Luteodorsum huanglongense (Gomphaceae, Gomphales), a New Genus and Species of Gomphoid Fungus from the Loess Plateau, Northwest China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. DNA Extraction, PCR Amplification, and DNA Sequencing
2.3. Phylogenetic Analyses
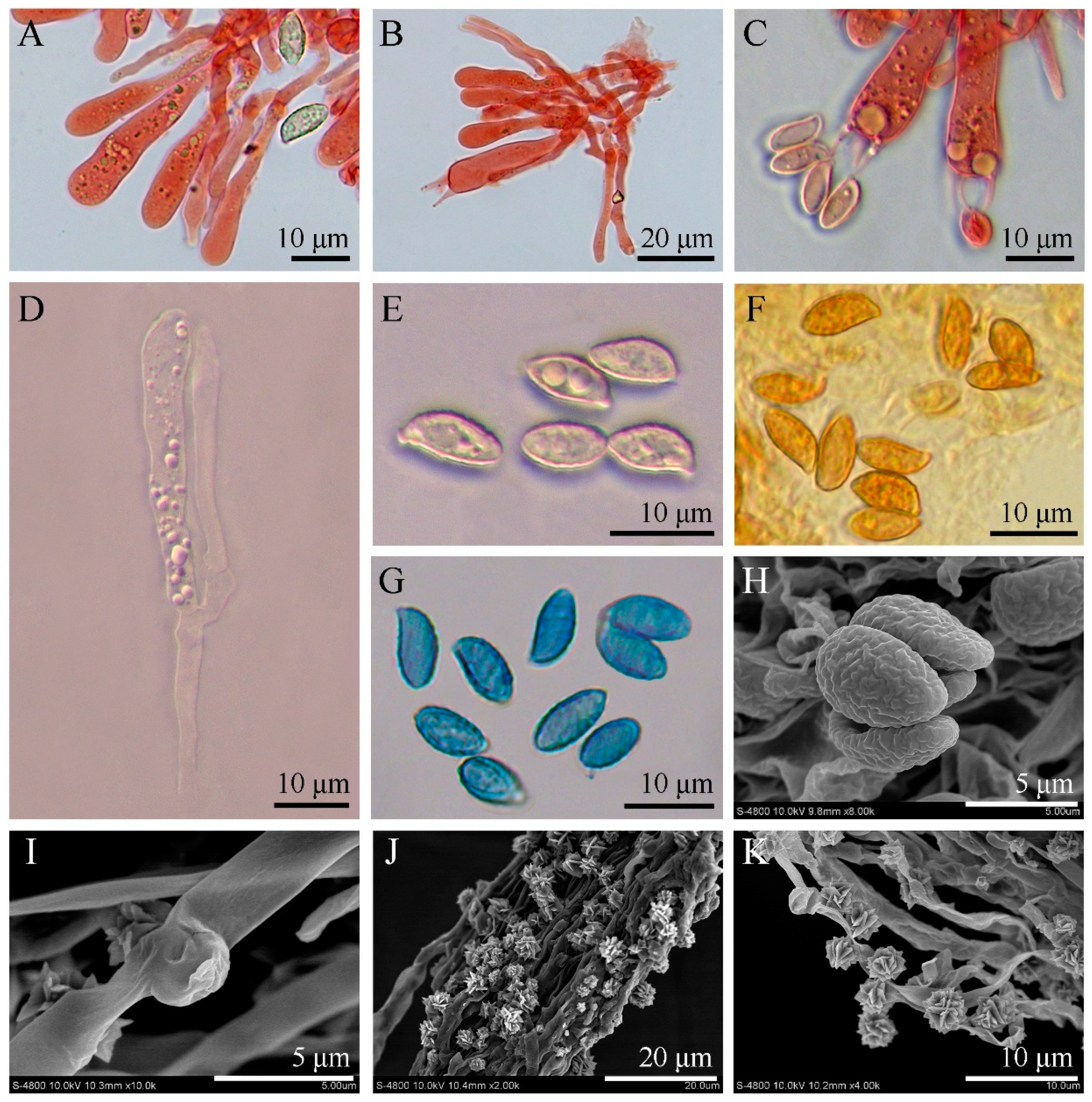
3. Results
3.1. Phylogenetic Analyses
3.2. SEM Observation and Qualitative X-ray Microanalysis
3.3. Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hosaka, K.; Bates, S.T.; Beever, R.E.; Castellano, M.A.; Colgan, W., 3rd; Domínguez, L.S.; Nouhra, E.R.; Geml, J.; Giachini, A.J.; Kenney, S.R.; et al. Molecular phylogenetics of the gomphoid-phalloid fungi with an establishment of the new subclass Phallomycetidae and two new orders. Mycologia 2006, 98, 949–959. [Google Scholar] [CrossRef]
- Giachini, A.J.; Hosaka, K.; Nouhra, E.; Spatafora, J.; Trappe, J.M. Phylogenetic relationships of the Gomphales based on nuc-25S-rDNA, mit-12S-rDNA, and mit-atp6-DNA combined sequences. Fungal Biol. 2010, 114, 224–234. [Google Scholar] [CrossRef]
- He, M.-Q.; Zhao, R.-L.; Hyde, K.D.; Begerow, D.; Kemler, M.; Yurkov, A.; McKenzie, E.H.C.; Raspé, O.; Kakishima, M.; Sánchez-Ramírez, S.; et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019, 99, 105–367. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.-Y.; Jian, S.-P.; Mao, N.; Yang, Z.-L.; Fan, L. Gomphocantharellus, a new genus of Gomphales. Mycologia 2022, 114, 748–756. [Google Scholar] [CrossRef]
- Kirk, P.M.; Cannon, P.F.; Minter, D.W.; Stalpers, J.A. Ainsworth and Bisby’s Dictionary of the Fungi, 10th ed.; CABI: Wallingford, UK, 2008; p. 289. ISBN 978-1-84593-933-5. [Google Scholar]
- Corner, E.J.H. Supplement to a monograph of Clavaria and allied genera. Beih. Nova Hedwig. 1970, 33, 1–299. [Google Scholar]
- Methven, A.S. The genus Clavariadelphus in North America. Bibl. Mycol. 1990, 138, 1–192. [Google Scholar]
- Huang, H.-Y.; Zhao, J.; Zhang, P.; Ge, Z.-W.; Li, X.; Tang, L.-P. The genus Clavariadelphus (Clavariadelphaceae, Gomphales) in China. Mycokeys 2020, 70, 89–121. [Google Scholar] [CrossRef]
- Chen, J.-J.; Shen, L.-L.; Cui, B. Morphological characters and molecular data reveal a new species of Hydnocristella (Gomphales, Basidiomycota) from southwestern China. Nova Hedwig. 2015, 101, 139–146. [Google Scholar] [CrossRef]
- Liu, L.-N.; Wu, L.; Chen, Z.-H.; Bau, T.; Zhang, P. The species of Lentaria (Gomphales, Basidiomycota) from China based on morphological and molecular evidence. Mycol. Prog. 2017, 16, 605–612. [Google Scholar] [CrossRef]
- Robledo, G.L.; Urcelay, C. Kavinia chacoserrana sp. nov. (Gomphales, Basidiomycota): A new species from South America based on morphological and molecular data. Mycosphere 2017, 8, 1028–1034. [Google Scholar] [CrossRef]
