Filamentous Fungi Are Potential Bioremediation Agents of Semi-Synthetic Textile Waste
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Culture and Colonization of Woodchips
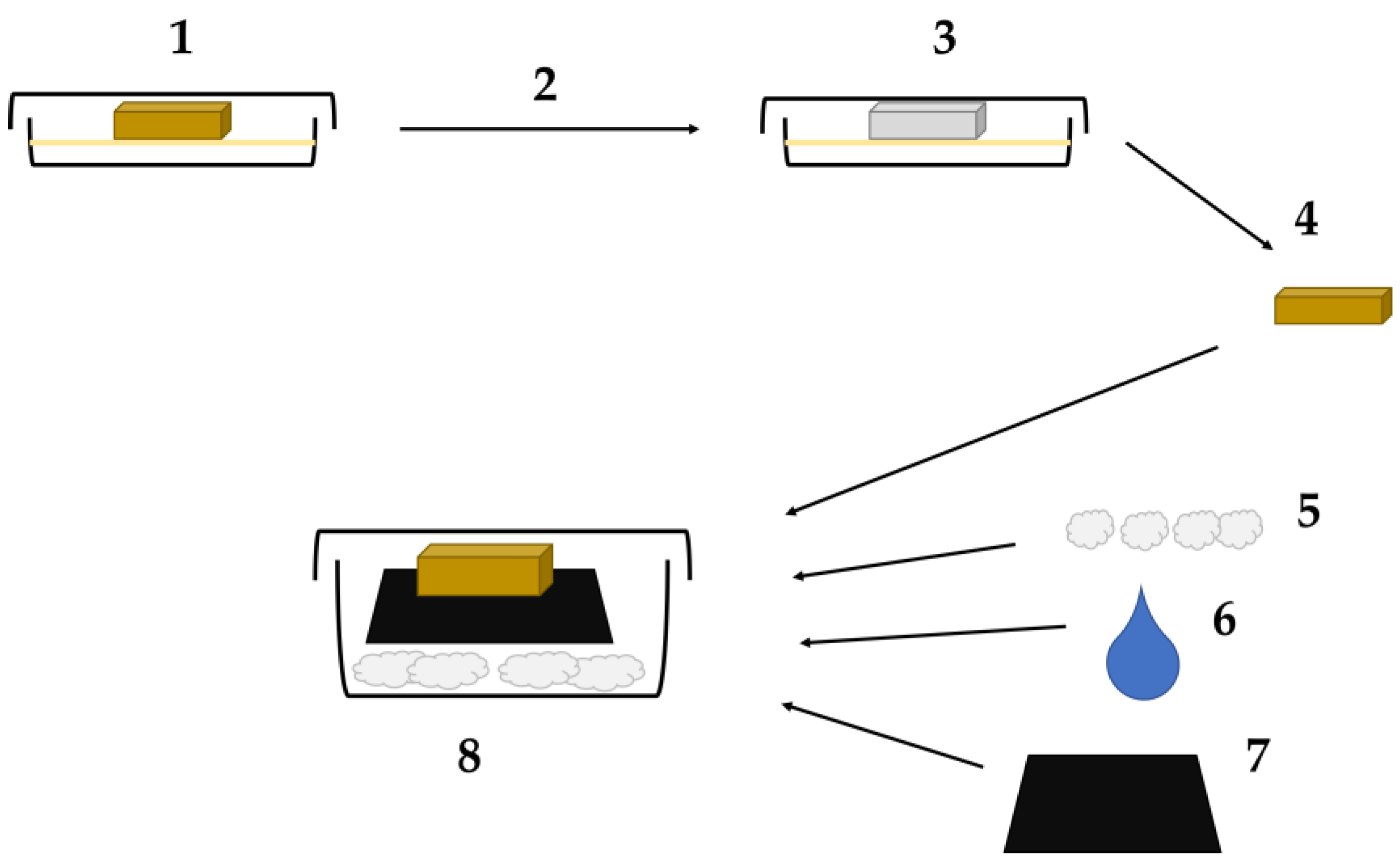
2.2. Microcosm Setup
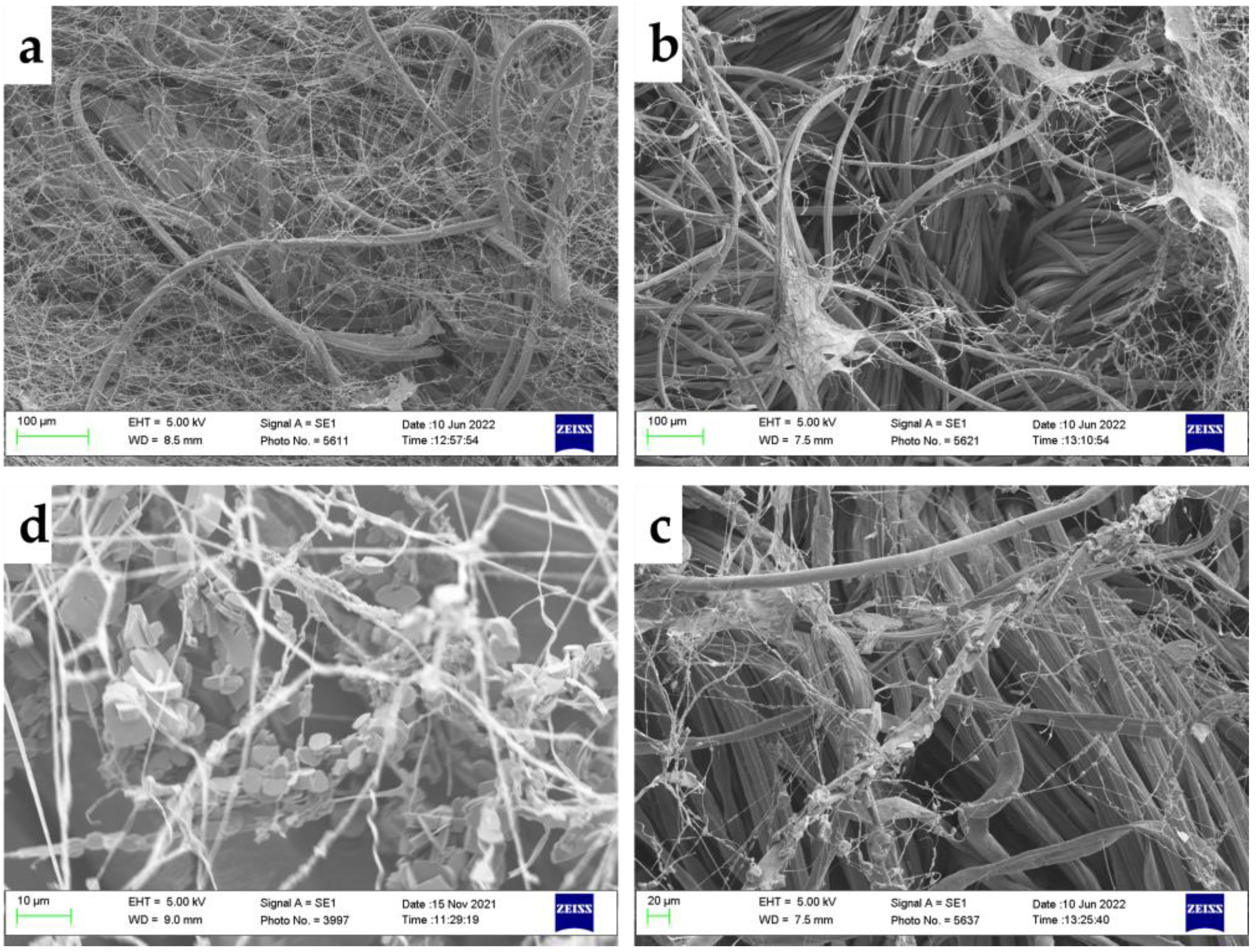
2.3. Imaging
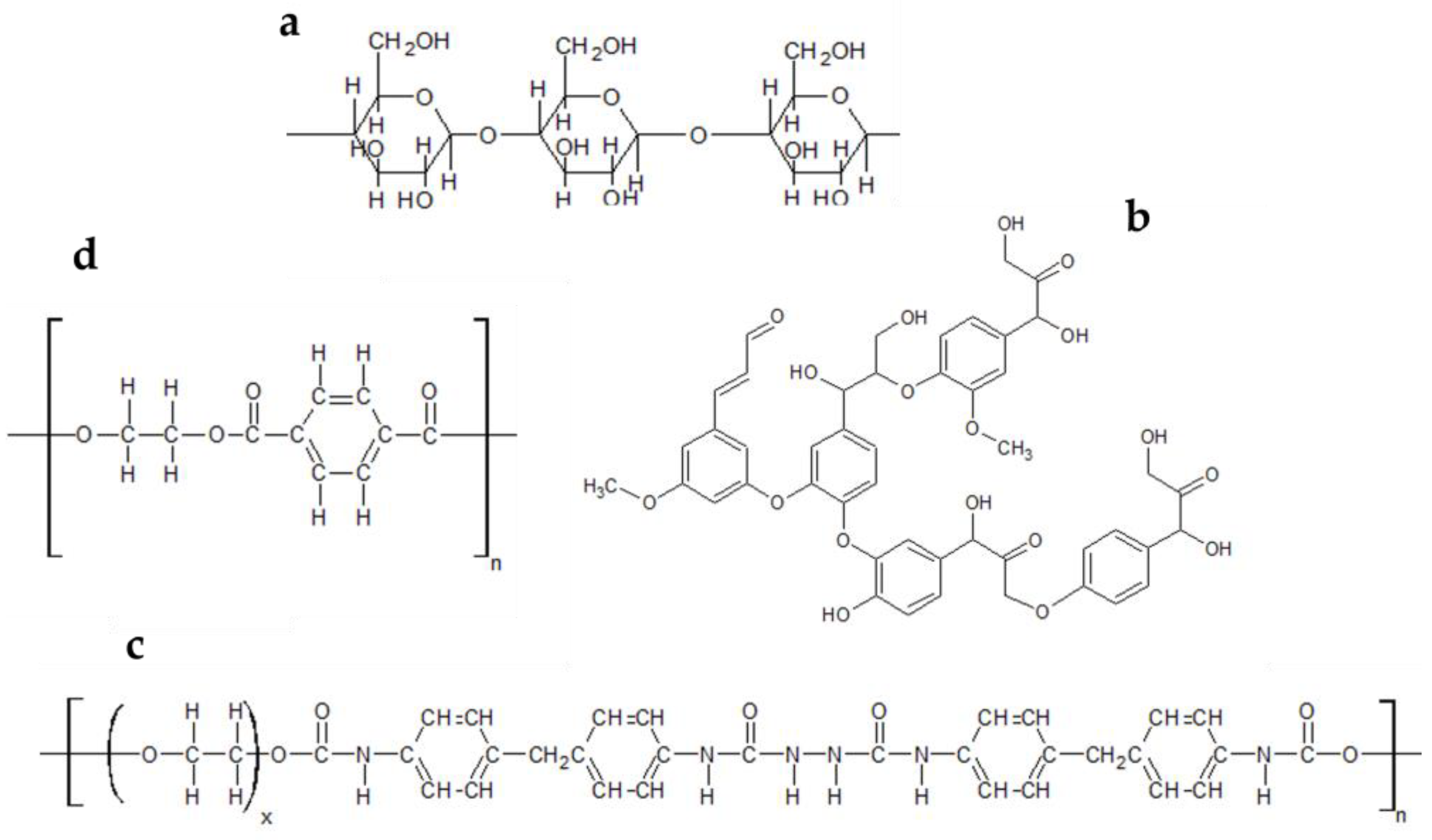
2.4. Determination of Dye Content
2.4.1. Thermogravimetric Analysis
2.4.2. Dye Extraction Essay
2.5. Gas Chromatography Mass Spectrometry of Volatiles
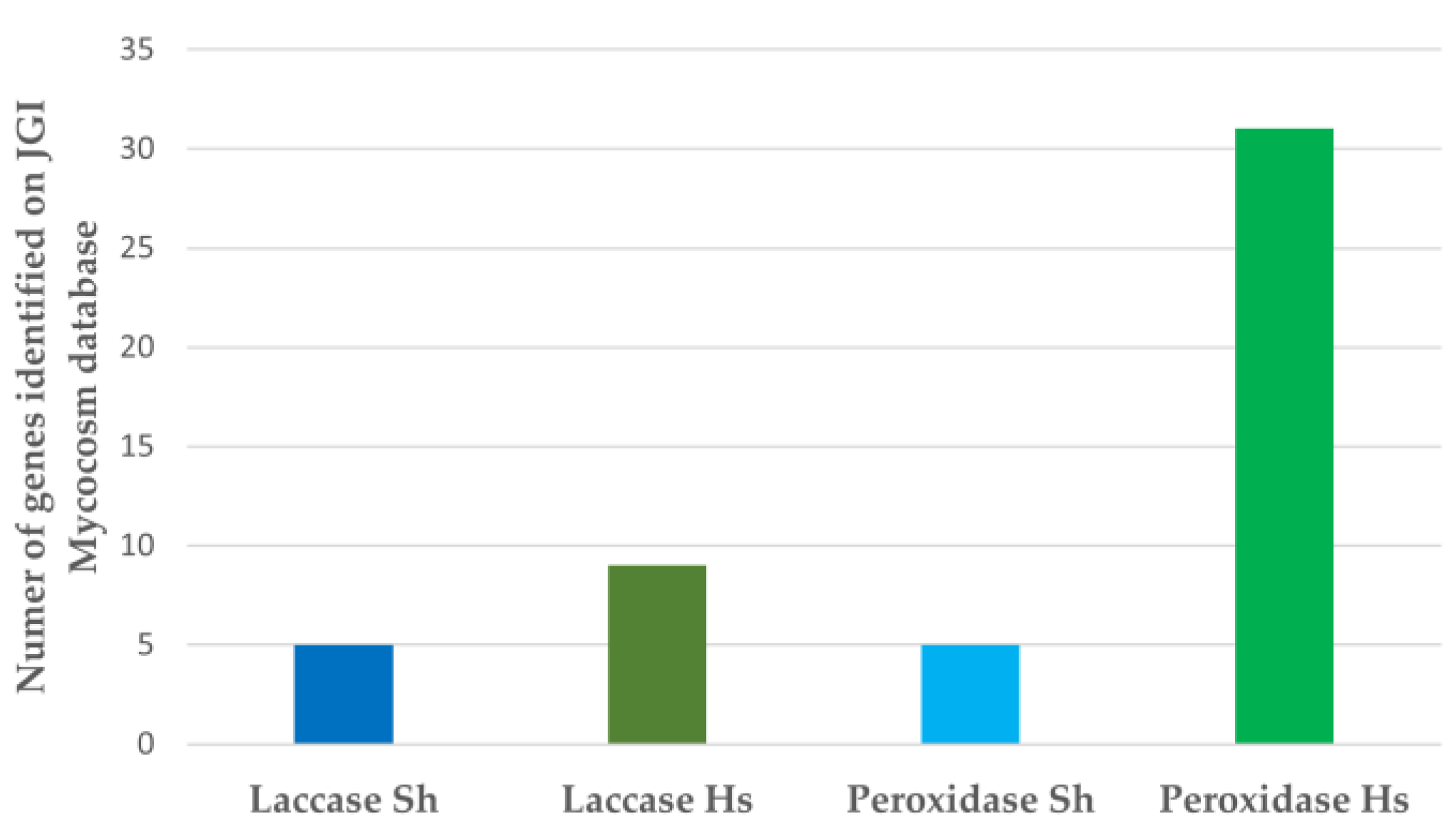
2.6. Bioinformatic Analysis of Genomes
3. Results
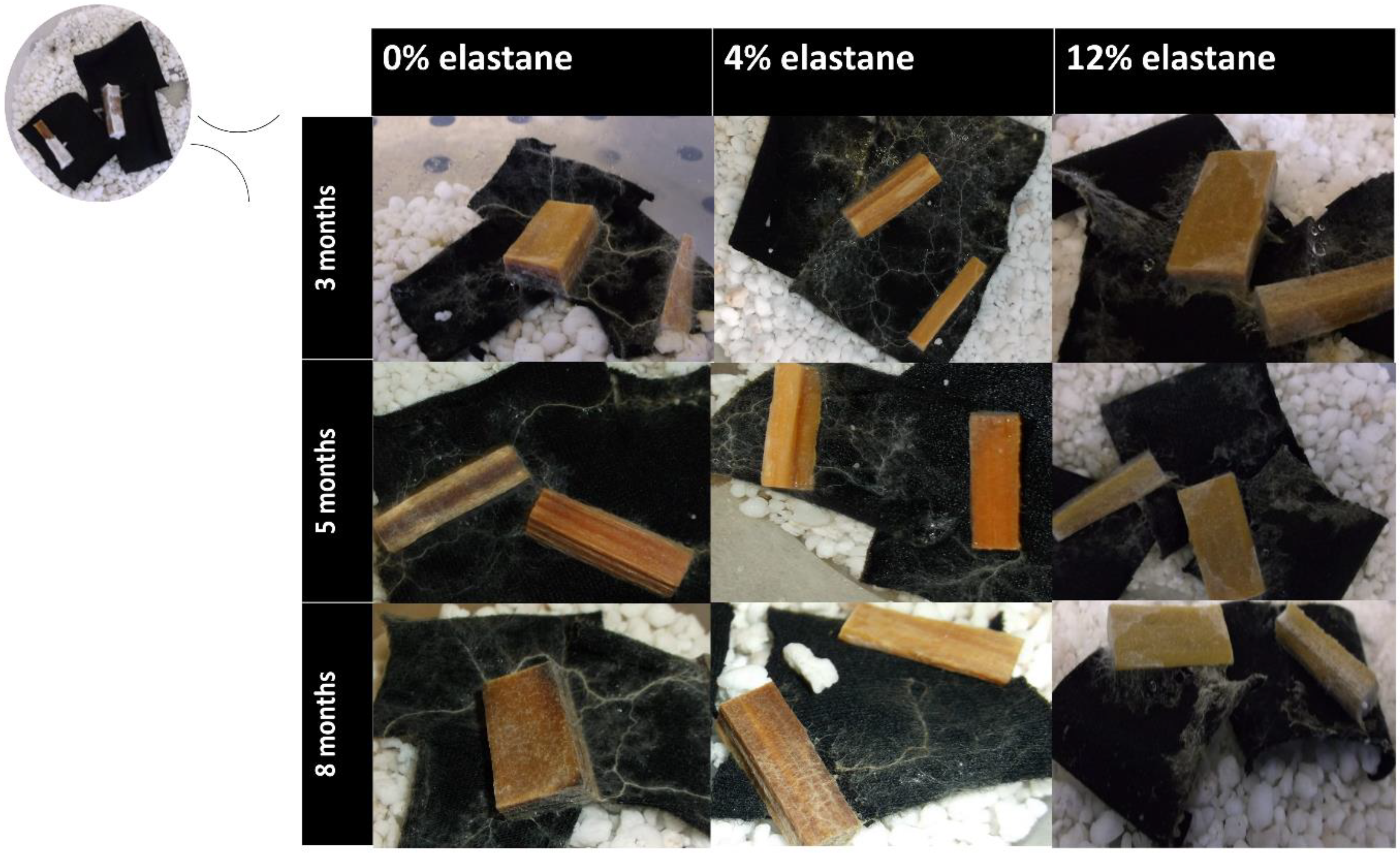
3.1. Fungal Growth on Textiles
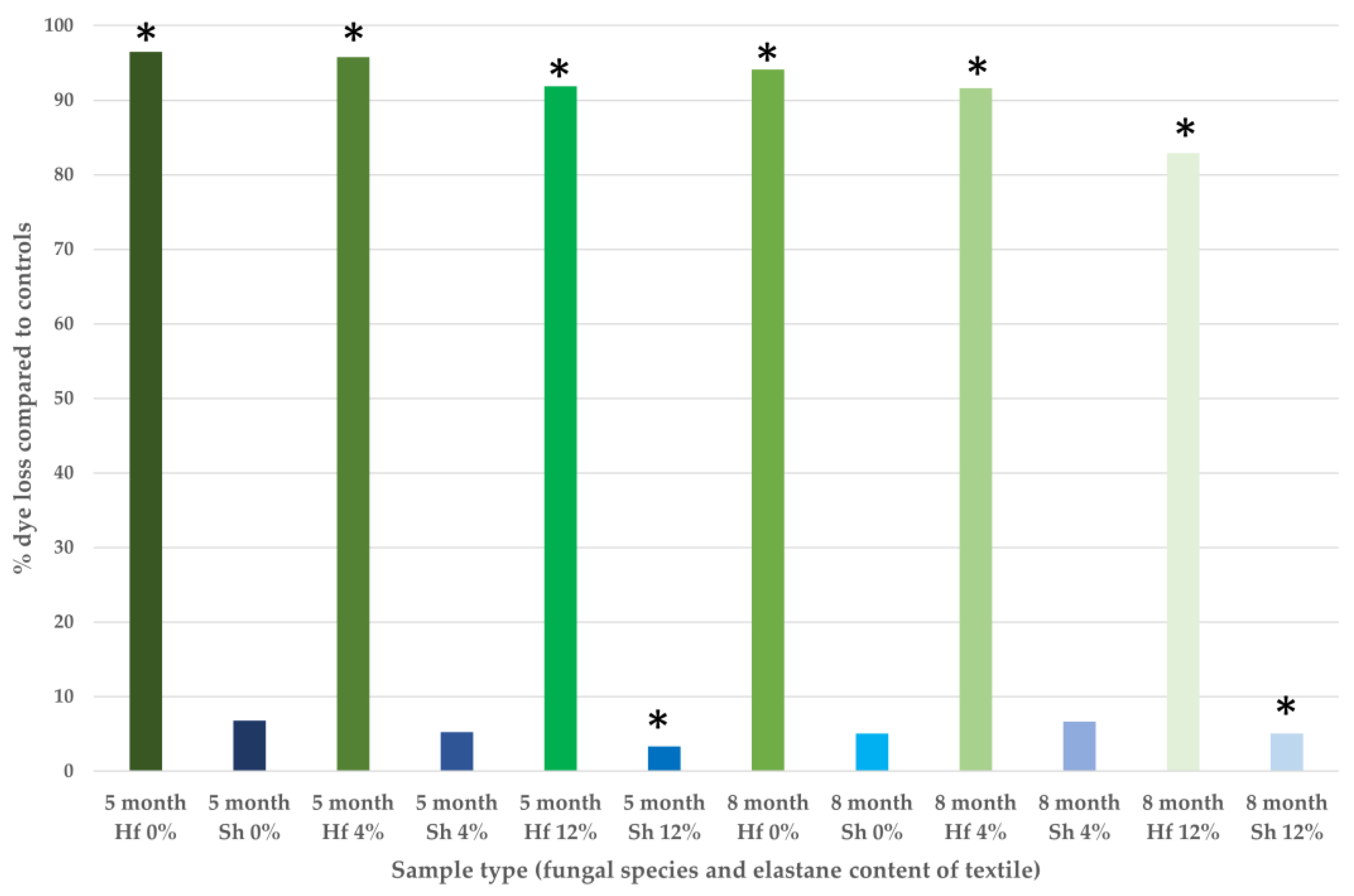
3.2. Quantification of Dye Remediation
3.2.1. Thermogravimetric Analysis (TGA)
3.2.2. Dye Extraction and Spectrophotometric Quantification
3.3. Bioinformatic Analysis of Enzyme Capacity for Dye Remediation
3.4. Gas Chromatography Mass Spectrometry (GCMS) Analysis of the Volatile Metabolome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Li, Y.; He, W. The energy footprint of China’s textile industry: Perspectives from decoupling and decomposition analysis. Energies 2017, 10, 1461. [Google Scholar] [CrossRef]
