Prediction of Suitable Habitat Distribution of Cryptosphaeria pullmanensis in the World and China under Climate Change
Abstract
:1. Introduction
2. Materials and Methods
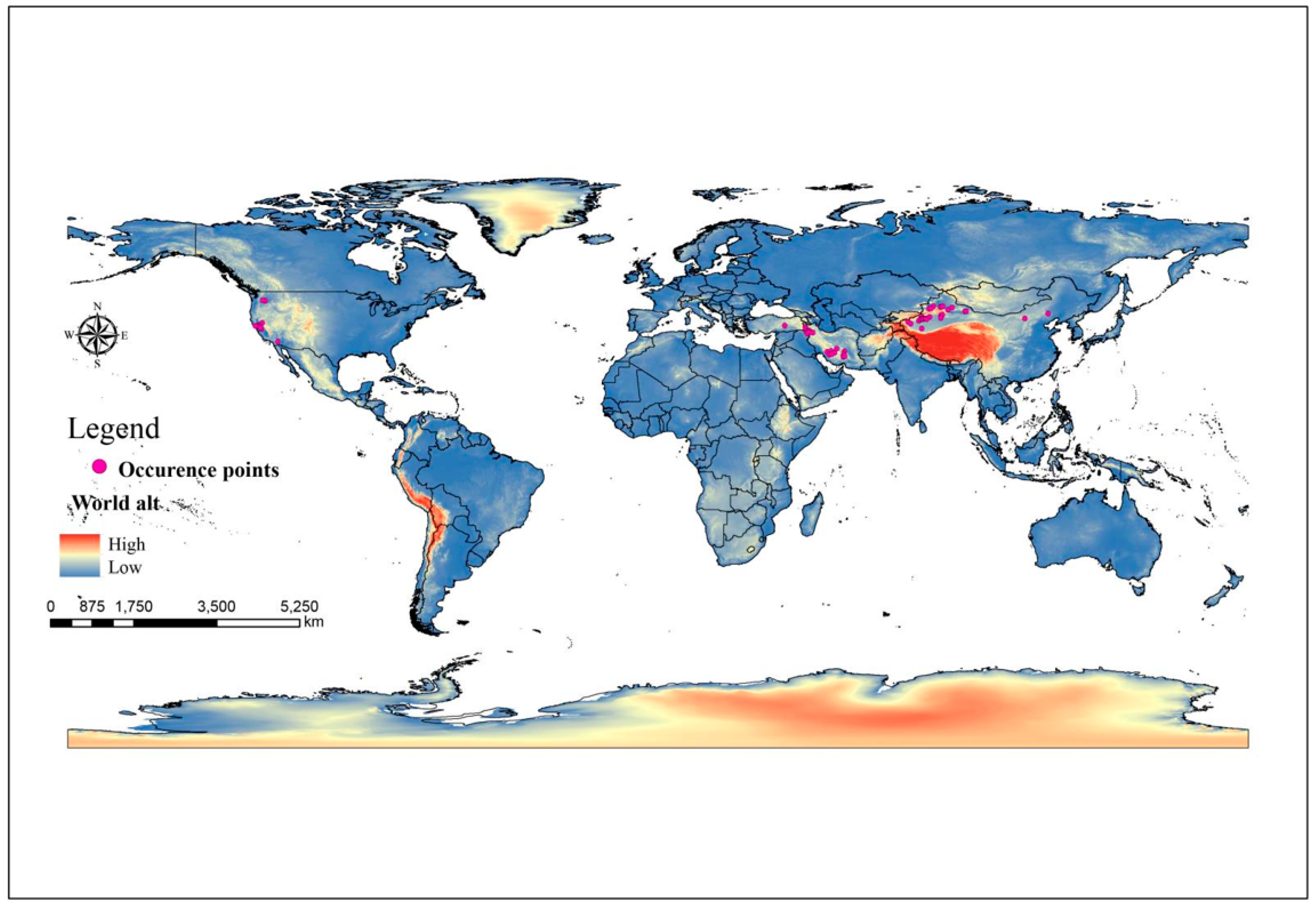
2.1. The Distribution Points of C. pullmanensis
2.2. Environmental Factor Variables
2.3. Construction and Accuracy Evaluation of the MaxEnt Model
2.3.1. Parameter Optimization of Maximum Entropy Model
2.3.2. Classification of Potentially Suitable Areas
2.4. Statistical and Spatial Analysis
2.4.1. Calculating Distribution Shifts
2.4.2. Centroid Migration
3. Results
3.1. Assessment of the Model’s Accuracy
3.2. Environmental Variable Analysis for the Identification of the Predicted Potentially Appropriate Area of C. pullmanensis
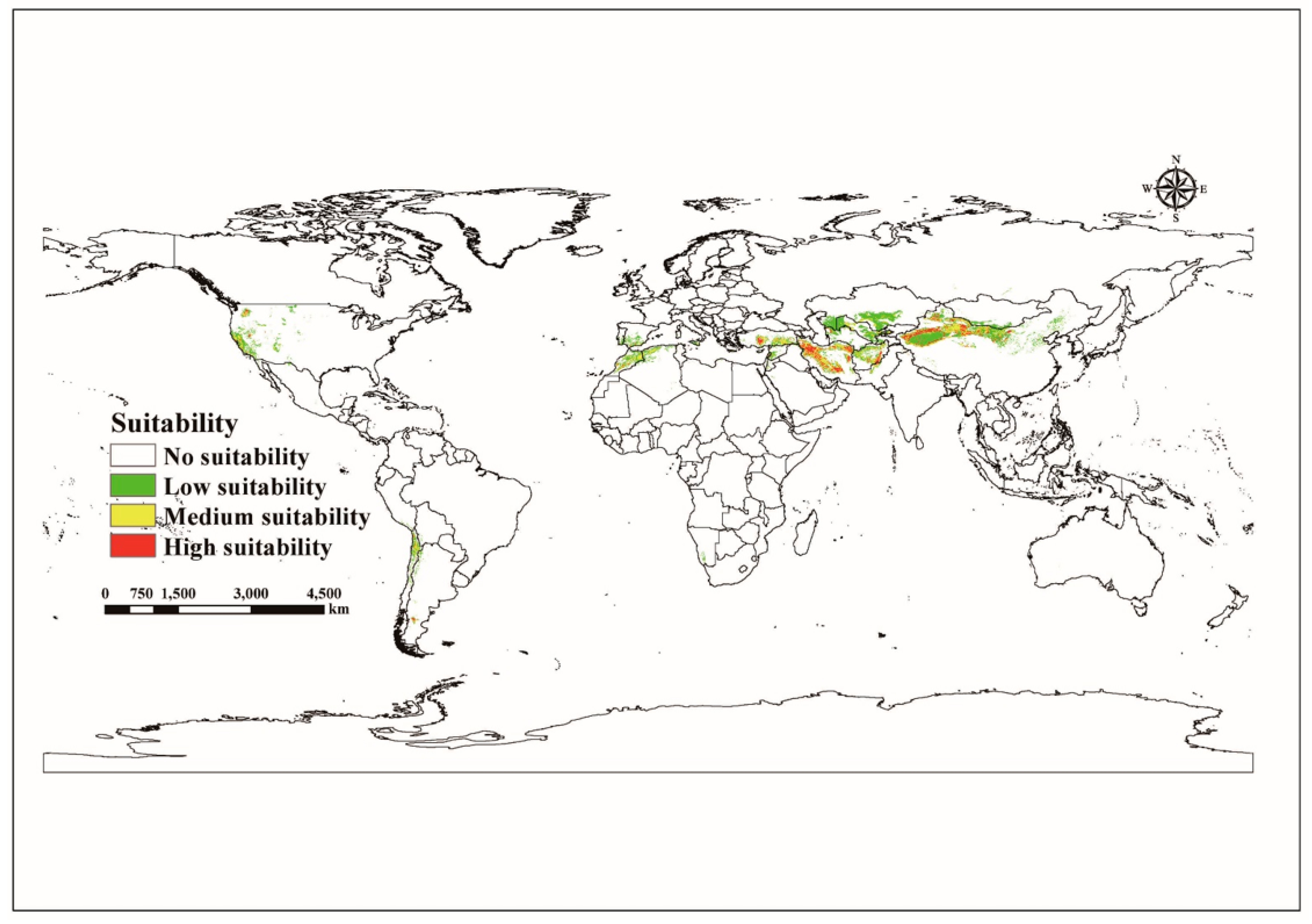
3.3. Current Potentially Suitable Habitats of C. pullmanensis in the World and China
3.3.1. Global Suitable Habitats under Current Climate Scenario Models
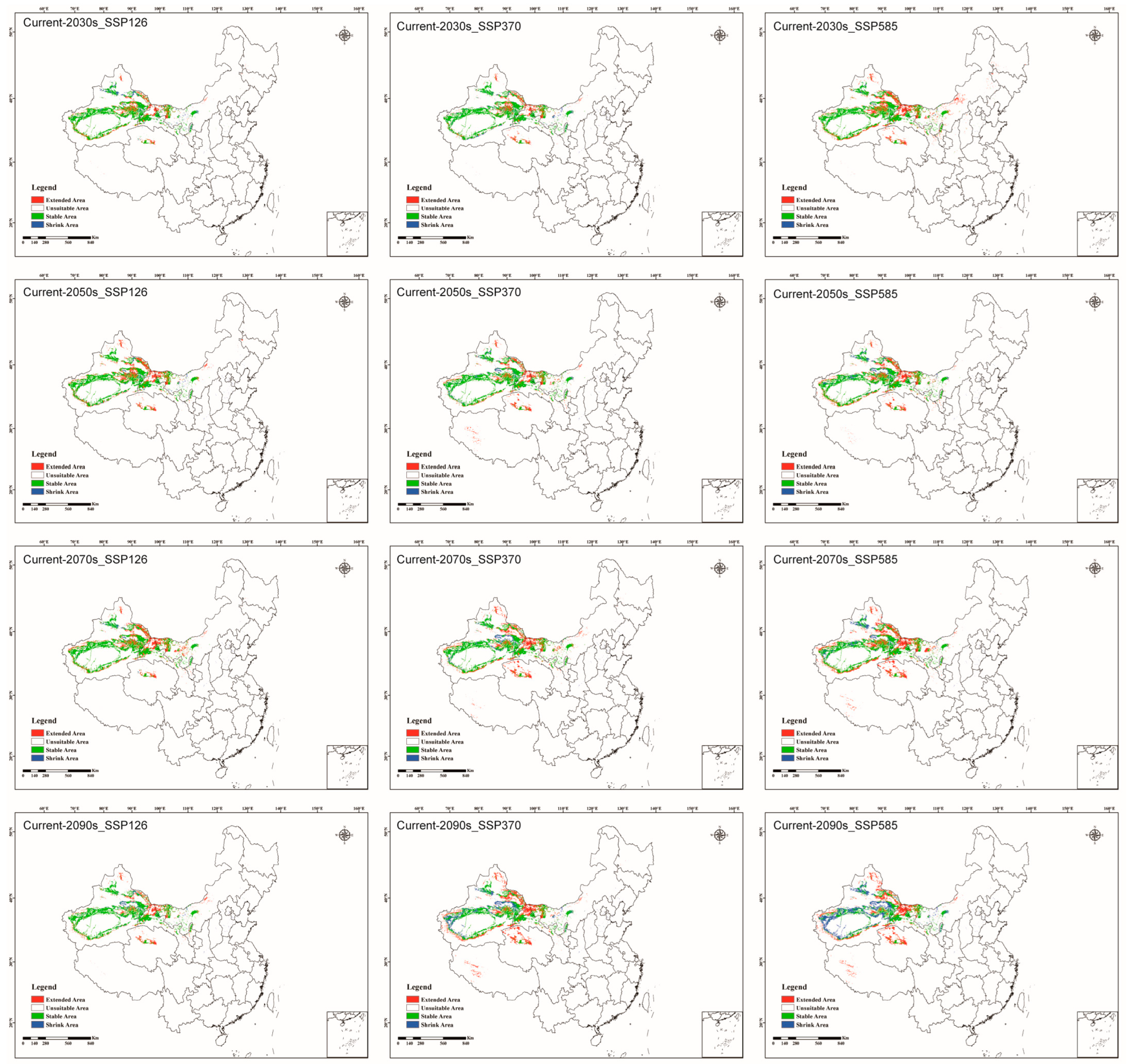
3.3.2. Current Potentially Suitable Habitats of C. pullmanensis in China
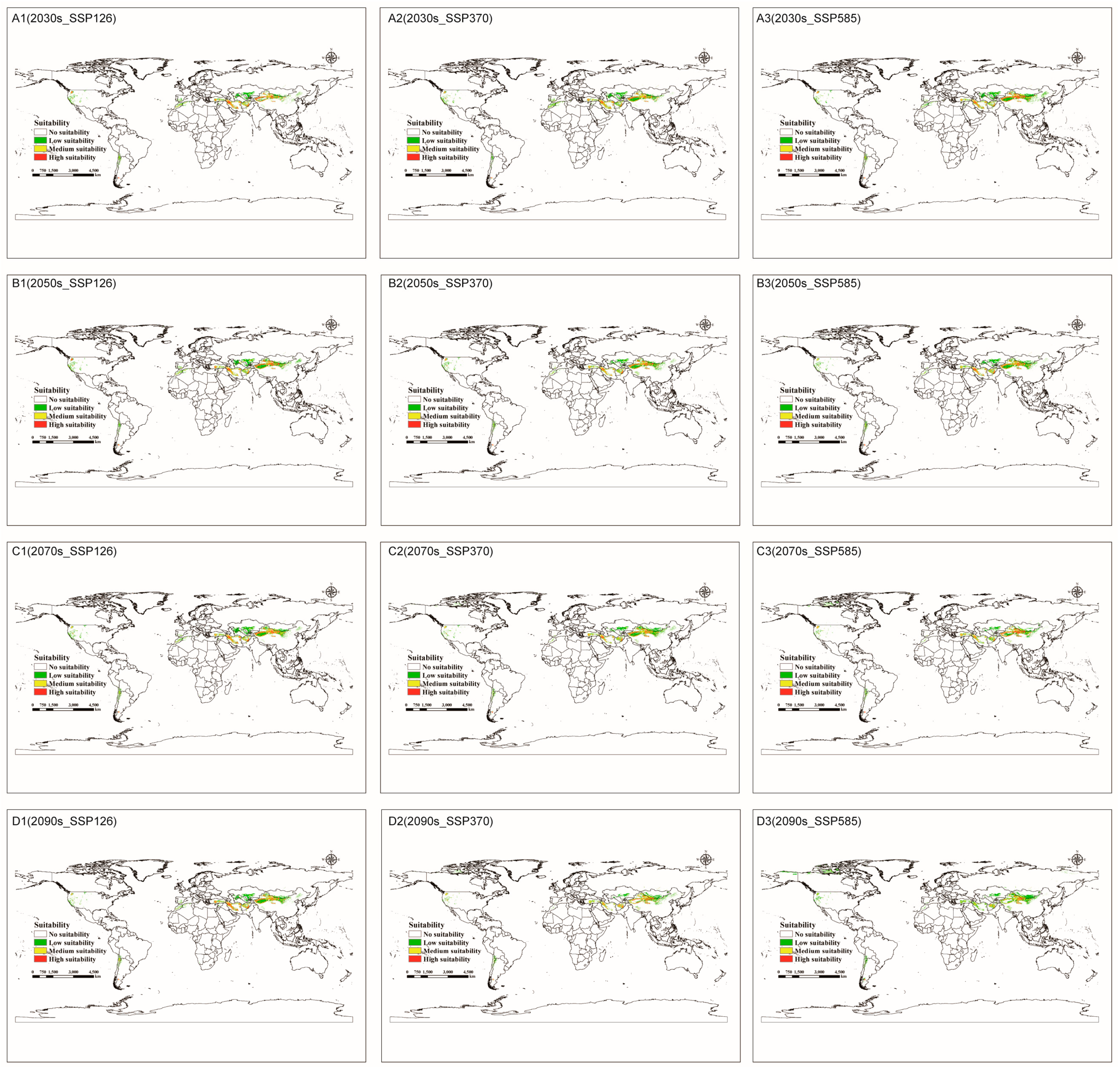
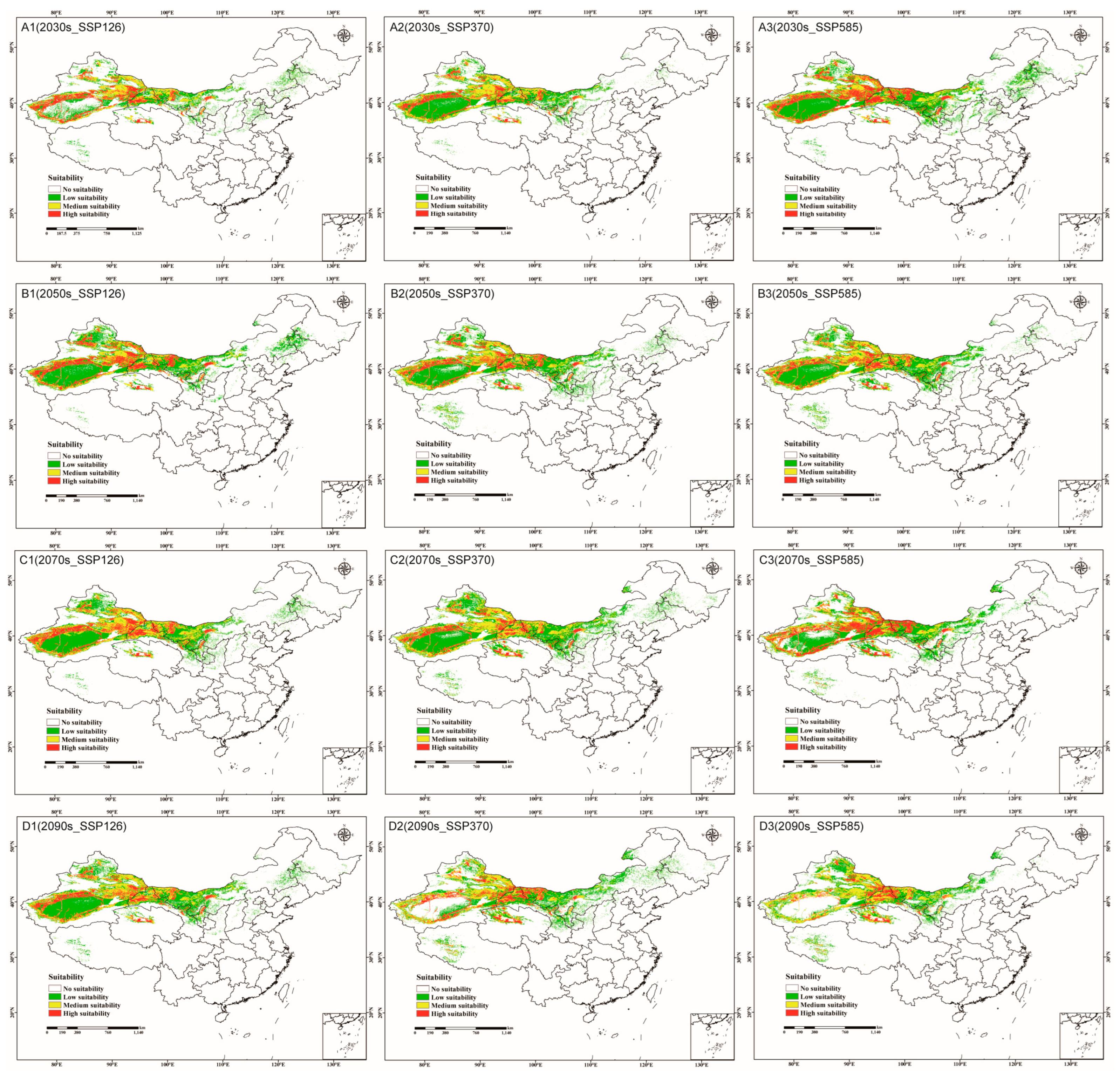
3.4. Future Potentially C. pullmanensis Habitats in the World and China
3.4.1. Global Suitable Areas under Future Climate Scenario Models
3.4.2. Potentially Suitable Habitats for C. pullmanensis Based on Future Climatic Scenarios in China
3.5. Centroid Migration of Potential Suitable areas for C. pullmanensis Based on Future Climatic Scenarios
4. Discussion
4.1. Accuracy of MaxEnt after Optimization
4.2. The Effect of Environmental Factors on the Distribution of C. pullmanensis
4.3. Changes in the Distribution of C. pullmanensis in the Future
4.4. Limitations of SDM in Predicting Species Distribution
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Y.; Xie, S.; Cao, J.; Zhao, H.; Yin, X.; Guo, Y.; Xu, C.; Guo, L.; Wu, H.; Zhang, M. Lasiodiplodia regiae sp. nov.: A new species causing canker and dieback of fruit trees in China. Phytopathology 2023. Online ahead of print. [Google Scholar] [CrossRef]
- Glawe, D.A.J.M. Cryptosphaeria pullmanensis, a new species from Washington state. Mycologia 1984, 76, 166–169. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Gubler, W.D. Cryptosphaeria Dieback of Fremont Cottonwood Caused by Cryptosphaeria pullmanensis and C. multicontinentalis in California. Plant Dis. 2016, 100, 777–783. [Google Scholar] [CrossRef] [Green Version]
