Coccidioidomycosis and Host Microbiome Interactions: What We Know and What We Can Infer from Other Respiratory Infections
Abstract
:1. Coccidioides Overview
2. Respiratory Tract Microbiome
3. Microbiome Changes during Perturbation
3.1. Influence of The Gut–Lung Axis during Viral and Bacterial Infections
3.2. COVID-19 and Lung Microbiome
3.3. Changes to Lung Microbiota Induced by Infection: Bacterial, Viral, Fungal, and Parasitic
4. Productive Immune Responses against Coccidioides
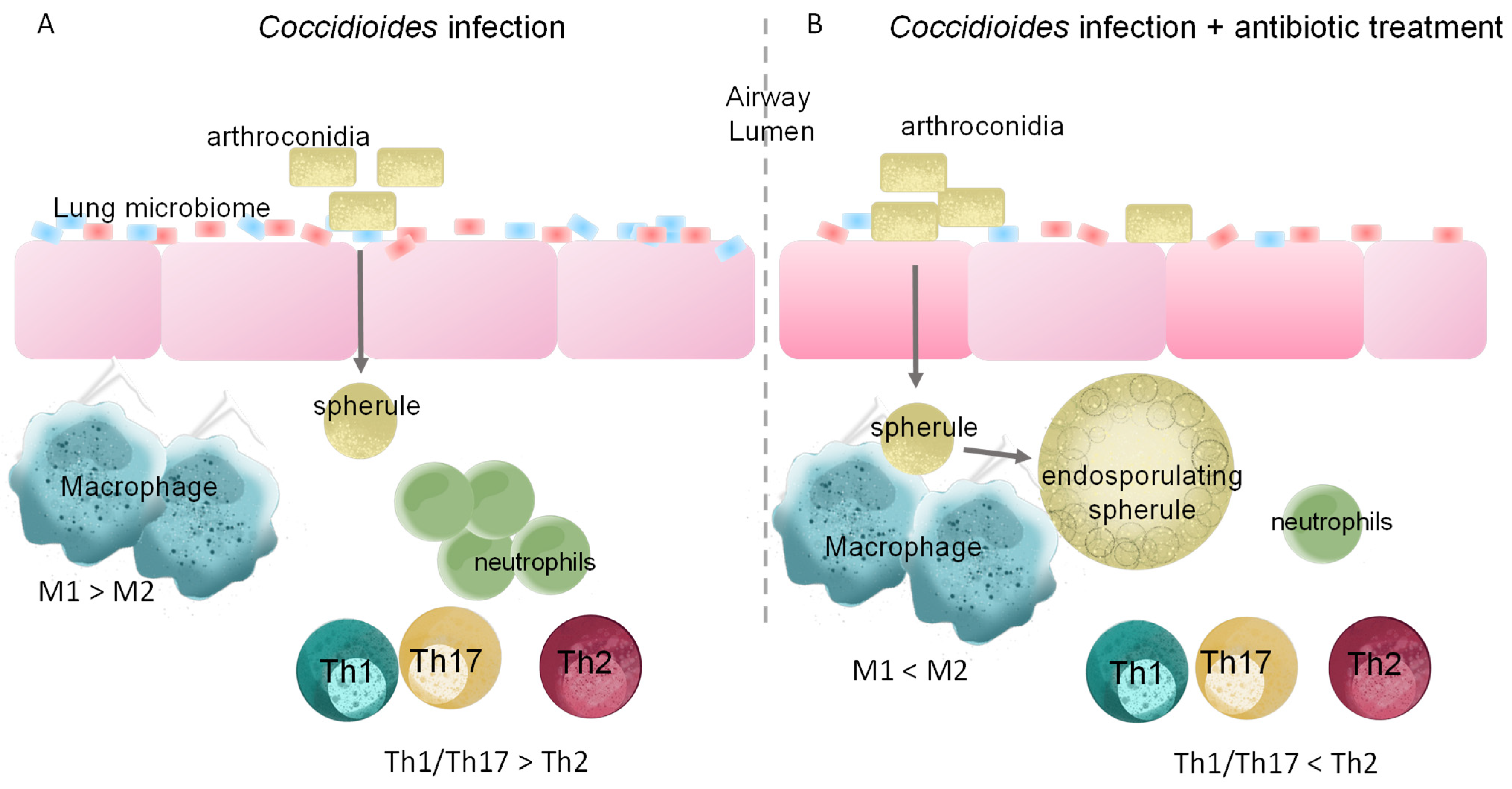
5. Changes to Lung Microbiota Induced by Antibiotic Treatment Alters Immune Response to Infection
6. Conclusions and Perspectives
6.1. Microbiota and Immune Features for Diagnostic Use
6.2. Probiotics and Bacterial Products as a Therapeutic Option
6.3. Future Directions for Coccidioidomycosis Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, S.L.; Chiller, T. Update on the Epidemiology, Diagnosis, and Treatment of Coccidioidomycosis. J. Fungi 2022, 8, 666. [Google Scholar] [CrossRef] [PubMed]
- Improving Diagnosis of Valley Fever to Improve Antibiotic Use|CDC. 2022. Available online: https://www.cdc.gov/antibiotic-use/stewardship-report/pdf/stewardship-report-valley-fever-factsheet-p.pdf (accessed on 28 February 2023).
- Gorris, M.E.; Treseder, K.K.; Zender, C.S.; Randerson, J.T. Expansion of Coccidioidomycosis Endemic Regions in the United States in Response to Climate Change. Geohealth 2019, 3, 308–327. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The microbiome of the lung. Transl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Moffatt, M.F.; Cookson, W.O. The lung microbiome in health and disease. Clin. Med. 2017, 17, 525–529. [Google Scholar] [CrossRef]
- Charlson, E.S.; Bittinger, K.; Haas, A.R.; Fitzgerald, A.S.; Frank, I.; Yadav, A.; Bushman, F.D.; Collman, R.G. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011, 184, 957–963. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Kim, E.; Cox, M.J.; Brodie, E.L.; Brown, R.; Wiener-Kronish, J.P.; Lynch, S.V. A persistent and diverse airway microbiota present during chronic obstructive pulmonary disease exacerbations. OMICS 2010, 14, 9–59. [Google Scholar] [CrossRef] [PubMed]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B.; et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011, 6, e16384. [Google Scholar] [CrossRef] [PubMed]