- Stone, C.; Windsor, F.M.; Munday, M.; Durance, I. Natural or synthetic—How global trends in textile usage threaten freshwater environments. Sci. Total Environ. 2020, 718, 134689. [Google Scholar] [CrossRef]
- De Falco, F.; di Pace, E.; Cocca, M.; Avella, M. The contribution of washing processes of synthetic clothes to microplastic pollution. Sci. Rep. 2019, 9, 6633. [Google Scholar] [CrossRef] [PubMed]
- Niinimäki, K.; Peters, G.; Dahlbo, H.; Perry, P.; Rissanen, T.; Gwilt, A. The environmental price of fast fashion. Nat. Rev. Earth Environ. 2020, 1, 189–200. [Google Scholar] [CrossRef]
- Bick, R.; Halsey, E.; Ekenga, C.C. The global environmental injustice of fast fashion. Environ. Health 2018, 17, 92. [Google Scholar] [CrossRef]
- Papamichael, I.; Chatziparaskeva, G.; Pedreno, J.N.; Voukkali, I.; Almendro Candel, M.B.; Zorpas, A.A. Building a new mind set in tomorrow fashion development through circular strategy models in the framework of waste management. Curr. Opin. Green Sustain. Chem. 2022, 36, 100638. [Google Scholar] [CrossRef]
