A Novel Effector, FSE1, Regulates the Pathogenicity of Fusarium oxysporum f. sp. cubense Tropical Race 4 to Banana by Targeting the MYB Transcription Factor MaEFM-Like
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis
2.2. Fungal Strains and Culture Conditions
2.3. Vector Construction and Protoplast Transformation
2.4. Inoculation of Banana Plantlets and Pathogenicity Assay
2.5. RT-qPCR
2.6. Yeast Two-Hybrid System
2.7. Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC)
3. Results
3.1. FSE1 Is a Special Candidate Effector Conserved in in Fusarium spp.
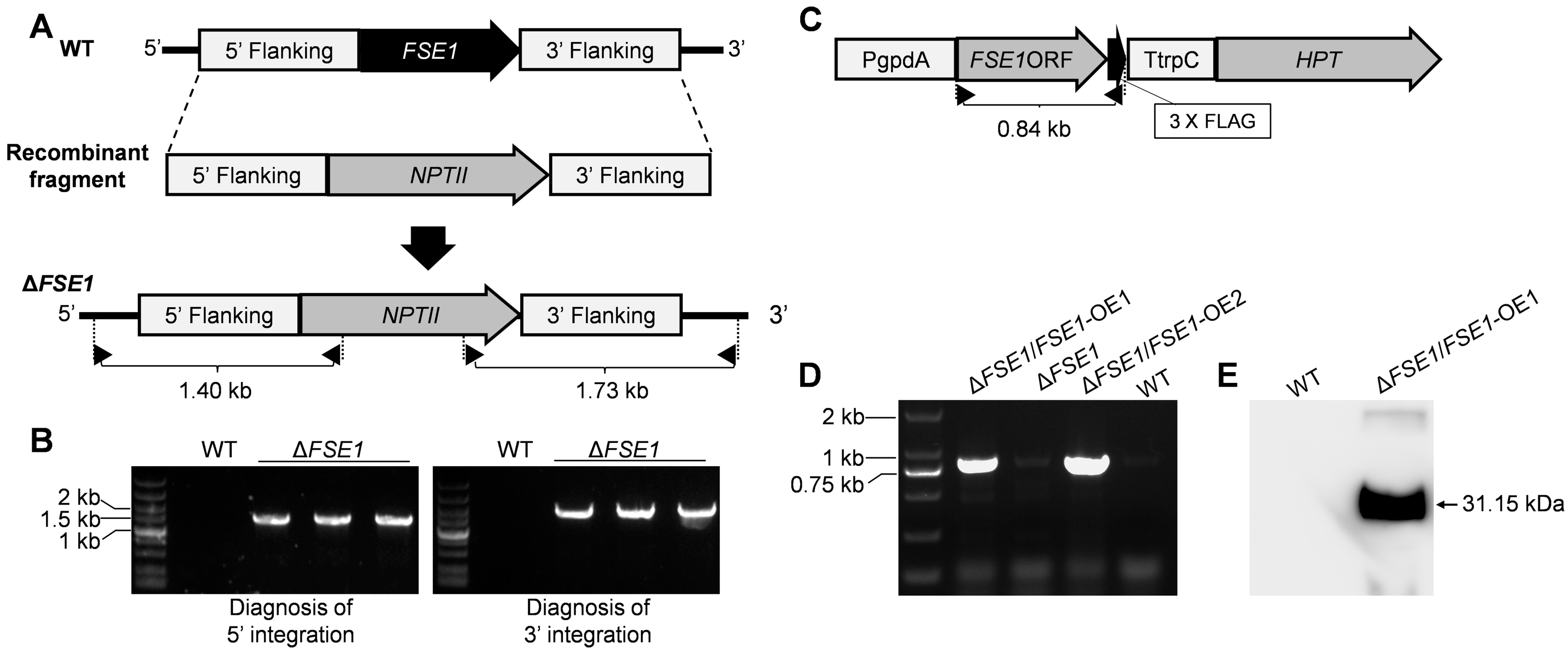
3.2. Construction of FSE1 Knock-Out and Over-Expression Strains
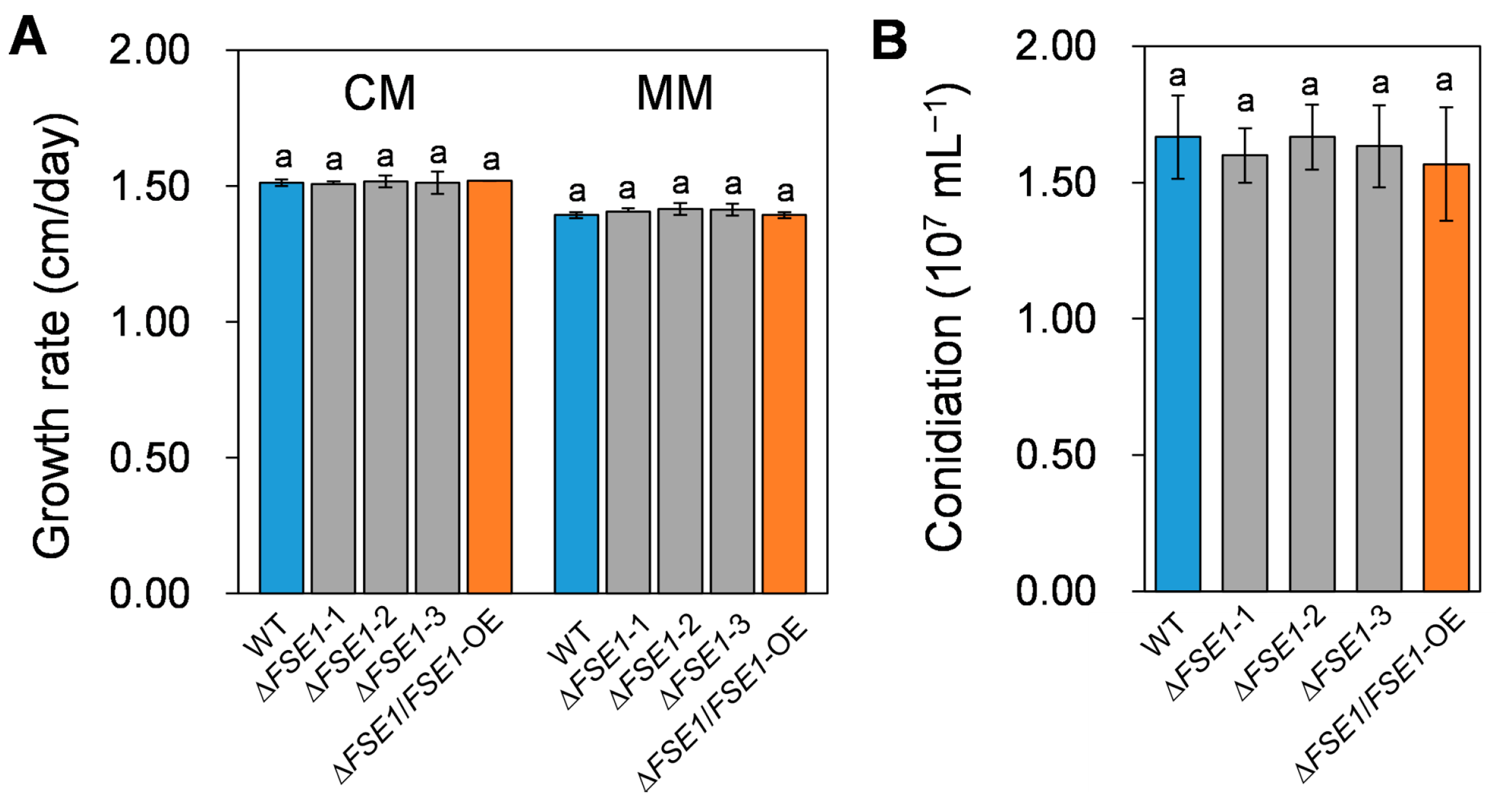
3.3. FSE1 Is Not Required for Vegetative Growth and Conidiation
3.4. FSE1 Is Involved in the Pathogenicity of Foc TR4
3.5. FSE1 Is Distributed in Vesicles of Foc TR4 and Localized in Cytoplasm and Nuclei of N. benthamiana Cells
3.6. FSE1 Interacted with MYB Family Transcription Factor EFM
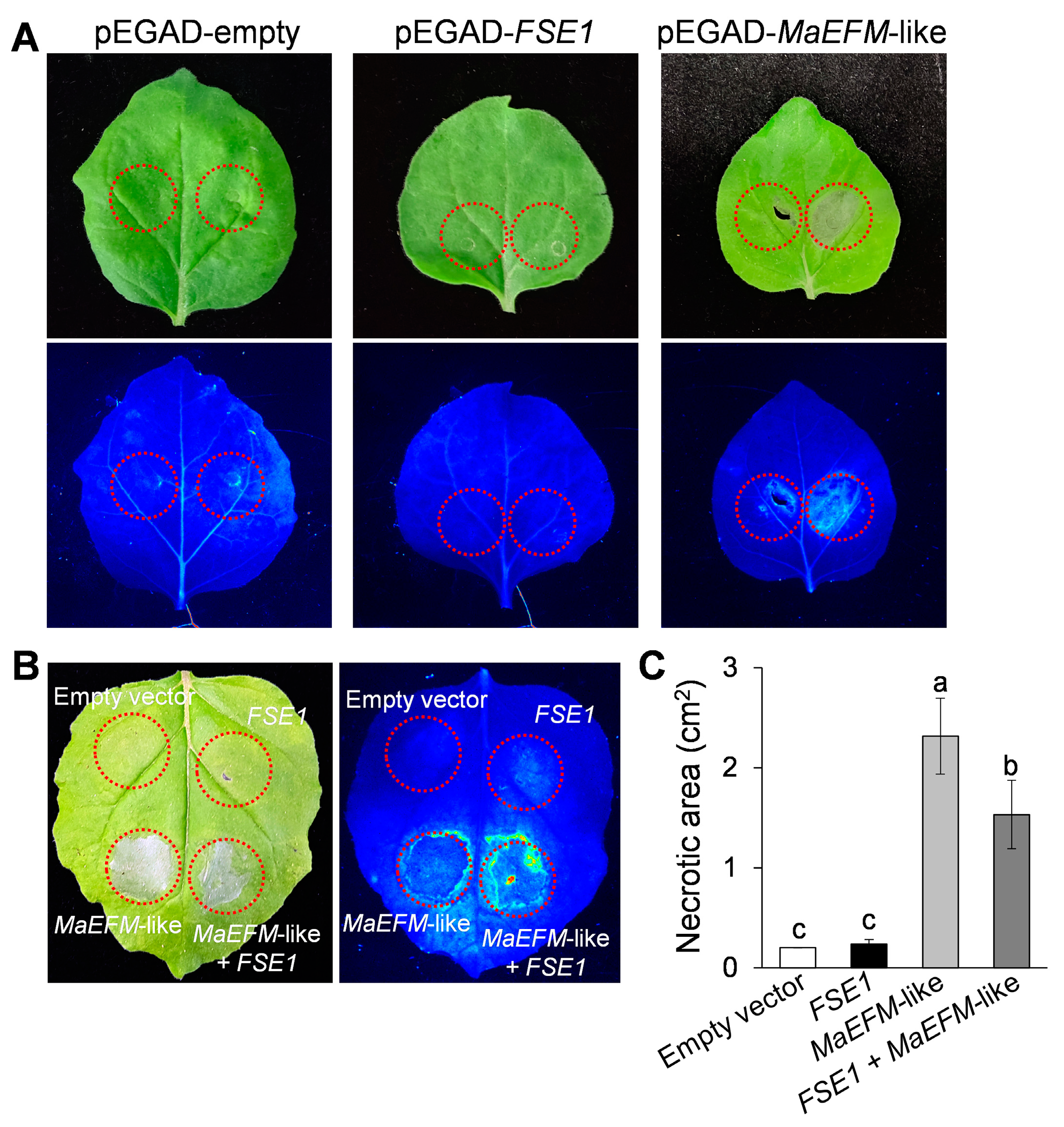
3.7. FSE1 Supressed the MaEFM-Like-Induced Cell Death
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Pegg, K.G.; Coates, L.M.; O’Neill, W.T.; Turner, D.W. The Epidemiology of Fusarium Wilt of Banana. Front. Plant. Sci. 2019, 10, 1395. [Google Scholar] [CrossRef] [Green Version]
- Gordon, T.R.; Martyn, R.D. The evolutionary biology of Fusarium oxysporum. Annu. Rev. Phytopathol. 1997, 35, 111–128. [Google Scholar] [CrossRef] [Green Version]
- Ploetz, R.C. Panama Disease: An Old Nemesis Rears Its Ugly Head: Part 1. The Beginnings of the Banana Export Trades. Plant Health Prog. 2005, 6, 18. [Google Scholar] [CrossRef]
- Ordonez, N.; Seidl, M.F.; Waalwijk, C.; Drenth, A.; Kilian, A.; Thomma, B.P.; Ploetz, R.C.; Kema, G.H. Worse comes to worst: Bananas and Panama disease-when plant and pathogen clones meet. PLoS Pathog. 2015, 11, e1005197. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Varden, F.A.; De la Concepcion, J.C.; Maidment, J.H.; Banfield, M.J. Taking the stage: Effectors in the spotlight. Curr. Opin. Plant Biol. 2017, 38, 25–33. [Google Scholar] [CrossRef] [PubMed]