- Trouillas, F.P.; Urbez-Torres, J.R.; Gubler, W.D.J.M. Diversity of diatrypaceous fungi associated with grapevine canker diseases in California. Mycologia 2010, 102, 319–336. [Google Scholar] [CrossRef] [PubMed]
- Rashid Aghdam, A.; Babai-Ahari, A.; Karimi, K.; Arzanlou, M. Cryptosphaeria Canker of Populus nigra Caused by Cryptosphaeria pullmanensis, a New Threat to Poplar Industry in Iran. J. Phytopathol. 2017, 165, 387–396. [Google Scholar] [CrossRef]
- Kaifa, G. Identification to Pathogens of Woods Canker, Genetic Differentiation Research to Its Main Species Valsa Mali and Rapid Detection to Primary Infection Source of V. Mali in Xinjiang. Ph.D. Thesis, Shihezi University, Shihezi, China, 2016. [Google Scholar]
- Ma, R.; Zhu, Y.F.; Fan, X.L.; Tian, C.M.J.F.P. Canker disease of willow and poplar caused by Cryptosphaeria pullmanensis recorded in China. For. Pathol. 2016, 46, 327–335. [Google Scholar] [CrossRef]
- Trouillas, F.P.; Hand, F.P.; Inderbitzin, P.; Gubler, W.D. The genus Cryptosphaeria in the western United States: Taxonomy, multilocus phylogeny and a new species, C. multicontinentalis. Mycologia 2015, 107, 1304–1313. [Google Scholar] [CrossRef] [PubMed]
- Yinming, L. Study of Ascomycetous Species on Branches Disease from Desert Plants in Xinjiang. Master’s Thesis, Xinjiang Agricultural University, Ürümqi, China, 2017. [Google Scholar]
- Sohrabi, M.; Mohammadi, H.; León, M.; Armengol, J.; Banihashemi, Z. Fungal pathogens associated with branch and trunk cankers of nut crops in Iran. Eur. J. Plant Pathol. 2020, 157, 327–351. [Google Scholar] [CrossRef]
- Yan, C.; Hao, H.; Feng, H.; Wang, Z.; Sha, S.; Li, M.; Wang, L.; Kang, Z.J.G. Whole genome sequence of Cryptosphaeria pullmanensis, an important pathogenic fungus potentially threatening crop and forestry production. Genomics 2023, 115, 110576. [Google Scholar] [CrossRef]
- Jia, W.; Ma, Q.; Su, J.; Li, Z.; Liu, Y.; Sun, P. Pathogen Identification of Poplar Stem Rot in Hohhot of Inner Mongolia. J. Northwest For. Univ. 2022, 37, 210–215, 237. [Google Scholar]
- Liu, B.; Zhang, Q.; Yang, X. Molecular Identification of Tree Rot Disease Pathogenic Fungi in Inner Mongolia. J. Chifeng Univ. 2022, 38, 91–95. [Google Scholar]