- Smith, G. 8—The Identification of Fungi Causing Mildew in Cotton Goods: The Genus Aspergillus. J. Text. Inst. Trans. 1928, 19, T92–T100. [Google Scholar] [CrossRef]
- Marsh, P.B.; Bollenbacher, K. The Fungi Concerned in Fiber Deterioration. Text. Res. J. 1949, 19, 313–324. [Google Scholar] [CrossRef]
- Liu, J.; Liang, J.; Ding, J.; Zhang, G.; Zeng, X.; Yang, Q.; Zhu, B.; Gao, W. Microfiber pollution: An ongoing major environmental issue related to the sustainable development of textile and clothing industry. Environ. Dev. Sustain. 2021, 23, 11240–11256. [Google Scholar] [CrossRef]
- Haslinger, S.; Hummel, M.; Anghelescu-Hakala, A.; Määttänen, M.; Sixta, H. Upcycling of cotton polyester blended textile waste to new man-made cellulose fibers. Waste Manag. 2019, 97, 88–96. [Google Scholar] [CrossRef]
- Lynch, J.M.; Moffat, A.J. Bioremediation—Prospects for the future application of innovative applied biological research. Ann. Appl. Biol. 2005, 146, 217–221. [Google Scholar] [CrossRef]
- Viswanath, B.; Rajesh, B.; Janardhan, A.; Kumar, A.P.; Narasimha, G. Fungal Laccases and Their Applications in Bioremediation. Enzym. Res. 2014, 2014, 163242. [Google Scholar] [CrossRef]
- Singh, R.K.; Tripathi, R.; Ranjan, A.; Srivastava, A.K. Fungi as potential candidates for bioremediation. In Abatement of Environmental Pollutants; Elsevier: Amsterdam, The Netherlands, 2020; pp. 177–191. [Google Scholar]
- Noman, E.; Al-Gheethi, A.; Mohamed, R.M.S.R.; Talip, B.A. Myco-Remediation of Xenobiotic Organic Compounds for a Sustainable Environment: A Critical Review. Top. Curr. Chem. 2019, 377, 17. [Google Scholar] [CrossRef]
- Chatterjee, S.; Sarma, M.K.; Deb, U.; Steinhauser, G.; Walther, C.; Gupta, D.K. Mushrooms: From nutrition to mycoremediation. Environ. Sci. Pollut. Res. 2017, 24, 19480–19493. [Google Scholar] [CrossRef]
