Proposal of Four New Aureobasidium Species for Exopolysaccharide Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Strain Isolation
2.2. DNA Isolation, PCR Amplification and Sequencing
2.3. Observation of Morphology
2.4. Phylogenetic Analyses
2.5. Exopolysaccharides Production
| Species | Strain | Date | Location | Latitude and Longitude | Source | GenBank No. | References | |
|---|---|---|---|---|---|---|---|---|
| ITS | D1/D2 | |||||||
| Aureobasidium acericola | CDH 2020−10 | June 2020 | South Korea | 37°45′49.50″ N, 127°11′3.8″ E | Acer pseudosieboldianum | MT863788 | MT863787 | [10] |
| Aureobasidium aerium | CFCC 50324 | April 2015 | Sennon, Beijing, China | NA | air | ON007058 | ON007081 | [11] |
| Aureobasidium castanea | CFCC 54591 * | November 2021 | Jinjing Town, Changsha Hunan, China | 28°58′52″ N, 113°34′38″ E | Castanea heryi | NR_177551 | MW364275 | [12] |
| Aureobasidium caulivorum | CBS 242.64 | NA | Oregon, America | NA | Trifolium incarnatum | FJ150871 | FJ150944 | [39] |
| Aureobasidium insectorum sp. nov. | KCL139 | September 2021 | Zhangjiakou, Hebei, China | 39°30′ N, 113°50′ E | spittle insects | OP856707 | OP857208 | This study |
| LPL−1C | September 2022 | Zhoushan, Zhejiang, China | 29°53′28.86″ N, 122°24′59.35″ E | leaf | OP856705 | OP857207 | This study | |
| XZY65−10 | October 2019 | Shannan City, Tibet, China | 29°14′9.68″ N, 91°45′59.50″ E | leaf | OP856706 | OP857206 | This study | |
| L2PL−7A | September 2022 | Zhoushan, Zhejiang, China | 29°53′28.86″ N, 122°24′59.35″ E | leaf | OP856715 | OP857216 | This study | |
| T1−27−2 | November 2021 | Motuo County, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | leaf | OP856714 | OP857215 | This study | |
| XZY249M1 | October 2019 | Nyingchi City, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | deadwood | OP856713 | OP857214 | This study | |
| XZY63−10 | October 2019 | Shannan City, Tibet, China | 29°14′9.68″ N, 91°45′59.50″ E | leaf | OP856712 | OP857213 | This study | |
| Aureobasidium intercalariosporum sp. nov. | MGL11−3 | September 2022 | Zhoushan, Zhejiang, China | 29°53′28.86″ N, 122°24′59.35″ E | leaf | OP856703 | OP857204 | This study |
| MQL9−100 | September 2022 | Zhoushan, Zhejiang, China | 29°53′28.86″ N, 122°24′59.35″ E | leaf | OP856703 | OP857205 | This study | |
| Aureobasidium iranianum | CCTU 268 | June 2009 | Southern parts of Iran | NA | bamboo stems | NR_137598 | NG_057049 | [13] |
| Aureobasidium khasianum | NFCCI 4275 | December 2016 | Meghalaya, India | NA | litter samples | MH188305 | MH188306 | [40] |
| Aureobasidium leucospermi | CBS 130593 | April 2008 | South Africa | NA | leaves and stems of Proteaceae with cankers or leaf spots | NR_156246 | MH877257 | [14] |
| Aureobasidium lini | CBS 125.21T | NA | UK | NA | Linum usitatissimum | FJ150897 | FJ150946 | [8] |
| Aureobasidium mangrovei | IBRCM 30265T | January 2016 | Qeshm Island, Iran | 26°47′ N, 55°45′ E | mangrove trees (Avicennia marina) | NR_174637 | NG_078639 | [15] |