- McDermott, J.E.; Corrigan, A.; Peterson, E.; Oehmen, C.; Niemann, G.; Cambronne, E.D.; Sharp, D.; Adkins, J.N.; Samudrala, R.; Heffron, F. Computational prediction of type III and IV secreted effectors in gram-negative bacteria. Infect. Immun. 2011, 79, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petre, B.; Kamoun, S. How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 2014, 12, e1001801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; Boer, A.D.D.; Cornelissen, B.J.C. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 2002, 130, 904–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houterman, P.M.; Cornelissen, B.J.C.; Rep, M. Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 2008, 4, e1000061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rep, M.; Van Der Does, H.C.; Meijer, M.; Van Wijk, R.; Houterman, P.M.; Dekker, H.L.; De Koster, C.G.; Cornelissen, B.J.C. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 2004, 53, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Houterman, P.M.; Ma, L.; Van Ooijen, G.; De Vroomen, M.J.; Cornelissen, B.J.C.; Takken, F.L.W.; Rep, M. The effector protein Avr2 of the xylem colonizing fungus Fusarium oxysporum activates the tomato resistance protein I-2 intracellularly. Plant J. 2009, 58, 970–978. [Google Scholar] [CrossRef]
- Takken, F.; Rep, M. The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 2010, 11, 309–314. [Google Scholar] [CrossRef]
- An, B.; Hou, X.; Guo, Y.; Zhao, S.; Luo, H.; He, C.; Wang, Q. The effector SIX8 is required for virulence of Fusarium oxysporum f.sp. cubense tropical race 4 to Cavendish banana. Fungal Biol. 2019, 123, 423–430. [Google Scholar] [CrossRef]
- Feng, Q.; Gao, X.; An, B.; He, C.; Wang, Q. Two cerato-platanin proteins FocCP1 interact with MaPR1 and contribute to virulence of Fusarium oxysporum f. sp. cubense to banana. J. Plant Interact. 2021, 16, 238–245. [Google Scholar] [CrossRef]