- Eastwood, D.C.; Floudas, D.; Binder, M.; Majcherczyk, A.; Schneider, P.; Aerts, A.; Asiegbu, F.O.; Baker, S.E.; Barry, K.; Bendiksby, M.; et al. The Plant Cell Wall–Decomposing Machinery Underlies the Functional Diversity of Forest Fungi. Science 2011, 333, 762–765. [Google Scholar] [CrossRef]
- Saraswat, P.; Yadav, K.; Gupta, A.; Prasad, M.; Ranjan, R. Physiological and molecular basis for remediation of polyaromatic hydrocarbons. In Handbook of Bioremediation; Elsevier: Amsterdam, The Netherlands, 2021; pp. 535–550. [Google Scholar]
- Błyskal, B. Gymnoascus arxii’s potential in deteriorating woollen textiles dyed with natural and synthetic dyes. Int. Biodeterior. Biodegrad. 2014, 86, 349–357. [Google Scholar] [CrossRef]
- Deguchi, T.; Kakezawa, M.; Nishida, T. Nylon biodegradation by lignin-degrading fungi. Appl. Environ. Microbiol. 1997, 63, 329–331. [Google Scholar] [CrossRef]
- Friedrich, J.; Zalar, P.; Mohorčič, M.; Klun, U.; Kržan, A. Ability of fungi to degrade synthetic polymer nylon-6. Chemosphere 2007, 67, 2089–2095. [Google Scholar] [CrossRef]