- Shasha, X. The Occurrence, Pathogenic Structural Composition and Genetic Diversity of Walnut Canker Disease in Xinjiang. Master’s Thesis, Tarim University, Alar, China, 2022. [Google Scholar]
- Guyon, J.; Hoffman, J. Survey of Aspen Disease in the Intermountain Region; USDA Forest Service Forest Health Protection: Ogden, UT, USA, 2011. [Google Scholar]
- Worrall, J.J.; Rehfeldt, G.E.; Hamann, A.; Hogg, E.H.; Marchetti, S.B.; Michaelian, M.; Gray, L.K. Recent declines of Populus tremuloides in North America linked to climate. For. Ecol. Manag. 2013, 299, 35–51. [Google Scholar] [CrossRef]
- Raoufi, F.; Banihashemi, Z.; Mirtalebi, M. First report of walnut canker caused by Cryptosphaeria pullmanensis in Iran. In Proceedings of the 22nd Iran Plant Protection Congress, Karaj, Iran, 27–30 August 2016; pp. 27–30. [Google Scholar]
- Trouillas, F.; Gubler, W.J.P.D. Pathogenicity of Diatrypaceae species in grapevines in California. Plant Dis. 2010, 94, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Naseri, B.; Alizadeh, M. Climate, powdery mildew, sainfoin resistance and yield. J. Plant Pathol. 2017, 99, 619–625. [Google Scholar]
- La Porta, N.; Capretti, P.; Thomsen, I.M.; Kasanen, R.; Hietala, A.M.; Von Weissenberg, K. Forest pathogens with higher damage potential due to climate change in Europe. Can. J. Plant Pathol. 2008, 30, 177–195. [Google Scholar] [CrossRef]
- Bebber, D.P.; Ramotowski, M.A.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Change 2013, 3, 985–988. [Google Scholar] [CrossRef]
- McKay, A. Pathogens in a warming world. Nat. Ecol. Evol. 2023, 7, 2. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zhao, Z.; Wei, H.; Gao, B.; Gu, W. Prediction of the potential geographic distribution of the ectomycorrhizal mushroom Tricholoma matsutake under multiple climate change scenarios. Nature 2017, 7, 46221. [Google Scholar] [CrossRef] [Green Version]
- Suz, L.M.; Barsoum, N.; Benham, S.; Cheffings, C.; Cox, F.; Hackett, L.; Jones, A.G.; Mueller, G.M.; Orme, D.; Seidling, W. Monitoring ectomycorrhizal fungi at large scales for science, forest management, fungal conservation and environmental policy. Ann. For. Sci. 2015, 72, 877–885. [Google Scholar] [CrossRef]