- González-Ávila, P.A.; Luna-Vega, I.; Ríos, M.V.; Saade, R.L.; Blanco, J.C. Current knowledge and importance of the order Gomphales (Fungi: Basidiomycota) in Mexico. Nova Hedwig. 2013, 97, 55–86. [Google Scholar] [CrossRef]
- Donk, M.A. Four New Families of Hymenomycetes. Persoonia 1961, 1, 405–407. [Google Scholar]
- Donk, M.A. A Conspectus of the Families of Aphyllophorales. Persoonia 1964, 3, 199–324. [Google Scholar]
- Petersen, R.H.; Pearman, W.R. Spore ornamentation in Ramaria as depicted by scanning electron micrographs. Persoonia 1973, 7, 289–292. [Google Scholar]
- Jülich, W.; Star, W. Ultrastructure of basidiospores I. Beenakia. Pers. 1983, 12, 67–74. [Google Scholar]
- Villegas, M.; Cifuentes, J.; Torres, A.E. Sporal characters in Gomphales and their significance for phylogenetics. Fungal Divers. 2005, 18, 157–175. [Google Scholar]
- Maneevun, A.; Dodgson, J.; Sanoamuang, N. Phaeoclavulina and Ramaria (Gomphaceae, Gomphales) from Nam Nao National Park, Thailand. Trop. Nat. Hist. 2012, 12, 147–164. [Google Scholar]
- Deng, C.Y.; Li, T.H. Gloeocantharellus persicinus, a new species from China. Mycotaxon 2008, 106, 449–453. [Google Scholar]
- Mifsud, S. Phaeoclavulina decurrens (Gomphales, Basidiomycetes)—The first record for a coral fungus for the Maltese Islands. Microb. Biosyst. 2019, 4, 55–59. [Google Scholar] [CrossRef] [Green Version]
- Dring, D.M. Techniques for microscopic preparation. In Methods in Microbiology; Booth, C., Ed.; Academic Press: New York, NY, USA, 1971; Volume 4, pp. 95–112. ISBN 978-0-12-521504-6. [Google Scholar] [CrossRef]
- Peng, Z.J.; Yu, A.; Luo, Z.Y.; Liu, X.Y.; Chen, W.F.; Yu, Z.D. Punctularia atropurpurascens (Punctulariaceae, Basidiomycota): A new record to China. Microbiol. China 2021, 48, 4232–4239. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Kretzer, A.M.; Bruns, T.D. Use of atp6 in Fungal Phylogenetics: An Example from the Boletales. Mol. Phylogenet. Evol. 1999, 13, 483–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.-L.; Li, G.-J.; Sánchez-Ramírez, S.; Stata, M.; Yang, Z.-L.; Wu, G.; Dai, Y.-C.; He, S.-H.; Cui, B.-K.; Zhou, J.-L.; et al. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Divers. 2017, 84, 43–74. [Google Scholar] [CrossRef]
- Edler, D.; Klein, J.; Antonelli, A.; Silvestro, D. raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol. Evol. 2021, 12, 373–377. [Google Scholar] [CrossRef]
- Swofford, D.L. PAUP*: Phylogenetic Analysis Using Parsimony (and Other Methods); Version 4.0b10; Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Hillis, D.M.; Bull, J.J. An Empirical Test of Bootstrapping as a Method for Assessing Confidence in Phylogenetic Analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or Bootstrap? A Simulation Study Comparing the Performance of Bayesian Markov Chain Monte Carlo Sampling and Bootstrapping in Assessing Phylogenetic Confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Carrière, E.A. Traité Conifer des Conifers, 2nd ed.; Chez l’Auteur: Paris, France, 1867; p. 510. [Google Scholar]