- Pereira de Almeida, A.; Macrae, A.; Dias Ribeiro, B.; Pires do Nascimento, R. Decolorization and Detoxification of Different Azo Dyes by Phanerochaete chrysosporium ME-446 under Submerged Fermentation. Braz. J. Microbiol. 2021, 52, 727–738. [Google Scholar] [CrossRef]
- Moody, S.C.; Dudley, E.; Hiscox, J.; Boddy, L.; Eastwood, D.C. Interdependence of primary metabolism and xenobiotic mitigation characterizes the proteome of Bjerkandera adusta during wood decomposition. Appl. Environ. Microbiol. 2018, 84, e01401–e01417. [Google Scholar] [CrossRef]
- Home, J.M.; Dudley, R.J. Thin-layer chromatography of dyes extracted from cellulosic fibres. Forensic Sci. Int. 1981, 17, 71–78. [Google Scholar] [CrossRef]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc database of metabolic pathways and enzymes—A 2019 update. Nucleic Acids Res. 2020, 48, D445–D453. [Google Scholar] [CrossRef] [PubMed]
- Bento da Silva, A.; Giacomoni, F.; Pavot, B.; Fillâtre, Y.; Rothwell, J.A.; Bartolomé Sualdea, B.; Veyrat, C.; Garcia-Villalba, R.; Gladine, C.; Kopec, R.; et al. PhytoHub V1.4: A new release for the online database dedicated to food phytochemicals and their human metabolites. In Proceedings of the International Conference on Food Bioactives & Health, Norwich, UK, 13 September 2016. [Google Scholar]
- Jewison, T.; Knox, C.; Neveu, V.; Djoumbou, Y.; Guo, A.C.; Lee, J.; Liu, P.; Mandal, R.; Krishnamurthy, R.; Sinelnikov, I.; et al. YMDB: The Yeast Metabolome Database. Nucleic Acids Res. 2012, 40, D815–D820. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Gaona, M.; Marcu, A.; Pon, A.; Guo, A.C.; Sajed, T.; Wishart, N.A.; Karu, N.; Djoumbou Feunang, Y.; Arndt, D.; Wishart, D.S. YMDB 2.0: A significantly expanded version of the yeast metabolome database. Nucleic Acids Res. 2017, 45, D440–D445. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Selbig, J.; Walther, D.; Kopka, J. The Golm Metabolome Database: A database for GC-MS based metabolite profiling. In Metabolomics: A Powerful Tool in Systems Biology; Springer: Berlin/Heidelberg, Germany, 2007; pp. 75–95. [Google Scholar]
- The Metabolomics Workbench. Available online: https://www.metabolomicsworkbench.org/search/index.php (accessed on 15 May 2022).