- Naseri, B.; Tabande, L. Patterns of Fusarium wilt epidemics and bean production determined according to a large-scale dataset from agro-ecosystems. Rhizosphere 2017, 3, 100–104. [Google Scholar] [CrossRef]
- Guo, Y.; Li, X.; Zhao, Z.; Nawaz, Z. Predicting the impacts of climate change, soils and vegetation types on the geographic distribution of Polyporus umbellatus in China. Sci. Total Environ. 2019, 648, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Alkhalifah, D.H.M.; Damra, E.; Melhem, M.B.; Hozzein, W.N. Fungus under a Changing Climate: Modeling the Current and Future Global Distribution of Fusarium oxysporum Using Geographical Information System Data. Microorganisms 2023, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-T.; Lu, Z.-P.; Gao, X.-Y.; Liu, M.-L.; Sa, W.; Liang, J.; Wang, L.; Yin, W.; Shang, Q.-H.; Li, Z.-H. Maximum entropy modeling the distribution area of Morchella dill. Ex Pers. species in China under changing climate. Biology 2022, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Yuan, Y.; Li, X.; Zhang, J. Maximum entropy modeling to predict the impact of climate change on pine wilt disease in China. Front. Plant Sci. 2021, 12, 652500. [Google Scholar] [CrossRef] [PubMed]
- Pattison, R.R.; Mack, R.N. Potential distribution of the invasive tree Triadica sebifera (Euphorbiaceae) in the United States: Evaluating CLIMEX predictions with field trials. Glob. Change Biol. 2008, 14, 813–826. [Google Scholar] [CrossRef]
- Beaumont, L.J.; Hughes, L.; Poulsen, M. Predicting species distributions: Use of climatic parameters in BIOCLIM and its impact on predictions of species’ current and future distributions. Ecol. Model. 2005, 186, 251–270. [Google Scholar] [CrossRef]
- Du, Z.; Wu, J.; Meng, X.; Li, J.; Huang, L. Predicting the global potential distribution of four endangered Panax species in middle-and low-latitude regions of china by the geographic information system for global medicinal plants (GMPGIS). Molecules 2017, 22, 1630. [Google Scholar] [CrossRef] [Green Version]
- Bie, J.; Zheng, K.; Gao, X.; Liu, B.; Ma, J.; Hayat, M.A.; Xiao, J.; Wang, H. Spatial risk analysis of Batrachochytrium dendrobatidis, a global emerging fungal pathogen. EcoHealth 2021, 18, 3–12. [Google Scholar] [CrossRef]
- Zhu, G.; Qiao, H. Effect of the Maxent model’s complexity on the prediction of species potential distributions. Bodiv. Sci. 2016, 24, 1189. [Google Scholar]