- Moskovskoe, O.L.P. Bulletin de la Société impériale des naturalists de Moscou. Soc. Imp. Nat. Moscou 1838, 11, 101. [Google Scholar]
- Ashburner, K.; McAllister, H.A. The Genus Betula: A Taxonomic Revision of Birches; Kew Publishing: London, UK, 2013; p. 291. ISBN 9781842461419. [Google Scholar]
- Linné, C.V.; Salvius, L. Species Plantarum; Impensis Laurentii Salvii: Stockholm, Sweden, 1753; pp. 982+994+1000. [Google Scholar]
- Pine, E.M.; Hibbett, D.S.; Donoghue, M.J. Phylogenetic relationships of cantharelloid and clavarioid Homobasidiomycetes based on mitochondrial and nuclear rDNA sequences. Mycologia 1999, 91, 944–963. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Pine, E.M.; Langer, E.; Langer, G.; Donoghue, M.J. Evolution of gilled mushrooms and puffballs inferred from ribosomal DNA sequences. Proc. Natl. Acad. Sci. USA 1997, 94, 12002–12006. [Google Scholar] [CrossRef] [Green Version]
- Nunez, M.P.; Ryvarden, L. A note on the genus Beenakia. Sydowia 1994, 46, 321–328. [Google Scholar]
- Singer, R. New genera of fungi II. Lloydia 1945, 8, 139–144. [Google Scholar]
- Giachini, A.J.; Castellano, M.A. A new taxonomic classification for species in Gomphus sensu lato. Mycotaxon 2011, 115, 183–201. [Google Scholar] [CrossRef]
- González-Ávila, A.; Martínez-González, C.R.; Espinosa, D.; Estrada-Torres, A. Phaeoclavulina liliputiana sp. nov. (Gomphaceae, Gomphales) a new endemic species from Tlaxcala, Mexico. Phytotaxa 2020, 470, 155–164. [Google Scholar] [CrossRef]
- Whitney, K.D.; Arnott, H.J. Calcium Oxalate Crystal Morphology and Development in Agaricus Bisporus. Mycologia 1987, 79, 180–187. [Google Scholar] [CrossRef]
- Webb, M.A. Cell-Mediated Crystallization of Calcium Oxalate in Plants. Plant Cell 1999, 11, 751–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agerer, R. Fungal relationships and structural identity of their ectomycorrhizae. Mycol. Prog. 2006, 5, 67–107. [Google Scholar] [CrossRef]
- Stodůlková, E.; Sulc, M.; Cisarova, I.; Novak, P.; Kolarik, M.; Flieger, M. Production of (+)-globulol needle crystals on the surface mycelium of Quambalaria cyanescens. Folia Microbiol. 2008, 53, 15–22. [Google Scholar] [CrossRef]
- Guggiari, M.; Bloque, R.; Aragno, M.; Verrecchia, E.; Job, D.; Junier, P. Experimental calcium-oxalate crystal production and dissolution by selected wood-rot fungi. Int. Biodeterior. Biodegrad. 2011, 65, 803–809. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Bleby, T.M.; Veneklaas, E.J.; Lambers, H.; Kuo, J. Morphologies and elemental compositions of calcium crystals in phyllodes and branchlets of Acacia robeorum (Leguminosae: Mimosoideae). Ann. Bot. 2012, 109, 887–896. [Google Scholar] [CrossRef] [Green Version]