- Zhao, S.; An, B.; Guo, Y.; Hou, X.; Luo, H.; He, C.; Wang, Q. Label free proteomics and systematic analysis of secretome reveals effector candidates regulated by SGE1 and FTF1 in the plant pathogen Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2020, 21, 275. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; An, B.; Luo, H.; He, C.; Wang, Q. The histone acetyltransferase FocGCN5 regulates growth, conidiation, and pathogenicity of the banana wilt disease causal agent Fusarium oxysporum f. sp. cubense tropical race 4. Res. Microbiol. 2022, 173, 103902. [Google Scholar] [CrossRef]
- Widinugraheni, S.; Nino-Sanchez, J.; van der Does, H.C.; van Dam, P.; Garcia-Bastidas, F.A.; Subandiyah, S.; Meijer, H.J.G.; Kistler, H.C.; Kema, G.H.J.; Rep, M. A SIX1 homolog in Fusarium oxysporum f. sp. cubense tropical race 4 contributes to virulence towards Cavendish banana. PLoS ONE 2018, 13, e0205896. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N.; Gardiner, D.M.; Singh, K.B.; Taylor, J.M. Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol. Plant Pathol. 2018, 19, 2094–2110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, D.; Yarden, O.; Hadar, Y. Seeking the roles for fungal small secreted proteins in affecting saprophytic lifestyles. Front. Microbiol. 2020, 11, 455. [Google Scholar] [CrossRef] [Green Version]
- Ito, K.; Tanaka, T.; Hatta, R.; Yamamoto, M.; Tsuge, T. Dissection of the host range of the fungal plant pathogen Alternaria alternata by modification of secondary metabolism. Mol. Microbiol. 2004, 52, 399–411. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. Apoplastic fungal effectors in historic perspective; a personal view. New Phytol. 2016, 212, 805–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, S.; Kahmann, R. Cell wall-associated effectors of plant-colonizing fungi. Mycologia 2021, 113, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Lyons, R.; Stiller, J.; Powell, J.; Rusu, A.; Manners, J.M.; Kazan, K. Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS ONE 2015, 10, e0121902. [Google Scholar] [CrossRef]