- Zhao, X.; Lei, M.; Wei, C.; Guo, X. Assessing the suitable regions and the key factors for three Cd-accumulating plants (Sedum alfredii, Phytolacca americana, and Hylotelephium spectabile) in China using MaxEnt model. Sci. Total Environ. 2022, 852, 158202. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, S.; Tang, S.; Pan, J.; Ren, L.; Tian, X.; Sun, Z.; Zhang, Z. Analysis of the prediction of the suitable distribution of Polygonatum kingianum under different climatic conditions based on the MaxEnt model. Front. Earth Sci. 2023, 11, 1111878. [Google Scholar] [CrossRef]
- Xu, W.; Sun, H.; Jin, J.; Cheng, J. Predicting the potential distribution of apple canker pathogen (Valsa mali) in China under climate change. Forests 2020, 11, 1126. [Google Scholar] [CrossRef]
- Hernández-Baz, F.; Romo, H.; González, J.M.; de Jesús Martínez Hernández, M.; Pastrana, R.G. Maximum entropy niche-based modeling (Maxent) of potential geographical distribution of Coreura albicosta (Lepidoptera: Erebidae: Ctenuchina) in Mexico. Fla. Entomol. 2016, 99, 376–380. [Google Scholar] [CrossRef] [Green Version]
- Krasnov, H.; Ezra, D.; Bahri, B.A.; Cacciola, S.O.; Meparishvili, G.; Migheli, Q.; Blank, L. Potential distribution of the citrus Mal Secco disease in the Mediterranean basin under current and future climate conditions. Plant Pathol. 2023, 72, 765–773. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.; Zhang, X.; Li, Q. Prediction of the Potential Distributions of Prunus salicina Lindl., Monilinia fructicola, and Their Overlap in China Using MaxEnt. J. Fungi 2023, 9, 189. [Google Scholar] [CrossRef] [PubMed]
- Al Ruheili, A.M.; Boluwade, A.; Al Subhi, A.M. Assessing the impact of climate change on the distribution of lime (16SRII-b) and alfalfa (16srii-d) phytoplasma disease using maxent. Plants 2021, 10, 460. [Google Scholar] [CrossRef]
- Naseri, B.; Marefat, A. Seasonal dynamics and prevalence of alfalfa fungal pathogens in Zanjan province, Iran. Int. J. Plant Prod. 2012, 2, 327–340. [Google Scholar]
- Warren, D.L.; Glor, R.E.; Turelli, M.J.E. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography 2010, 33, 607–611. [Google Scholar] [CrossRef]