- Schilling, J.S.; Jellison, J. Extraction and translocation of calcium from gypsum during wood biodegradation by oxalate-producing fungi. Int. Biodeter. Biodegr. 2007, 60, 8–15. [Google Scholar] [CrossRef]
- Wang, J.B.; Yang, P. Analysis of Physical and Chemical Properties of Forest Soil in Huanglongshan Crossptilon Nature Reserve in Hancheng, Shaanxi Province. For. Resour. Manag. 2014, 105–109. (In Chinese) [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Zhang, M.; Cui, Y.; Bai, X.; Song, B.; Zhang, J.; Yu, X. Dynamic microbial community composition, co-occurrence pattern and assembly in rhizosphere and bulk soils along a coniferous plantation chronosequence. Catena 2023, 223, 106914. [Google Scholar] [CrossRef]
- Jia, H.Y. The physical and chemical character of forest soil in loess regions. Sci. Silvae Sin. 1990, 26, 74–78. (In Chinese) [Google Scholar]
- Lamus, V.; Franco, S.; Montoya, L.; Endara, A.R.; Caballero, L.A.; Bandala, V.M. Mycorrhizal synthesis of the edible mushroom Turbinellus floccosus with Abies religiosa from central Mexico. Mycoscience 2015, 56, 622–626. [Google Scholar] [CrossRef]
- Liu, J.-W.; Luangharn, T.; Wan, S.-P.; Wang, R.; Yu, F.-Q. A new edible species of Gomphus (Gomphaceae) from southwestern China. Mycoscience 2022, 63, 293–297. [Google Scholar] [CrossRef]




| Fungal Taxon | Specimen Voucher | LSU | atp6 | mtSSU |
|---|---|---|---|---|
| Beenakia fricta | K2083 | AY574693 | AY574833 | AY574766 |
| Clavariadelphus ligula | OSC67068 | AY574650 | AY574793 | AY574723 |
| Clavariadelphus lovejoyae | OSC61068 | AY577827 | AY577865 | AY577854 |
| Clavariadelphus occidentalis | OSC37018 | AY574648 | AY574791 | AY574721 |
| Clavariadelphus truncatus | OSC67280 | AY574649 | AY574792 | AY574722 |
| Gautieria caudata | OSC59201 | DQ218483 | DQ218767 | DQ218658 |
| Gautieria crispa | OSC61308 | DQ218484 | DQ218768 | DQ218659 |
| Gautieria monticola | OSC65121 | AY574651 | AY574794 | AY574724 |
| Gautieria pterosperma | OSC69649 | DQ218614 | DQ218900 | DQ218747 |
| Gautieria parksiana | OSC58907 | AY574652 | AY574795 | AY574725 |
| Gautieria otthii | REG636 | — | EU339254 | AF393085 |
| Gloeocantharellus albidocarneus | FCME14883 | — | MH537976 | MT271764 |
| Gloeocantharellus calakmulensis | FCME19868 | — | MH537977 | MT271765 |
| Gloeocantharellus dingleyae | PDD:30179 | AY574668 | — | AY574741 |
| Gloeocantharellus novae-zelandiae | PDD:44960 | AY574666 | AY574809 | AY574739 |
| Gloeocantharellus pallidus | BPI54917 | AY574673 | AY574815 | — |
| Gloeocantharellus papuanus | PERTH4549 | AY574667 | AY574810 | AY574740 |
| Gloeocantharellus pleurobrunnescens | 1924 | MT261811 | MH537978 | MT271766 |
| Gloeocantharellus purpurascens | TENN12793 | AY574683 | AY574823 | AY574756 |
| Gloeocantharellus purpurascens | TENN14265 | AY574684 | AY574824 | AY574757 |
| Gomphus clavatus | UPS | AY574665 | AY574808 | AY574738 |
| Gomphus clavatus | OSC97616 | AY574664 | AY574807 | AY574737 |