- Willner, D.; Haynes, M.R.; Furlan, M.; Schmieder, R.; Lim, Y.W.; Rainey, P.B.; Rohwer, F.; Conrad, D. Spatial distribution of microbial communities in the cystic fibrosis lung. ISME J. 2012, 6, 471–474. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.; Zhao, N.; Zheng, R.; Wang, D.; Liu, W.; Liu, B. Oral administration of Clostridium butyricum rescues streptomycin-exacerbated respiratory syncytial virus-induced lung inflammation in mice. Virulence 2021, 12, 2133–2148. [Google Scholar] [CrossRef]
- Dessein, R.; Bauduin, M.; Grandjean, T.; Le Guern, R.; Figeac, M.; Beury, D.; Faure, K.; Faveeuw, C.; Guery, B.; Gosset, P.; et al. Antibiotic-related gut dysbiosis induces lung immunodepression and worsens lung infection in mice. Crit. Care 2020, 24, 611. [Google Scholar] [CrossRef] [PubMed]
- Merenstein, C.; Liang, G.; Whiteside, S.A.; Cobián-Güemes, A.G.; Merlino, M.S.; Taylor, L.J.; Glascock, A.; Bittinger, K.; Tanes, C.; Graham-Wooten, J.; et al. Signatures of COVID-19 Severity and Immune Response in the Respiratory Tract Microbiome. mBio 2021, 12, e0177721. [Google Scholar] [CrossRef] [PubMed]
- Bernard-Raichon, L.; Venzon, M.; Klein, J.; Axelrad, J.E.; Zhang, C.; Sullivan, A.P.; Hussey, G.A.; Casanovas-Massana, A.; Noval, M.G.; Valero-Jimenez, A.M.; et al. Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nat. Commun. 2022, 13, 5926. [Google Scholar] [CrossRef] [PubMed]
- Schuijt, T.J.; Lankelma, J.M.; Scicluna, B.P.; de Sousa e Melo, F.; Roelofs, J.J.; de Boer, J.D.; Hoogendijk, A.J.; de Beer, R.; de Vos, A.; Belzer, C.; et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016, 65, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Fagundes, C.T.; Amaral, F.A.; Vieira, A.T.; Soares, A.C.; Pinho, V.; Nicoli, J.R.; Vieira, L.Q.; Teixeira, M.M.; Souza, D.G. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 2012, 188, 1411–1420. [Google Scholar] [CrossRef]