- van Proosdij, A.S.; Sosef, M.S.; Wieringa, J.J.; Raes, N.J.E. Minimum required number of specimen records to develop accurate species distribution models. Ecography 2016, 39, 542–552. [Google Scholar] [CrossRef]
- Guisan, A.; Thuiller, W.; Zimmermann, N.E. Habitat Suitability and Distribution Models: With Applications in R; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Lu, Y.; Wu, T.; Li, Y.; Yang, B. Mitigation of the double ITCZ syndrome in BCC-CSM2-MR through improving parameterizations of boundary-layer turbulence and shallow convection. Geosci. Model Dev. 2021, 14, 5183–5204. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, H.; Zhao, Z.; Liu, J.; Ran, Q.; Yu, J.; Gu, W. Optimization of the fuzzy matter element method for predicting species suitability distribution based on environmental data. Sustainability 2018, 10, 3444. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.-Q.; Kushwaha, S.; Saran, S.; Xu, J.; Roy, P. Maxent modeling for predicting the potential distribution of medicinal plant, Justicia adhatoda L. in Lesser Himalayan foothills. Ecol. Eng. 2013, 51, 83–87. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Dong, L.; Li, C.; Zhang, L.; Wang, B.; Ma, D.; Mu, Q.; Wang, J.; Hou, H. Predicting global potential distribution of Peromyscopsylla hesperomys and Orchopeas sexdentatus and risk assessment for invading China under climate change. Front. Public Health 2023, 10, 1018327. [Google Scholar] [CrossRef]
- Zhao, Z.; Xiao, N.; Shen, M.; Li, J. Comparison between optimized MaxEnt and random forest modeling in predicting potential distribution: A case study with Quasipaa boulengeri in China. Sci. Total Environ. 2022, 842, 156867. [Google Scholar] [CrossRef] [PubMed]
- Cobos, M.E.; Peterson, A.T.; Barve, N.; Osorio-Olvera, L. kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ 2019, 7, e6281. [Google Scholar] [CrossRef] [Green Version]
- Akaike, H. Maximum likelihood identification of Gaussian autoregressive moving average models. Biometrika 1973, 60, 255–265. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Dai, X.; Wu, W.; Ji, L.; Tian, S.; Yang, B.; Guan, B.; Wu, D. MaxEnt model-based prediction of potential distributions of Parnassiawightiana (Celastraceae) in China. Biodivers. Data J. 2022, 10, e81073. [Google Scholar] [CrossRef]