| Gomphus clavatus | OSC97588 | AY577836 | AY577874 | AY577863 |
| Gomphus clavatus | OSC97587 | DQ218487 | DQ218771 | DQ218662 |
| Gomphocantharellus cylindrosporus | BJTCFM109 | OK660766 | OK665160 | OK660767 |
| Gomphocantharellus cylindrosporus | BJTCFM375 | OK660768 | OK665161 | OK660770 |
| Gomphocantharellus cylindrosporus | HSA335 | OK660772 | OK665162 | OK660771 |
| Luteodorsum huanglongense | HMAS256997 | OQ801490 | OQ790052 | OQ801494 |
| Luteodorsum huanglongense | HMAS256998 | OQ801491 | OQ790053 | OQ801495 |
| Luteodorsum huanglongense | MNWAFU-CF-P209 | OQ929933 | OQ924518 | OQ929931 |
| Luteodorsum huanglongense | MNWAFU-CF-P210 | OQ929934 | OQ924519 | OQ929932 |
| Hydnocristella himantia | O102156 | AY574691 | AY574831 | AY574764 |
| Kavinia alboviridis | O102140 | AY574692 | AY574832 | AY574765 |
| Lentaria pinicola | SUCM89 | AY574688 | — | AY574761 |
| Lentaria pinicola | SUCM560 | AY574690 | AY574830 | AY574763 |
| Lentaria pinicola | SUCM46 | AY574689 | AY574829 | AY574762 |
| Phaeoclavulina africana | TENN39621 | AY574653 | AY574796 | AY574726 |
| Phaeoclavulina cokeri | TENN36030 | AY574701 | AY574843 | AY574774 |
| Phaeoclavulina curta | OSC8711 | AY574713 | AY574858 | — |
| Phaeoclavulina cyanocephala | TENN37827 | AY574710 | AY574854 | AY574779 |
| Phaeoclavulina eumorpha | TENN37842 | — | AY574857 | AY574782 |
| Phaeoclavulina eumorpha | TENN36218 | AY574712 | AY574856 | AY574781 |
| Phaeoclavulina gigantea | FH109 | AY574703 | AY574845 | AY574776 |
| Phaeoclavulina grandis | BR079158-06 | AY574678 | AY574820 | AY574751 |
| Phaeoclavulina guadelupensis | FH120 | AY574682 | — | AY574755 |
| Phaeoclavulina guyanensis | FH84 | AY574706 | AY574848 | — |
| Phaeoclavulina insignis | FH104 | AY574704 | AY574846 | — |
| Phaeoclavulina longicaulis | TENN33826 | AY574700 | AY574842 | AY574773 |
| Phaeoclavulina ochraceovirens | OSC23475 | AY574714 | AY574859 | — |
| Phaeoclavulina pancaribbea | TENN31836 | AY574707 | AY574849 | — |
| Phaeoclavulina subclaviformis | BR079159-07 | AY574679 | — | AY574752 |
| Phaeoclavulina viridis | PERTH4302 | AY574677 | AY574819 | AY574750 |
| Phaeoclavulina viridis | OSC97708 | AY574675 | AY574817 | AY574748 |
| Phaeoclavulina viridis | FH1853 | AY574676 | AY574818 | — |
| Ramaria apiculata | OSC23549 | AY574695 | AY574836 | AY574768 |
| Ramaria apiculate var. brunnea | TENN53935 | AY574696 | AY574837 | AY574769 |
| Ramaria araiospora var. araiospora | SUCM739 | AF213068 | AY574838 | AF213141 |
| Ramaria araiospora var. araiospora | SUCM556 | AY574697 | AY574839 | AY574770 |
| Ramaria botrytis var. botrytis | SUCM740 | AY574699 | AY574841 | AY574772 |
| Ramaria botrytis var. botrytis | SUCM457 | AY574698 | AY574840 | AY574771 |
| Ramaria circinans var. anceps | SUCM615 | AY574711 | AY574855 | AY574780 |