- Wu, S.; Jiang, Z.Y.; Sun, Y.F.; Yu, B.; Chen, J.; Dai, C.Q.; Wu, X.L.; Tang, X.L.; Chen, X.Y. Microbiota regulates the TLR7 signaling pathway against respiratory tract influenza A virus infection. Curr. Microbiol. 2013, 67, 414–422. [Google Scholar] [CrossRef]
- Antunes, K.H.; Fachi, J.L.; de Paula, R.; da Silva, E.F.; Pral, L.P.; Dos Santos, A.; Dias, G.B.M.; Vargas, J.E.; Puga, R.; Mayer, F.Q.; et al. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat. Commun. 2019, 10, 3273. [Google Scholar] [CrossRef]
- Yang, F.; Yang, Y.; Chen, L.; Zhang, Z.; Liu, L.; Zhang, C.; Mai, Q.; Chen, Y.; Chen, Z.; Lin, T.; et al. The gut microbiota mediates protective immunity against tuberculosis. Gut Microbes 2022, 14, 2029997. [Google Scholar] [CrossRef]
- Miranda, N.; Hoyer, K.K. Valley fever and other infectious granulomas: Advancements, gaps, and challenges.
- Kim, Y.J.; Lee, J.Y.; Lee, J.J.; Jeon, S.M.; Silwal, P.; Kim, I.S.; Kim, H.J.; Park, C.R.; Chung, C.; Han, J.E.; et al. Arginine-mediated gut microbiome remodeling promotes host pulmonary immune defense against nontuberculous mycobacterial infection. Gut Microbes 2022, 14, 2073132. [Google Scholar] [CrossRef]
- Wang, J.; Li, F.; Wei, H.; Lian, Z.X.; Sun, R.; Tian, Z. Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 2014, 211, 2397–2410. [Google Scholar] [CrossRef]
- Cuthbertson, L.; James, P.; Habibi, M.S.; Thwaites, R.S.; Paras, A.; Chiu, C.; Openshaw, P.J.M.; Cookson, W.O.C.; Moffatt, M.F. Resilience of the respiratory microbiome in controlled adult RSV challenge study. Eur. Respir. J. 2022, 59, 2101932. [Google Scholar] [CrossRef] [PubMed]
- Krcméry, V.; Matejicka, F.; Pichnová, E.; Jurga, L.; Sulcova, M.; Kunová, A.; West, D. Documented fungal infections after prophylaxis or therapy with wide spectrum antibiotics: Relationship between certain fungal pathogens and particular antimicrobials? J. Chemother. 1999, 11, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Impact of Fungal Diseases in the United States|CDC. 2022. Available online: https://www.cdc.gov/fungal/cdc-and-fungal/burden.html#:~:text=More%20than%2075%2C000%20hospitalizations%20and,fungal%20diseases%20(Table%201) (accessed on 28 February 2023).