- Krol, P.; Iqielski, R.; Pollmann, S.; Kepczynska, E. Priming of seeds with methyl jasmonate induced resistance to hemi-biotroph Fusarium oxysporum f. sp. lycopersici in tomato via 12-oxo-phytodienoic acid, salicylic acid and flavonol accumulation. J. Plant Physiol. 2015, 179, 122–132. [Google Scholar] [CrossRef]
- Galán, J.E.; Lara-Tejero, M.; Marlovits, T.C.; Wagner, S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu. Rev. Microbiol. 2014, 68, 415–438. [Google Scholar] [CrossRef] [Green Version]
- Mitchum, M.G.; Hussey, R.S.; Baum, T.J.; Wang, X.; Elling, A.A.; Wubben, M.; Davis, E.L. Nematode effector proteins: An emerging paradigm of parasitism. New Phytol. 2013, 199, 879–894. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, L.; Kahmann, R. How filamentous plant pathogen effectors are translocated to host cells. Curr. Opin. Plant Biol. 2017, 38, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol. Mol. Biol. Rev. 2018, 82, e00068-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toruño, T.Y.; Stergiopoulos, I.; Coaker, G. Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 2016, 54, 419–441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Dröge-Laser, W.; Snoek, B.L.; Snel, B.; Weiste, C. The Arabidopsis bZIP transcription factor family—An update. Curr. Opin. Plant Biol. 2018, 45, 36–49. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Qi, T.; Guo, J.; Liu, P.; He, F.; Wan, C.; Islam, M.A.; Tyler, B.M.; Kang, Z.; Guo, J. Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant. 2019, 12, 1624–1638. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ning, Y.; Shi, X.; He, F.; Zhang, C.; Fan, J.; Jiang, N.; Zhang, Y.; Zhang, T.; Hu, Y.; et al. Immunity to rice blast disease by suppression of effector-triggered necrosis. Curr. Biol. 2016, 26, 2399–2411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, G.; Yang, J.; Zhang, Q.; Wang, W.; Feng, L.; Zhao, L.; An, B.; Wang, Q.; He, C.; Luo, H. The effector protein CgNLP1 of Colletotrichum gloeosporioides affects invasion and disrupts nuclear localization of necrosis-induced transcription factor HbMYB8-Like to suppress plant defense signaling. Front. Microbiol. 2022, 13, 911479. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Vailleau, F.; Léger, A.; Joubès, J.; Miersch, O.; Huard, C.; Blée, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Yu, X.; Thompson, A.; Guo, M.; Yoshida, S.; Asami, T.; Chory, J.; Yin, Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009, 58, 275–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; An, B.; Guo, Y.; Luo, H.; He, C.; Wang, Q. A Novel Effector, FSE1, Regulates the Pathogenicity of Fusarium oxysporum f. sp. cubense Tropical Race 4 to Banana by Targeting the MYB Transcription Factor MaEFM-Like. J. Fungi 2023, 9, 472. https://doi.org/10.3390/jof9040472
Yang Y, An B, Guo Y, Luo H, He C, Wang Q. A Novel Effector, FSE1, Regulates the Pathogenicity of Fusarium oxysporum f. sp. cubense Tropical Race 4 to Banana by Targeting the MYB Transcription Factor MaEFM-Like. Journal of Fungi. 2023; 9(4):472. https://doi.org/10.3390/jof9040472
Chicago/Turabian StyleYang, Yongbao, Bang An, Yunfeng Guo, Hongli Luo, Chaozu He, and Qiannan Wang. 2023. "A Novel Effector, FSE1, Regulates the Pathogenicity of Fusarium oxysporum f. sp. cubense Tropical Race 4 to Banana by Targeting the MYB Transcription Factor MaEFM-Like" Journal of Fungi 9, no. 4: 472. https://doi.org/10.3390/jof9040472
APA StyleYang, Y., An, B., Guo, Y., Luo, H., He, C., & Wang, Q. (2023). A Novel Effector, FSE1, Regulates the Pathogenicity of Fusarium oxysporum f. sp. cubense Tropical Race 4 to Banana by Targeting the MYB Transcription Factor MaEFM-Like. Journal of Fungi, 9(4), 472. https://doi.org/10.3390/jof9040472