| Aureobasidium melanogenum | CBS 105.22 | NA | NA | NA | leaf | NR_159598 | NG_056960 | [8] |
| Aureobasidium microstictum | CBS 342.66 | NA | Germany | NA | dying or dead leaves | KT693743 | FJ150945 | [8] |
| Aureobasidium microstictum | CBS 114.64 | NA | Wageningen, The Netherlands | NA | Hemerocallis sp. | KT693744 | KT693986 | [8] |
| Aureobasidium microtermitis | NA | NA | NA | NA | NA | MW276135 | MW276136 | NA |
| Aureobasidium motuoense sp. nov. | E82−2 | October 2019 | Motuo County, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | soil | OP856702 | OP857203 | This study |
| XZY411−4 | August 2019 | Motuo County, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | leaf | OP856710 | OP857211 | This study | |
| E31−1 | October 2019 | Motuo County, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | soil | OP856709 | OP857210 | This study | |
| E26−4 | October 2019 | Motuo County, Tibet, China | 29°19′37.128″ N, 95°19′53.76″ E | soil | OP856708 | OP857209 | This study | |
| Aureobasidium mustum | AWRI 4233 CO−2020 | NA | South Australia | NA | grape juice | NA | NA | [17] |
| Aureobasidium namibiae | CBS 147.97 | 1997 | Namib Desert, Namibia | NA | dolomitic marble | FJ150875 | FJ150937 | [8] |
| Aureobasidium pini | CFCC 52778 | May 2018 | Miyun District, Beijing, China | 40°41′18″ N, 116°55′21″ E | pine needles covered with mycelium | MK184533 | MK184535 | [18] |
| Aureobasidium planticola sp. nov. | MDSC−10 | September 2022 | Zhoushan, Zhejiang, China | 29°53′28.86″ N, 122°24′59.35″ E | leaf | OP856711 | OP857212 | This study |
| Aureobasidium proteae | CBS 114273 | February 2006 | Netherlands | NA | Protea sp. | JN712491 | JN712557 | [15] |
| Aureobasidium proteae | CPC 13701 | July 1998 | Hilly Lands Farm, Somerset West, South Africa | NA | Protea cv. ‘Sylvia’ | JN712490 | JN712556 | [15] |
| Aureobasidium pullulans | CBS 584.75 | 1974 | France | NA | fruit of Vitis vinifera | FJ150906 | FJ150942 | [8] |
| Aureobasidium pullulans | CBS 146.30 | NA | Germany, Ohlsdorf near Hamburg | NA | slime flux of Quercus sp. | FJ150902 | FJ150916 | [8] |
| Aureobasidium subglaciale | EXF−2481 | June and August 2001 | Norway, Svalbard, Kongsvegen | 79° N, 12° E | subglacial ice from seawater | FJ150895 | FJ150913 | [8] |
| Aureobasidium thailandense | NRRL 58539T | 2006 | Nakhonratchasima, Thailand | NA | leaf of Cerbera odollum | JX462674 | JX462674 | [19] |
| Aureobasidium thailandense | NRRL 58543 | 2006 | Prachuapkhirikhan, Thailand | NA | wood surface | JX462675 | JX462675 | [19] |
| Aureobasidium tremulum | UN 1 | NA | NA | NA | NA | MK503657 | MK503660 | NA |
| Aureobasidium uvarum | AWRI 4620 CO−2020 | NA | NA | NA | NA | NA | NA | [17] |
| Aureobasidium vineae | AWRI4619 CO−2020 | NA | NA | NA | NA | NA | NA | [17] |
| Selenophoma mahoniae | CBS 388.92 | NA | Colorado, America | NA | Mahonia repens, leaf | FJ150872 | FJ150943 | [8] |
| Sydowia polyspora | CBS 750.71 | September 1969 | Quebec, Lac Normand, Canada | NA | Pinus strobus, twig | MH872085 | MH872085 | [41] |