- Osorio-Olvera, L.; Lira-Noriega, A.; Soberón, J.; Peterson, A.T.; Falconi, M.; Contreras-Díaz, R.G.; Martínez-Meyer, E.; Barve, V.; Barve, N. ntbox: An r package with graphical user interface for modelling and evaluating multidimensional ecological niches. Methods Ecol. Evol. 2020, 11, 1199–1206. [Google Scholar] [CrossRef]
- Hosni, E.M.; Nasser, M.G.; Al-Ashaal, S.A.; Rady, M.H.; Kenawy, M.A. Modeling current and future global distribution of Chrysomya bezziana under changing climate. Nature 2020, 10, 4947. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Wu, F.; Guo, W.; Liu, N. Identification of potential cultivation region for Santalum album in China by the MaxEnt ecologic niche model. Sci. Silvae Sin. 2014, 50, 27–33. [Google Scholar]
- Hanley, J.A.; Mcneil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Zhang, M.; Yang, Q.; Ye, L.; Liu, Y.; Zhang, G.; Chen, S.; Lai, W.; Wen, G.; Zheng, S. Prediction of suitable distribution of a critically endangered plant Glyptostrobus pensilis. Forests 2022, 13, 257. [Google Scholar] [CrossRef]
- Roberts, D.R.; Bahn, V.; Ciuti, S.; Boyce, M.S.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef] [Green Version]
- Moya, W.; Jacome, G.; Yoo, C. Past, current, and future trends of red spiny lobster based on PCA with MaxEnt model in Galapagos Islands, Ecuador. Ecol. Evol. 2017, 7, 4881–4890. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; Yue, J.; Xie, T.; Xu, Y.; Tian, Y. Predicting shifts in the suitable climatic distribution of walnut (Juglans regia L.) in China: Maximum entropy model paves the way to forest management. Forest 2018, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.L. SDM toolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014, 5, 694–700. [Google Scholar] [CrossRef]
- Hu, X.-G.; Jin, Y.; Wang, X.-R.; Mao, J.-F.; Li, Y. Predicting Impacts of Future Climate Change on the Distribution of the Widespread Conifer Platycladus orientalis. PLoS ONE 2015, 10, e0132326. [Google Scholar] [CrossRef]
- Gebrewahid, Y.; Abrehe, S.; Meresa, E.; Eyasu, G.; Abay, K.; Gebreab, G.; Kidanemariam, K.; Adissu, G.; Abreha, G.; Darcha, G. Current and future predicting potential areas of Oxytenanthera abyssinica (A. Richard) using MaxEnt model under climate change in Northern Ethiopia. Ecol. Process. 2020, 9, 6. [Google Scholar] [CrossRef] [Green Version]
