammalian model hosts of cryptococcal infection. Comp. Med. 2007, 57, 9–17. [Google Scholar]
- Franzot, S.P.; Mukherjee, J.; Cherniak, R.; Chen, L.C.; Hamdan, J.S.; Casadevall, A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect. Immun. 1998, 66, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Maziarz, E.K.; Perfect, J.R. Cryptococcosis. Infect. Dis. Clin. N. Am. 2016, 30, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Frazão, S.O.; Sousa, H.R.; Silva, L.G.D.; Folha, J.D.S.; Gorgonha, K.C.M.; Oliveira, G.P., Jr.; Felipe, M.S.S.; Silva-Pereira, I.; Casadevall, A.; Nicola, A.M.; et al. Laccase affects the rate of Cryptococcus neoformans nonlytic exocytosis from macrophages. mBio 2020, 11, e02085-20. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Desnos-Ollivier, M.; Dromer, F. Dynamics of Cryptococcus neoformans-macrophage interactions reveal that fungal background influences outcome during cryptococcal meningoencephalitis in humans. mBio 2011, 2, e00158-11. [Google Scholar] [CrossRef] [PubMed]
- Sabiiti, W.; Robertson, E.; Beale, M.A.; Johnston, S.A.; Brouwer, A.E.; Loyse, A.; Jarvis, J.N.; Gilbert, A.S.; Fisher, M.C.; Harrison, T.S.; et al. Efficient phagocytosis and laccase activity affect the outcome of HIV-associated cryptococcosis. J. Clin. Investig. 2014, 124, 2000–2008. [Google Scholar] [CrossRef]
- Hansakon, A.; Mutthakalin, P.; Ngamskulrungroj, P.; Chayakulkeeree, M.; Angkasekwinai, P. Cryptococcus neoformans and Cryptococcus gattii clinical isolates from Thailand display diverse phenotypic interactions with macrophages. Virulence 2019, 10, 26–36. [Google Scholar] [CrossRef]
- Xie, Q.; Kawakami, K.; Kudeken, N.; Zhang, T.; Qureshi, M.H.; Saito, A. Different susceptibility of three clinically isolated strains of Cryptococcus neoformans to the fungicidal effects of reactive nitrogen and oxygen intermediates: Possible relationships with virulence. Microbiol. Immunol. 1997, 41, 725–731. [Google Scholar] [CrossRef]
- Robertson, E.J.; Najjuka, G.; Rolfes, M.A.; Akampurira, A.; Jain, N.; Anantharanjit, J.; von Hohenberg, M.; Tassieri, M.; Carlsson, A.; Meya, D.B.; et al. Cryptococcus neoformans ex vivo capsule size is associated with intracranial pressure and host immune response in HIV-associated cryptococcal meningitis. J. Infect. Dis. 2014, 209, 74–82. [Google Scholar] [CrossRef]
- Nishikaku, A.S.; Soldá, M.V.; Ricci, G.; Ponzio, V.; Pagliari, C.; Medina-Pestana, J.O.; de Franco, M.F.; Colombo, A.L. Correlation between clinical outcome and tissue inflammatory response in kidney transplant recipients with cryptococcosis. Pathog. Dis. 2020, 78, ftaa054. [Google Scholar] [CrossRef]
- Desnos-Ollivier, M.; Patel, S.; Spaulding, A.R.; Charlier, C.; Garcia-Hermoso, D.; Nielsen, K.; Dromer, F. Mixed infections and in vivo evolution in the human fungal pathogen Cryptococcus neoformans. mBio 2010, 1, e00091-10. [Google Scholar] [CrossRef]
- Garcia-Hermoso, D.; Dromer, F.; Janbon, G. Cryptococcus neoformans capsule structure evolution in vitro and during murine infection. Infect. Immun. 2004, 72, 3359–3365. [Google Scholar] [CrossRef]
- Amos, B.; Aurrecoechea, C.; Barba, M.; Barreto, A.; Basenko, E.Y.; Bażant, W.; Belnap, R.; Blevins, A.S.; Böhme, U.; Brestelli, J.; et al. VEuPathDB: The eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 2022, 50, D898–D911. [Google Scholar] [CrossRef] [PubMed]
- Meyer, W.; Aanensen, D.M.; Boekhout, T.; Cogliati, M.; Diaz, M.R.; Esposto, M.C.; Fisher, M.; Gilgado, F.; Hagen, F.; Kaocharoen, S.; et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med. Mycol. 2009, 47, 561–570. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, K.M.; Ding, M.; Nielsen, K. Importance of Clinical Isolates in Cryptococcus neoformans Research. J. Fungi 2023, 9, 364. https://doi.org/10.3390/jof9030364
Jackson KM, Ding M, Nielsen K. Importance of Clinical Isolates in Cryptococcus neoformans Research. Journal of Fungi. 2023; 9(3):364. https://doi.org/10.3390/jof9030364
Chicago/Turabian StyleJackson, Katrina M., Minna Ding, and Kirsten Nielsen. 2023. "Importance of Clinical Isolates in Cryptococcus neoformans Research" Journal of Fungi 9, no. 3: 364. https://doi.org/10.3390/jof9030364
APA StyleJackson, K. M., Ding, M., & Nielsen, K. (2023). Importance of Clinical Isolates in Cryptococcus neoformans Research. Journal of Fungi, 9(3), 364. https://doi.org/10.3390/jof9030364