- Valley Fever Statistics|Coccidioidomycosis|Types of Fungal Diseases|Fungal|CDC. 2022. Available online: https://www.cdc.gov/fungal/diseases/coccidioidomycosis/statistics.html (accessed on 28 February 2023).
- Heaney, A.K.; Head, J.R.; Broen, K.; Click, K.; Taylor, J.; Balmes, J.R.; Zelner, J.; Remais, J.V. Coccidioidomycosis and COVID-19 Co-Infection, United States, 2020. Emerg. Infect. Dis. 2021, 27, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Casalini, G.; Giacomelli, A.; Ridolfo, A.; Gervasoni, C.; Antinori, S. Invasive Fungal Infections Complicating COVID-19: A Narrative Review. J. Fungi 2021, 7, 921. [Google Scholar] [CrossRef] [PubMed]
- Sous, R.; Levkiavska, Y.; Sharma, R.; Jariwal, R.; Amodio, D.; Johnson, R.H.; Heidari, A.; Kuran, R. Two Cases of Miliary and Disseminated Coccidioidomycosis Following Glucocorticoid Therapy and Literature Review. J. Investig. Med. High Impact. Case Rep. 2022, 10, 23247096211051928. [Google Scholar] [CrossRef]
- Shen, Z.; Xiao, Y.; Kang, L.; Ma, W.; Shi, L.; Zhang, L.; Zhou, Z.; Yang, J.; Zhong, J.; Yang, D.; et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 713–720. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Gao, Y.; Zhou, J.; Wang, S.; Huang, B.; Wu, J.; Cao, Q.; Chen, Y.; Wang, Z.; et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Infect. 2020, 81, e64–e67. [Google Scholar] [CrossRef]
- Cui, Z.; Zhou, Y.; Li, H.; Zhang, Y.; Zhang, S.; Tang, S.; Guo, X. Complex sputum microbial composition in patients with pulmonary tuberculosis. BMC Microbiol. 2012, 12, 276. [Google Scholar] [CrossRef]
- Macleod, T.; Ainscough, J.S.; Hesse, C.; Konzok, S.; Braun, A.; Buhl, A.L.; Wenzel, J.; Bowyer, P.; Terao, Y.; Herrick, S.; et al. The Proinflammatory Cytokine IL-36γ Is a Global Discriminator of Harmless Microbes and Invasive Pathogens within Epithelial Tissues. Cell Rep. 2020, 33, 108515. [Google Scholar] [CrossRef]
- de Mello, T.P.; Aor, A.C.; Branquinha, M.H.; Dos Santos, A.L.S. Insights into the interaction of Scedosporium apiospermum, Scedosporium aurantiacum, Scedosporium minutisporum, and Lomentospora prolificans with lung epithelial cells. Braz. J. Microbiol. 2020, 51, 427–436. [Google Scholar] [CrossRef]
- Cadena, A.M.; Ma, Y.; Ding, T.; Bryant, M.; Maiello, P.; Geber, A.; Lin, P.L.; Flynn, J.L.; Ghedin, E. Profiling the airway in the macaque model of tuberculosis reveals variable microbial dysbiosis and alteration of community structure. Microbiome 2018, 6, 180. [Google Scholar] [CrossRef]
- Lozupone, C.; Cota-Gomez, A.; Palmer, B.E.; Linderman, D.J.; Charlson, E.S.; Sodergren, E.; Mitreva, M.; Abubucker, S.; Martin, J.; Yao, G.; et al. Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 2013, 187, 1110–1117. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhu, M.; Zhang, L.; Peng, M.; Li, C.; Wang, L.; Wang, W.; Ma, Z.; Li, S.; Zeng, W.; et al. Differences in microbiome of healthy Sprague Dawley rats with Paragonimus proliferus infection and potential pathogenic role of microbes in paragonimiasis. Acta Trop. 2022, 233, 106578. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Gilchrist, C.A.; Uddin, M.J.; Burgess, S.L.; Abhyankar, M.M.; Moonah, S.N.; Noor, Z.; Donowitz, J.R.; Schneider, B.N.; Arju, T.; et al. Microbiome-mediated neutrophil recruitment via CXCR2 and protection from amebic colitis. PLoS Pathog. 2017, 13, e1006513. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; El-Fahmawi, A.; Christian, D.A.; Fang, Q.; Radaelli, E.; Chen, L.; Sullivan, M.C.; Misic, A.M.; Ellringer, J.A.; Zhu, X.Q.; et al. Infection-Induced Intestinal Dysbiosis Is Mediated by Macrophage Activation and Nitrate Production. mBio 2019, 10, e00935-19. [Google Scholar] [CrossRef]
- Samuelson, D.R.; Charles, T.P.; de la Rua, N.M.; Taylor, C.M.; Blanchard, E.E.; Luo, M.; Shellito, J.E.; Welsh, D.A. Analysis of the intestinal microbial community and inferred functional capacities during the host response to Pneumocystis pneumonia. Exp. Lung Res. 2016, 42, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Costa, M.C.; Santos, J.R.; Ribeiro, M.J.; Freitas, G.J.; Bastos, R.W.; Ferreira, G.F.; Miranda, A.S.; Arifa, R.D.; Santos, P.C.; Martins, F.o.S.; et al. The absence of microbiota delays the inflammatory response to Cryptococcus gattii. Int. J. Med. Microbiol. 2016, 306, 187–195. [Google Scholar] [CrossRef]
- Hérivaux, A.; Willis, J.R.; Mercier, T.; Lagrou, K.; Gonçalves, S.M.; Gonçales, R.A.; Maertens, J.; Carvalho, A.; Gabaldón, T.; Cunha, C. Lung microbiota predict invasive pulmonary aspergillosis and its outcome in immunocompromised patients. Thorax 2022, 77, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Diep, A.L.; Hoyer, K.K. Host Response to Coccidioides Infection: Fungal Immunity. Front. Cell. Infect. Microbiol. 2020, 10, 581101. [Google Scholar] [CrossRef]
- Davini, D.; Naeem, F.; Phong, A.; Al-Kuhlani, M.; Valentine, K.M.; McCarty, J.; Ojcius, D.M.; Gravano, D.M.; Hoyer, K.K. Elevated regulatory T cells at diagnosis of Coccidioides infection associates with chronicity in pediatric patients. J. Allergy Clin. Immunol. 2018, 142, 1971–1974.e1977. [Google Scholar] [CrossRef]
- Diep, A.L.; Tejeda-Garibay, S.; Miranda, N.; Hoyer, K.K. Macrophage and Dendritic Cell Activation and Polarization in Response to Coccidioides posadasii infection. J. Fungi 2021, 7, 630. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.Y.; Yu, J.J.; Seshan, K.R.; Reichard, U.; Cole, G.T. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory Fungal pathogen. Infect. Immun. 2002, 70, 3443–3456. [Google Scholar] [CrossRef] [PubMed]
- Richards, J.O.; Ampel, N.M.; Galgiani, J.N.; Lake, D.F. Dendritic cells pulsed with Coccidioides immitis lysate induce antigen-specific naive T cell activation. J. Infect. Dis. 2001, 184, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.Y.; Wozniak, K.L.; Cole, G.T. Flow Cytometric Analysis of Protective T-Cell Response Against Pulmonary Coccidioides Infection. Methods Mol. Biol. 2016, 1403, 551–566. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G. Interleukin-12 and its role in the generation of TH1 cells. Immunol. Today 1993, 14, 335–338. [Google Scholar] [CrossRef]
- Magee, D.M.; Cox, R.A. Interleukin-12 regulation of host defenses against Coccidioides immitis. Infect. Immun. 1996, 64, 3609–3613. [Google Scholar] [CrossRef]