3. Results
3.1. Phylogeny
3.2. Taxonomy
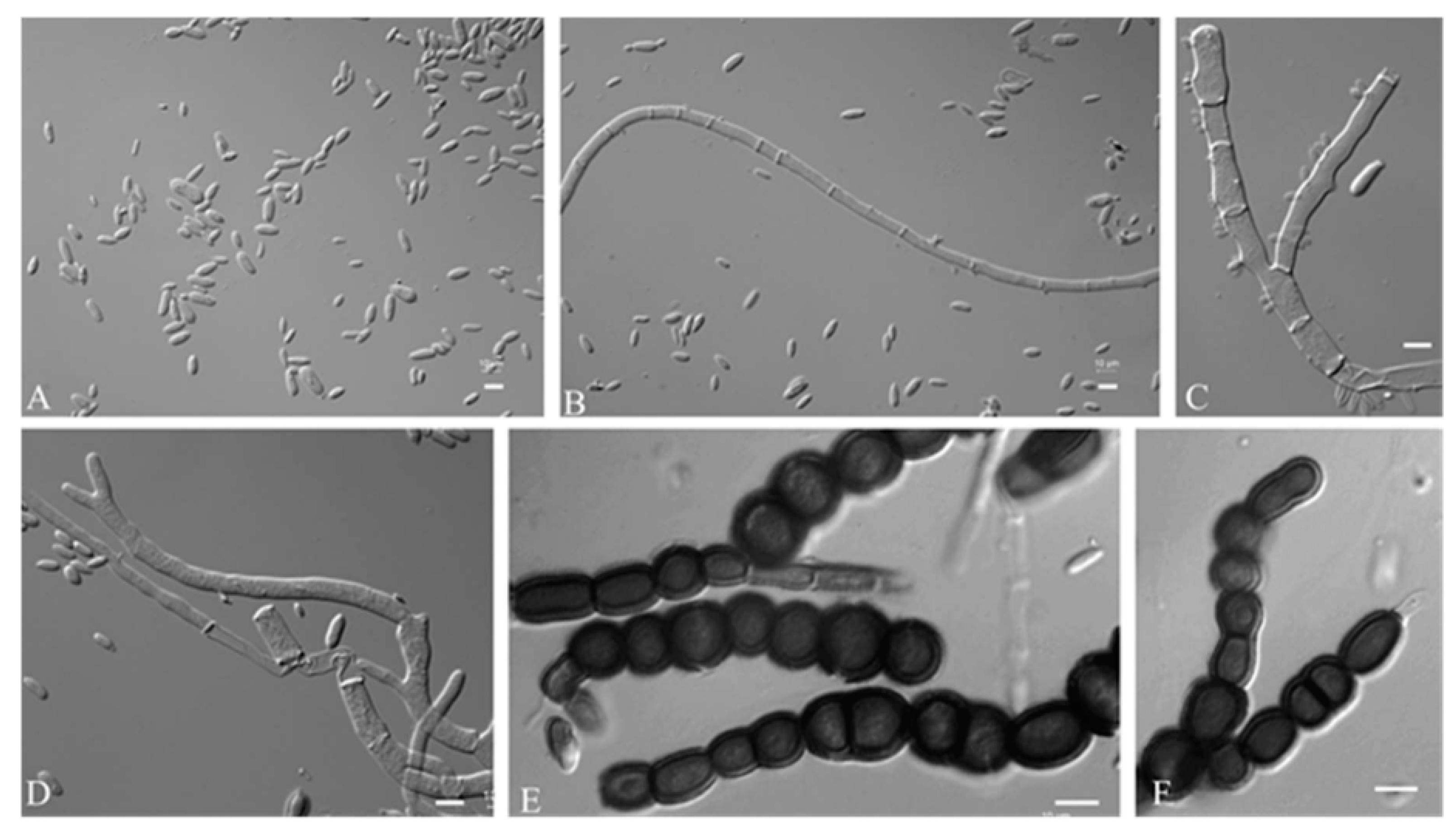
3.2.1. Aureobasidium insectorum Q.M. Wang, F. Wu & M.M. Wang sp. nov.
3.2.2. Aureobasidium planticola Q.M. Wang, F. Wu & M.M. Wang sp. nov.
3.2.3. Aureobasidium motuoense Q.M. Wang, F. Wu & M.M. Wang sp. nov.
3.2.4. Aureobasidium intercalariosporum Q.M. Wang, F. Wu & M.M. Wang sp. nov.
3.3. Exopolysaccharides Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hymphries, Z.; Seifert, K.A.; Hirooka, Y.; Visagie, M. A new family and genus in Dothideales for Aureobasidium-like species isolated from house dust. IMA Fungus 2017, 8, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Thambugala, K.M.; Ariyawansa, H.A.; Li, Y.M.; Boonmee, S.; Hongsanan, S.; Tian, Q.; Singtripop, C.; Bhat, D.J.; Camporesi, E.; Jayawardena, R.; et al. Dothideales. Fungal Divers. 2014, 68, 105–158. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Crous, P.W.; Kirk, P.M.; Hawksworth, D.L.; Boonmee, S.; Braun, U.; Dai, D.Q.; D’souza, M.J.; Diederich, P.; Dissanayake, A.; et al. Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Divers. 2014, 69, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Hermanides-Nijhof, E.J. Aureobasidium and allied genera. Stud. Mycol. 1977, 15, 141–177. [Google Scholar]
- De Hoog, G.S.; Yurlova, N.A. Conidiogenesis, nutritional physiology and taxonomy of Aureobasidium and Hormonema. Antonie Van Leeuwenhoek 1994, 65, 41–54. [Google Scholar] [CrossRef] [PubMed]
- De Hoog, G.S.; Zalar, P.; Urzi, C.; De Leo, F.; Yurlova, N.A.; Sterflinger, K. Relationships of dothideaceous black yeasts and meristematic fungi based on 5.8 S and ITS2 rDNA sequence comparison. Stud. Mycol. 1999, 43, 31–37. [Google Scholar]
- Yurlova, N.A.; Uijthof, J.M.J.; De Hoog, G.S. Distinction of species in Aureobasidium and related genera by PCR-ribotyping. Antonie Van Leeuwenhoek 1996, 69, 323. [Google Scholar] [CrossRef]
- Zalar, P.; Gostinčar, C.; De Hoog, G.S.; Uršič, V.; Sudhadham, M.; Gunde-Cimerman, N. Redefinition of Aureobasidium pullulans and its varieties. Stud. Mycol. 2008, 61, 21–38. [Google Scholar] [CrossRef]
- Gostinčar, C.; Ohm, R.A.; Kogej, T.; Sonjak, S.; Turk, M.; Zajc, J.; Zalar, P.; Grube, M.; Sun, H.; Han, J.; et al. Genome sequencing of four Aureobasidium pullulans varieties: Biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014, 15, 549. [Google Scholar] [CrossRef]
- Lee, D.H.; Cho, S.E.; Oh, J.Y.; Cho, E.J.; Kwon, S. A novel species of Aureobasidium (Dothioraceae) recovered from Acer pseudosieboldianum in Korea. J. Asia-Pac. Biodivers. 2021, 14, 657–661. [Google Scholar] [CrossRef]