| Ramaria circinans | NYS1 | AY574702 | AY574844 | AY574775 |
| Ramaria rainierensis | SUCM431 | AY574694 | AY574835 | AY574767 |
| Ramaria rainierensis | SUCM231 | AF213115 | AY574834 | AF213135 |
| Ramaria rubribrunnescens | SUCM844 | AF213098 | AY574852 | AF213142 |
| Ramaria stuntzii | SUCM214 | AF213102 | AY574850 | AF213134 |
| Ramaria suecica | BPI1 | AY574705 | AY574847 | — |
| Ramaria vinosimaculans | OSC23287 | AY574709 | AY574853 | AY574778 |
| Turbinellus floccosus | MICH5588 | AY574660 | AY574803 | AY574733 |
| Turbinellus floccosus | OSC69167 | AY574656 | AY574799 | AY574729 |
| Turbinellus floccosus | OSA-MY-1839 | AY574654 | AY574797 | AY574727 |
| Turbinellus floccosus | OSA-MY-1840 | AY574655 | AY574798 | AY574728 |
| Turbinellus floccosus | TENN33233 | AY574657 | AY574800 | AY574730 |
| Turbinellus floccosus | SFSU21238 | AY574658 | AY574801 | AY574731 |
| Turbinellus floccosus | TENN33295 | AY574659 | AY574802 | AY574732 |
| Turbinellus floccosus | MICH10721 | AY574661 | AY574804 | AY574734 |
| Turbinellus floccosus | UC759902 | AY574662 | AY574805 | AY574735 |
| Turbinellus floccosus | UC924302 | AY574663 | AY574806 | AY574736 |
| Turbinellus fujisanensis | OSA-MY-1841 | AY574670 | AY574812 | AY574743 |
| Turbinellus fujisanensis | OSA-MY-1842 | AY574669 | AY574811 | AY574742 |
| Turbinellus kauffmanii | OSC97590 | AY574672 | AY574814 | AY574745 |
| Turbinellus kauffmanii | MICH10069 | AY574671 | AY574813 | AY574744 |
| Mutinus elegans | OSC107657 | AY574643 | AY574785 | AY574717 |
| Phallus impudicus | OSC107655 | AY574642 | AY574784 | AY574716 |
| Pseudocolus fusiformis | DSH96-033 | AF518641 | — | AF026666 |
| Element | Weight% | Atomic% | Weight% (Corrected) | Atomic% (Corrected) |
|---|---|---|---|---|
| CK | 5.83 | 18.18 | 6.69 | 13.58 |
| OK | 16.89 | 39.53 | 32.41 | 49.38 |
| PtM | 40.32 | 7.74 | — | — |
| CaK | 36.97 | 34.55 | 60.90 | 37.04 |
| Matrix | Correction | ZAF | Correction | ZAF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Z.; Wu, Y.; Luo, Z.; Xiong, C.; Liu, X.; Wang, B.; Ma, B.; Wei, J.; Yu, Z. Luteodorsum huanglongense (Gomphaceae, Gomphales), a New Genus and Species of Gomphoid Fungus from the Loess Plateau, Northwest China. J. Fungi 2023, 9, 664. https://doi.org/10.3390/jof9060664
Peng Z, Wu Y, Luo Z, Xiong C, Liu X, Wang B, Ma B, Wei J, Yu Z. Luteodorsum huanglongense (Gomphaceae, Gomphales), a New Genus and Species of Gomphoid Fungus from the Loess Plateau, Northwest China. Journal of Fungi. 2023; 9(6):664. https://doi.org/10.3390/jof9060664
Chicago/Turabian StylePeng, Zijia, Yiming Wu, Zeyu Luo, Chaowei Xiong, Xiaoyong Liu, Bin Wang, Baoyou Ma, Jianxian Wei, and Zhongdong Yu. 2023. "Luteodorsum huanglongense (Gomphaceae, Gomphales), a New Genus and Species of Gomphoid Fungus from the Loess Plateau, Northwest China" Journal of Fungi 9, no. 6: 664. https://doi.org/10.3390/jof9060664