- Hung, C.Y.; Gonzalez, A.; Wüthrich, M.; Klein, B.S.; Cole, G.T. Vaccine immunity to coccidioidomycosis occurs by early activation of three signal pathways of T helper cell response (Th1, Th2, and Th17). Infect. Immun. 2011, 79, 4511–4522. [Google Scholar] [CrossRef]
- Laiman, V.; Lo, Y.C.; Chen, H.C.; Yuan, T.H.; Hsiao, T.C.; Chen, J.K.; Chang, C.W.; Lin, T.C.; Li, S.J.; Chen, Y.Y.; et al. Effects of antibiotics and metals on lung and intestinal microbiome dysbiosis after sub-chronic lower-level exposure of air pollution in ageing rats. Ecotoxicol. Environ. Saf. 2022, 246, 114164. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef]
- Grayson, M.H.; Camarda, L.E.; Hussain, S.A.; Zemple, S.J.; Hayward, M.; Lam, V.; Hunter, D.A.; Santoro, J.L.; Rohlfing, M.; Cheung, D.S.; et al. Intestinal Microbiota Disruption Reduces Regulatory T Cells and Increases Respiratory Viral Infection Mortality Through Increased IFNγ Production. Front. Immunol. 2018, 9, 1587. [Google Scholar] [CrossRef]
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012, 37, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.B. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect. Immun. 2014, 82, 4596–4606. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.W.; Chen, P.H.; Hsu, C.M. Commensal microflora contribute to host defense against Escherichia coli pneumonia through Toll-like receptors. Shock 2011, 36, 67–75. [Google Scholar] [CrossRef]
- Rakoff-Nahoum, S.; Paglino, J.; Eslami-Varzaneh, F.; Edberg, S.; Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 2004, 118, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Burwell, L.A.; Park, B.J.; Wannemuehler, K.A.; Kendig, N.; Pelton, J.; Chaput, E.; Jinadu, B.A.; Emery, K.; Chavez, G.; Fridkin, S.K. Outcomes among inmates treated for coccidioidomycosis at a correctional institution during a community outbreak, Kern County, California, 2004. Clin. Infect. Dis. 2009, 49, e113–e119. [Google Scholar] [CrossRef]
- Chi, G.C.; Benedict, K.; Beer, K.D.; Jackson, B.R.; McCotter, O.; Xie, F.; Lawrence, J.M.; Tartof, S.Y. Antibiotic and antifungal treatment among persons with confirmed coccidioidomycosis—Southern California, 2011. Med. Mycol. 2020, 58, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.W.; Johnson, R.H.; Einstein, H.E.; Welch, G. Evaluation of response to early azole treatment in primary coccidioidomycosis. In Proceedings of the Coccidioidomycosis: Proceedings of the 5th International Conference, Bethesda, MD, USA, 24–27 August 1994; National Foundation for Infectious Diseases: Bethesda, MD, USA, 1996; pp. 275–284. [Google Scholar]
- Pfeiffer, S.; Jatzlauk, G.; Lund, J.V.; Boateng, E.; Kovacevic, D.; Hylkema, M.N.; Bartel, S.; Schloter, M.; Krauss-Etschmann, S. Oral application of vancomycin alters murine lung microbiome and pulmonary immune responses. Immun. Inflamm. Dis. 2022, 10, e675. [Google Scholar] [CrossRef]