- Wang, C.B.; Jiang, N.; Zhu, Y.Q.; Xue, H.; Li, Y. Aureobasidium aerium (Saccotheciaceae, Dothideales), a new yeast-like fungus from the air in Beijing, China. Phytotaxa 2022, 544, 185–192. [Google Scholar] [CrossRef]
- Jiang, N.; Fan, X.; Tian, C. Identification and Characterization of Leaf-Inhabiting Fungi from Castanea Plantations in China. J. Fungi 2021, 18, 64. [Google Scholar] [CrossRef]
- Arzanlou, M. Aureobasidium iranianum, a new species on bamboo from Iran. Mycosphere 2012, 3, 404–408. [Google Scholar] [CrossRef]
- Crous, P.W.; Summerell, B.A.; Swart, L.; Denman, S.; Taylor, J.E.; Bezuidenhout, C.M.; Palm, M.E.; Marincowitz, S.; Groenewald, J.Z. Fungal pathogens of Proteaceae. Persoonia 2011, 27, 20–45. [Google Scholar] [CrossRef]
- Nasr, S.; Mohammadimehr, M.; Geranpayeh Vaghei, M.; Amoozegar, M.A.; Shahzadeh Fazeli, S.A. Aureobasidium mangrovei sp. nov., an ascomycetous species recovered from Hara protected forests in the Persian Gulf, Iran. Antonie Van Leeuwenhoek 2018, 111, 1697–1705. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Thangavel, R.; Wingfield, M.J.; Noordeloos, M.E.; Dima, B.; Brandrud, T.; Jansen, G.M.; et al. Fungal Planet description sheets. Persoonia 2021, 46, 313–528. [Google Scholar] [CrossRef]
- Onetto, C.A.; Schmidt, S.A.; Roach, M.J.; Borneman, A.R. Comparative genome analysis proposes three new Aureobasidium species isolated from grape juice. FEMS Yeast Res. 2020, 20, 52. [Google Scholar] [CrossRef]
- Jiang, N.; Liang, Y.M.; Tian, C.M. Aureobasidium pini sp. nov. from pine needle in China. Phytotaxa 2019, 402, 10. [Google Scholar] [CrossRef]
- Peterson, S.; Manitchotpisit, P.; Leathers, T. Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. Int. J. Syst. Evol. Microbiol. 2012, 63, 790–795. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal Planet description sheets. Persoonia 2019, 44, 291–473. [Google Scholar] [CrossRef]
- Prasongsuk, S.; Lotrakul, P.; Ali, I.; Bankeeree, W.; Punnapayak, H. The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 2018, 63, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Manitchotpisit, P.; Leathers, T.D.; Peterson, S.W.; Kurtzman, C.P.; Li, X.L.; Eveleigh, D.E.; Lotrakul, P.; Prasongsuk, S.; Dunlap, C.A.; Vermillion, K.E.; et al. Multilocus phylogenetic analyses, pullulan production and xylanase activity of tropical isolates of Aureobasidium pullulans. Mycol. Res. 2009, 113, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Kutleša, M.; Mlinarić-Missoni, E.; Hatvani, L.; Voncina, D.; Simon, S.; Lepur, D.; Baršić, B. Chronic fungal meningitis caused by Aureobasidium proteae. Diagn. Microbiol. Infect. Dis. 2012, 73, 271–272. [Google Scholar] [CrossRef] [PubMed]