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Grigoriev, I.V.; Nordberg, H.; Shabalov, I.; Aerts, A.; Cantor, M.; Goodstein, D.; Kuo, A.; Minovitsky, S.; Nikitin, R.; Ohm, R.A.; et al. The Genome Portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012, 40, D26–D32. [Google Scholar] [CrossRef]
- Dowson, C.G.; Springham, P.; Rayner Adm Boddy, L. Resource relationships of foraging mycelial systems of Phanerochaete velutina and Hypholoma fasciculare in soil. New Phytol. 1989, 111, 501–509. [Google Scholar] [CrossRef]
- Boddy, L. Interspecific combative interactions between wood-decaying basidiomycetes. FEMS Microbiol. Ecol. 2000, 31, 185–194. [Google Scholar] [CrossRef]
- Salem, M.Z.M. EDX measurements and SEM examination of surface of some imported woods inoculated by three mold fungi. Measurement 2016, 86, 301–309. [Google Scholar] [CrossRef]
- Anh, D.H.; Dumri, K.; Yen, L.T.H.; Punyodom, W. The earth-star basidiomycetous mushroom Astraeus odoratus produces polyhydroxyalkanoates during cultivation on malt extract. Arch. Microbiol. 2023, 205, 34. [Google Scholar] [CrossRef]
- A New Textiles Economy: Redesigning Fashion’s Future. Ellen MacArthur Foundation. 2017. Available online: http://www.ellenmacarthurfoundation.org/publications (accessed on 4 April 2023).
- Schumacher, K.; Forster, A. Facilitating a Circular Economy for Textiles Workshop Report. 2022. Available online: https://nvlpubs.nist.gov/nistpubs/SpecialPublications/NIST.SP.1500-207.pdf (accessed on 22 April 2023).
- Alam, R.; Mahmood, R.A.; Islam, S.; Ardiati, F.C.; Solihat, N.N.; Alam, M.B.; Lee, S.H.; Yanto, D.H.Y.; Kim, S. Understanding the biodegradation pathways of azo dyes by immobilized white-rot fungus, Trametes hirsuta D7, using UPLC-PDA-FTICR MS supported by in silico simulations and toxicity assessment. Chemosphere 2023, 313, 137505. [Google Scholar] [CrossRef]
- Gadd, G.M.; Bahri-Esfahani, J.; Li, Q.; Rhee, Y.J.; Wei, Z.; Fomina, M.; Liang, X. Oxalate production by fungi: Significance in geomycology, biodeterioration and bioremediation. In Fungal Biology Reviews; Elsevier Ltd.: Amsterdam, The Netherlands, 2014; Volume 28, pp. 36–55. [Google Scholar]
- Girometta, C.; Zeffiro, A.; Malagodi, M.; Savino, E.; Doria, E.; Nielsen, E.; Buttafava, A.; Dondi, D. Pretreatment of alfalfa stems by wood decay fungus Perenniporia meridionalis improves cellulose degradation and minimizes the use of chemicals. Cellulose 2017, 24, 3803–3813. [Google Scholar] [CrossRef]
- Baptista, P.; Guedes de Pinho, P.; Moreira, N.; Malheiro, R.; Reis, F.; Padrão, J.; Tavares, R.; Lino-Neto, T. In vitro interactions between the ectomycorrhizal Pisolithus tinctorius and the saprotroph Hypholoma fasciculare fungi: Morphological aspects and volatile production. Mycology 2021, 12, 216–229. [Google Scholar] [CrossRef]
- Singh, R.L.; Singh, P.K.; Singh, R.P. Enzymatic decolorization and degradation of azo dyes—A review. In International Biodeterioration and Biodegradation; Elsevier Ltd.: Amsterdam, The Netherlands, 2015; Volume 104, pp. 21–31. [Google Scholar]
- Gul, R.; Sharma, P.; Kumar, R.; Umar, A.; Ibrahim, A.A.; Alhamami, M.A.M.; Jaswal, V.S.; Kumar, M.; Dixit, A.; Baskoutas, S. A sustainable approach to the degradation of dyes by fungal species isolated from industrial wastewaters: Performance, parametric optimization, kinetics and degradation mechanism. Environ. Res. 2023, 216, 114407. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harper, R.; Moody, S.C. Filamentous Fungi Are Potential Bioremediation Agents of Semi-Synthetic Textile Waste. J. Fungi 2023, 9, 661. https://doi.org/10.3390/jof9060661
Harper R, Moody SC. Filamentous Fungi Are Potential Bioremediation Agents of Semi-Synthetic Textile Waste. Journal of Fungi. 2023; 9(6):661. https://doi.org/10.3390/jof9060661
Chicago/Turabian StyleHarper, Rachel, and Suzy Clare Moody. 2023. "Filamentous Fungi Are Potential Bioremediation Agents of Semi-Synthetic Textile Waste" Journal of Fungi 9, no. 6: 661. https://doi.org/10.3390/jof9060661
APA StyleHarper, R., & Moody, S. C. (2023). Filamentous Fungi Are Potential Bioremediation Agents of Semi-Synthetic Textile Waste. Journal of Fungi, 9(6), 661. https://doi.org/10.3390/jof9060661