- Nguyen, P.T.N.; Le, N.V.; Dinh, H.M.N.; Nguyen, B.Q.P.; Nguyen, T.V.A. Lung penetration and pneumococcal target binding of antibiotics in lower respiratory tract infection. Curr. Med. Res. Opin. 2022, 38, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Cavalcante, G.G.; Guimarães, A.G.; Queiroz-Glauss, C.P.; Gonçalves Pereira, M.H.; Dias, A.S.L.; Horta, L.S.; de Oliveira, J.S.; Cangussú, S.D.; Magalhães, P.P.; Russo, R.C.; et al. Treatment with Distinct Antibiotic Classes Causes Different Pulmonary Outcomes on Allergic Airway Inflammation Associated with Modulation of Symbiotic Microbiota. J. Immunol. Res. 2022, 2022, 1466011. [Google Scholar] [CrossRef]
- Lauer, A.; Baal, J.D.; Mendes, S.D.; Casimiro, K.N.; Passaglia, A.K.; Valenzuela, A.H.; Guibert, G. Valley Fever on the Rise-Searching for Microbial Antagonists to the Fungal Pathogen Coccidioides immitis. Microorganisms 2019, 7, 31. [Google Scholar] [CrossRef]
- Garcia-Nuñez, M.; Millares, L.; Pomares, X.; Ferrari, R.; Pérez-Brocal, V.; Gallego, M.; Espasa, M.; Moya, A.; Monsó, E. Severity-related changes of bronchial microbiome in chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 4217–4223. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Walker, A.W.; Oliver, A.E.; Rogers, G.B.; Rivett, D.W.; Hampton, T.H.; Ashare, A.; Elborn, J.S.; De Soyza, A.; Carroll, M.P.; et al. Lung function and microbiota diversity in cystic fibrosis. Microbiome 2020, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Pan, B.; Xu, S.; Xu, Z.; Zhang, T.; Zhang, Q.; Bao, Y.; Wang, Y.; Zhang, J.; Xu, C.; et al. A meta-analysis reveals the effectiveness of probiotics and prebiotics against respiratory viral infection. Biosci. Rep. 2021, 41, BSR20203638. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Moon, A.; Huang, J.; Sun, Y.; Qiu, H.J. Antiviral Effects and Underlying Mechanisms of Probiotics as Promising Antivirals. Front. Cell. Infect. Microbiol. 2022, 12, 928050. [Google Scholar] [CrossRef] [PubMed]
- Debnath, N.; Kumar, A.; Yadav, A.K. Probiotics as a biotherapeutics for the management and prevention of respiratory tract diseases. Microbiol. Immunol. 2022, 66, 277–291. [Google Scholar] [CrossRef]
- Ribeiro, F.C.; Rossoni, R.D.; de Barros, P.P.; Santos, J.D.; Fugisaki, L.R.O.; Leão, M.P.V.; Junqueira, J.C. Action mechanisms of probiotics on Candida spp. and candidiasis prevention: An update. J. Appl. Microbiol. 2020, 129, 175–185. [Google Scholar] [CrossRef]
- Sam, Q.H.; Yew, W.S.; Seneviratne, C.J.; Chang, M.W.; Chai, L.Y.A. Immunomodulation as Therapy for Fungal Infection: Are We Closer? Front. Microbiol. 2018, 9, 1612. [Google Scholar] [CrossRef]
| Infection | Antibiotic(s) | Δ Intestinal Microbiota | Δ Respiratory Microbiota | Δ Immune | Infection Outcome | Reference |
|---|---|---|---|---|---|---|
| Respiratory Syncytial Virus | Streptomycin sulfate | ↓ Lactobacillus, Clostridium_XlVa, Alistipes ↑ Bacteroides | N/A | ↑ eosinophils, lymphocytes, neutrophils (inflammatory cells) ↑ IFN- γ, IL-17A ↓ IL-10, IL-3, IL-4, IL-5 | No effect on viral load Enhanced pulmonary inflammation Dysregulated immune response | [11] |