- Leathers, T.D. Biotechnological production and applications of pullulan. Appl. Microbiol. Biotechnol. 2002, 62, 468–473. [Google Scholar] [CrossRef]
- Cheng, K.C.; Demirci, A.; Catchmark, J.M. Pullulan: Biosynthesis, production, and applications. Appl. Microbiol. Biotechnol. 2011, 92, 29–44. [Google Scholar] [CrossRef]
- Manitchotpisit, P.; Skory, C.D.; Peterson, S.W.; Price, N.P.J.; Vermillion, K.E.; Leathers, T.D. Poly (β-L-malic acid) production by diverse phylogenetic clades of Aureobasidium pullulans. J. Ind. Microbiol. Biotechnol. 2012, 39, 125–132. [Google Scholar] [CrossRef]
- Muthusamy, S.; Anandharaj, S.J.; Kumar, P.S. Microbial pullulan for food, biomedicine, cosmetic, and water treatment: A review. Environ. Chem. Lett. 2022, 20, 3199–3234. [Google Scholar] [CrossRef]
- Kang, X.X.; Jia, S.L.; Wei, X.; Zhang, M.; Liu, G.L.; Hu, Z.; Chi, Z.; Chi, Z.M. Liamocins biosynthesis, its regulation in Aureobasidium spp., and their bioactivities. Crit. Rev. Biotechnol. 2022, 42, 93–105. [Google Scholar] [CrossRef]
- Suzuki, T.; Kusano, K.; Kondo, N.; Nishikawa, K.; Kuge, T.; Ohno, N. Biological Activity of High-Purity β-1,3-1,6-Glucan Derived from the Black Yeast Aureobasidium pullulans: A Literature Review. Nutrients 2021, 13, 242. [Google Scholar] [CrossRef]
- Xin, Z.; Chen, J. A high throughput DNA extraction method with high yield and quality. Plant Methods 2012, 8, 26. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification, and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Lin, D.; Wu, L.C.; Rinaldi, M.G.; Lehmann, P.F. Three distinct genotypes within Candida parapsilosis from clinical sources. J. Clin. Microbiol. 1995, 33, 1815–1821. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef]
- Wang, Q.M.; Begerow, D.; Groenewald, M.; Liu, X.; Theelen, B.; Bai, F.Y.; Boekhout, T. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Stud. Mycol. 2015, 81, 55–83. [Google Scholar] [CrossRef]
- Haghighatpanah, N.; Mirzaee, H.; Khodaiyan, F.; Kennedy, J.F.; Aghakhani, A.; Hosseini, S.S.; Jahanbin, K. Optimization and characterization of pullulan produced by a newly identified strain of Aureobasidium pullulans. Int. J. Biol. Macromol. 2020, 152, 305–313. [Google Scholar] [CrossRef]
- Buksa, K.; Kowalczyk, M.; Boreczek, J. Extraction, purification and characterisation of exopolysaccharides produced by newly isolated lactic acid bacteria strains and the examination of their influence on resistant starch for-mation. Food Chem. 2021, 362, 130221. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Prabhugaonkar, A.; Jalmi, P. Aureobasidium khasianum (Aureobasidiaceae) a novel species with distinct morphology. Phytotaxa 2018, 374, 257. [Google Scholar] [CrossRef]
- Verkley, G.J.; Starink-Willemse, M.; van Iperen, A.; Abeln, E.C. Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia 2004, 96, 558–571. [Google Scholar] [CrossRef] [PubMed]
- Groenewald, M.; Boundy-Mills, K.; Čadež, N.; Endoh, R.; Jindamorakot, S.; Pohl-Albertyn, C.; Rosa, C.A.; Turchetti, B.; Yurkov, A. Census of yeasts isolated from natural ecosystem and conserved in worldwide collections. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin, Germany, 2017; pp. 455–476. [Google Scholar]
- Blackwell, M. Yeasts in Insects and Other Invertebrates. In Yeasts in Natural Ecosystems: Diversity; Buzzini, P., Lachance, M.A., Yurkov, A., Eds.; Springer: Berlin, Germany, 2017; pp. 397–433. [Google Scholar] [CrossRef]
- Boekhout, T.; Amend, A.S.; EI Baidouri, F.; Gabaldón, T.; GemI, J.; Mittelbach, M.; Rober, V.; Tan, C.S.; Turchetti, B.; Vu, D.; et al. Trends in yeast diversity discovery. Fungal Divers. 2022, 114, 491–537. [Google Scholar] [CrossRef]
- Boekhout, T. Biodiversity: Gut feeling for yeasts. Nature 2005, 434, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Madden, A.A.; Epps, M.J.; Fukami, T.; Irwin, R.E.; Sheppard, J.; Sorger, D.M.; Dunn, R.R. The ecology of insect-yeast relationships and its relevance to human industry. Proc. Biol. Sci. 2018, 285, 20172733. [Google Scholar] [CrossRef]
- Zou, X.; Cheng, C.; Feng, J.; Song, X.; Lin, M.; Yang, S.T. Biosynthesis of polymalic acid in fermentation: Advances and prospects for industrial application. Crit. Rev. Biotechnol. 2019, 39, 408–421. [Google Scholar] [CrossRef]





| Strain | Species | Fermentation Liquid Color | Exopolysaccharides Yield (g/L) | Average Weight (g/L) |
|---|---|---|---|---|
| PTSL5−5 | A. thailandense | Light yellow | 8.47 | 6.17 |
| PTSL4−6 | A. thailandense | Light yellow | 8.09 | |
| PTSL5−3 | A. thailandense | Light yellow | 2.71 | |
| PTSL11−5 | A. thailandense | Light yellow | 5.42 | |
| PTSL9−106 | A. melanogenum | Pink | 1.53 | 39.06 |
| PTSL19−101 | A. melanogenum | Light yellow | 32.53 | |
| PTSL6−101 | A. melanogenum | Light yellow | 34.50 | |
| PTSL19−107 | A. melanogenum | Yellow | 41.84 | |
| PTSL19−104 | A. melanogenum | Yellow | 54.58 | |