| Influenza A virus | Vancomycin (V), Neomycin (N), Metronidazole(M), or ampicillin(A) | ↓ bacterial load and dominated by Sphingomonas species(N) ↓ Gram+ bacteria(A) ↑ Enterobacter species | Dominated by Lactobacillus species (V & M) ↓ Gram+ bacteria (A) ↑ Enterobacter species | ↓ CD 8 T cell responses (neomycin) | ↑ viral load (V, N, M, A) ↓ pro-IL-1β, pro-IL-18, NLRP3 (V, N, M, A) ↓ mLN total #, proliferation, differentiation, activation and migration of DCs (V, N, M, A) Impaired immune responses (N) | [53] |
| Sendai virus | Streptomycin | ↓ alpha diversity ↑ Bacillales | No significant differences | ↑ IL-6, IFN- γ, CCL2, CCL11 ↑ %NK1.1-expressing lymphocytes ↓ Foxp3+ Tregs | ↑ mortality | [54] |
| Lymphocytic Choriomeningitis Virus | Ampicillin, Gentamicin, Metronidazole, Neomycin, Vancomycin, and sucralose | ↓ commensal bacteria | N/A | ↓ CD8+ T cell responses ↓ IgG antibody titers ↑ T cell exhaustion (PD-1, 2B4 CD160, LAG-3) ↓ innate antiviral immune responses | ↑ viral titer in kidneys Delayed viral clearance | [55] |
| Streptococcus pneumoniae | Ampicillin, Neomycin, Metronidazole, and Vancomycin | ↓ microbial diversity | N/A | ↑ IL-1β, IL-6, and CXCL1 ↓ TNF-α and IL-10 | ↑ mortality rate ↑ bacterial load ↑ lung neutrophil influx ↑ tissue inflammation, liver damage, and hepatic injury ↓ alveolar macrophage phagocytosis capacity | [15] |
| Klebsiella pneumoniae | Ampicillin, Neomycin sulfate, Metronidazole, and Vancomycin | N/A | N/A | ↓ IL-6, TNF-α ↓ bacterial killing by alveolar macrophages ↓ H2O2 | ↑ bacterial burden | [56] |
| Escherichia coli | Ampicillin, Vancomycin, Neomycin sulfate, and Metronidazole | Not detectable (no sequencing) | N/A | ↓ myeloperoxidase activity ↓ TNF-α production ↓ alveolar macrophage bacterial killing ↑ IL-6 and IL-1β ↓ NF-kβ DNA-binding activity in intestinal mucosa and lung ↓ TLR4, TNF-α, KC, ICAM, and CXCR2 expression in intestinal mucosa ↑ IL-1β, KC, and MIP-2 in the lung | ↑ bacterial burden ↑ bacteria in blood ↑ mortality ↑ interstitial edema, septal edema, and alveolar edema in lung | [57,58] |
| Pseudomonas aeruginosa | Vancomycin-colistin | ↓ diversity ↓ Muribaculaceae, Prevotellaceae, and Lachnospiraceae ↑ Burkholderiaceae, Clostridiales, Lactobacillaceae | No change | ↓ macrophages, cDC2, inflammatory monocytes, neutrophils, iNKT cells ↓ Flt3-ligand | ↑ P. aeruginosa load in lung and spleen ↑ lung injury ↓ survival | [12] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tejeda-Garibay, S.; Hoyer, K.K. Coccidioidomycosis and Host Microbiome Interactions: What We Know and What We Can Infer from Other Respiratory Infections. J. Fungi 2023, 9, 586. https://doi.org/10.3390/jof9050586
Tejeda-Garibay S, Hoyer KK. Coccidioidomycosis and Host Microbiome Interactions: What We Know and What We Can Infer from Other Respiratory Infections. Journal of Fungi. 2023; 9(5):586. https://doi.org/10.3390/jof9050586
Chicago/Turabian StyleTejeda-Garibay, Susana, and Katrina K. Hoyer. 2023. "Coccidioidomycosis and Host Microbiome Interactions: What We Know and What We Can Infer from Other Respiratory Infections" Journal of Fungi 9, no. 5: 586. https://doi.org/10.3390/jof9050586
APA StyleTejeda-Garibay, S., & Hoyer, K. K. (2023). Coccidioidomycosis and Host Microbiome Interactions: What We Know and What We Can Infer from Other Respiratory Infections. Journal of Fungi, 9(5), 586. https://doi.org/10.3390/jof9050586