| PTSL20−102 | A. melanogenum | Light yellow | 48.13 | |
| PTSL20−104 | A. melanogenum | Yellow | 52.33 | |
| PTSL19−104 | A. melanogenum | Light yellow | 45.71 | |
| PTSL19−104 | A. melanogenum | Light yellow | 45.71 | |
| PTSL17−4 | A. melanogenum | Light yellow | 34.36 | |
| PTSL9−100 | A. melanogenum | Light yellow | 38.45 | |
| LF75−2 | A. leucospermi | Light yellow | 0.92 | 17.24 |
| SXY35−16 | A. leucospermi | Light yellow | 28.94 | |
| SXY35−15 | A. leucospermi | Light yellow | 23.37 | |
| LF45−2 | A. leucospermi | Light yellow | 15.75 | |
| LPL−7A | A. insectorum | Light yellow | 27.67 | 14.7 |
| KCL139 | A. insectorum | Dark yellow | 8.64 | |
| XZY65−10 | A. insectorum | Dark yellow | 7.80 | |
| E26−4 | A. motuoense | Yellow | 15.74 | 26.57 |
| E31−1 | A. motuoense | Dark yellow | 21.39 | |
| XZY411−4 | A. motuoense | Dark yellow | 31.72 | |
| E82−2 | A. motuoense | Dark yellow | 37.43 | |
| MGL11−3 | A. intercalariosporum | Light yellow | 29.43 | 31.79 |
| MQL9−100 | A. intercalariosporum | Light yellow | 34.15 | |
| MDSC−10 | A. planticola | Black | 2.10 | 2.1 |
| Aureobasidium planticola | Aureobasidium intercalariosporum | Aureobasidium motuoense | Aureobasidium insectorum | |
|---|---|---|---|---|
| PDA | 22–25 | 32–36 | 34–38 | 32–35 |
| M40Y | 33–40 | 35–40 | 41–43 | 32–34 |
| M60Y | 27–34 | 29–34 | 36–40 | 25–27 |
| MEA + 5% NaCl | 14–15 | 14–17 | 13–15 | 9–12 |
| MEA + 10% NaCl | 7–8 | 8–11 | 10–11 | 8–8 |
| MEA + 15% NaCl | 0 | 9–13 | 5–8 | 8–9 |
| MEA + 20% NaCl | 0 | 0 | 0 | 0 |
| MEA at 4 °C | 0 | 0 | 0 | 5–5 |
| MEA at 17 °C | 10–13 | 13–15 | 8–13 | 12–15 |
| MEA at 28 °C | 27–27 | 24–25 | 32–35 | 28–28 |
| MEA at 30 °C | 11–10 | 13–14 | 38–44 | 7–8 |
| MEA at 37 °C | 0 | 0 | 7–8 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, F.; Feng, Z.; Wang, M.; Wang, Q. Proposal of Four New Aureobasidium Species for Exopolysaccharide Production. J. Fungi 2023, 9, 447. https://doi.org/10.3390/jof9040447
Wu F, Feng Z, Wang M, Wang Q. Proposal of Four New Aureobasidium Species for Exopolysaccharide Production. Journal of Fungi. 2023; 9(4):447. https://doi.org/10.3390/jof9040447
Chicago/Turabian StyleWu, Feng, Zixuan Feng, Manman Wang, and Qiming Wang. 2023. "Proposal of Four New Aureobasidium Species for Exopolysaccharide Production" Journal of Fungi 9, no. 4: 447. https://doi.org/10.3390/jof9040447
APA StyleWu, F., Feng, Z., Wang, M., & Wang, Q. (2023). Proposal of Four New Aureobasidium Species for Exopolysaccharide Production. Journal of Fungi, 9(4), 447. https://doi.org/10.3390/jof9040447