- Kong, W.Y.; Li, X.H.; Zou, H.F. Optimizing MaxEnt model in the prediction of species distribution. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2019, 30, 2116–2128. [Google Scholar]
- Radosavljevic, A.; Anderson, R.P. Making better Maxent models of species distributions: Complexity, overfitting and evaluation. J. Biogeogr. 2014, 41, 629–643. [Google Scholar] [CrossRef]
- Zhang, J.; Nielsen, S.E.; Chen, Y.; Georges, D.; Qin, Y.; Wang, S.S.; Svenning, J.C.; Thuiller, W. Extinction risk of North American seed plants elevated by climate and land-use change. J. Appl. Ecol. 2017, 54, 303–312. [Google Scholar] [CrossRef]
- Zheng, S.; Zhang, B.; Peng, D.; Yu, L.; Lin, B.; Pan, Y.; Xie, Q. The trend towards a warmer and wetter climate observed in arid and semi-arid areas of northwest China from 1959 to 2019. Environ. Res. Commun. 2021, 3, 115011. [Google Scholar] [CrossRef]
- Min, X.J.; Zang, Y.X.; Sun, W.; Ma, J.Y. Contrasting water sources and water-use efficiency in coexisting desert plants in two saline-sodic soils in northwest China. Plant Biol. 2019, 21, 1150–1158. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Rusuli, Y. Assessment of Soil Salinization Risk by Remote Sensing-Based Ecological Index (RSEI) in the Bosten Lake Watershed, Xinjiang in Northwest China. Sustainability 2022, 14, 7118. [Google Scholar] [CrossRef]
- Arshad, A.; Raza, M.A.; Zhang, Y.; Zhang, L.; Wang, X.; Ahmed, M.; Habib-ur-Rehman, M. Impact of climate warming on cotton growth and yields in China and Pakistan: A regional perspective. Agriculture 2021, 11, 97. [Google Scholar] [CrossRef]
- Jiang, Z.; Chen, W.; Ji, W.; Jie, S. Projection of Extreme Climate Events in China in the 21st Century Based on AR4/IPCC Models. 2023. [Google Scholar]
- Shen, J.; Zhang, Z.; Liu, R.; Wang, Z. Ecological restoration of eroded karst utilizing pioneer moss and vascular plant species with selection based on vegetation diversity and underlying soil chemistry. Int. J. Phytoremediat. 2018, 20, 1369–1379. [Google Scholar] [CrossRef]
- Pesendorfer, M.B.; Ascoli, D.; Bogdziewicz, M.; Hacket-Pain, A.; Pearse, I.S.; Vacchiano, G. The Ecology and Evolution of Synchronized Reproduction in Long-Lived Plants; The Royal Society: London, UK, 2021; Volume 376, p. 20200369. [Google Scholar]

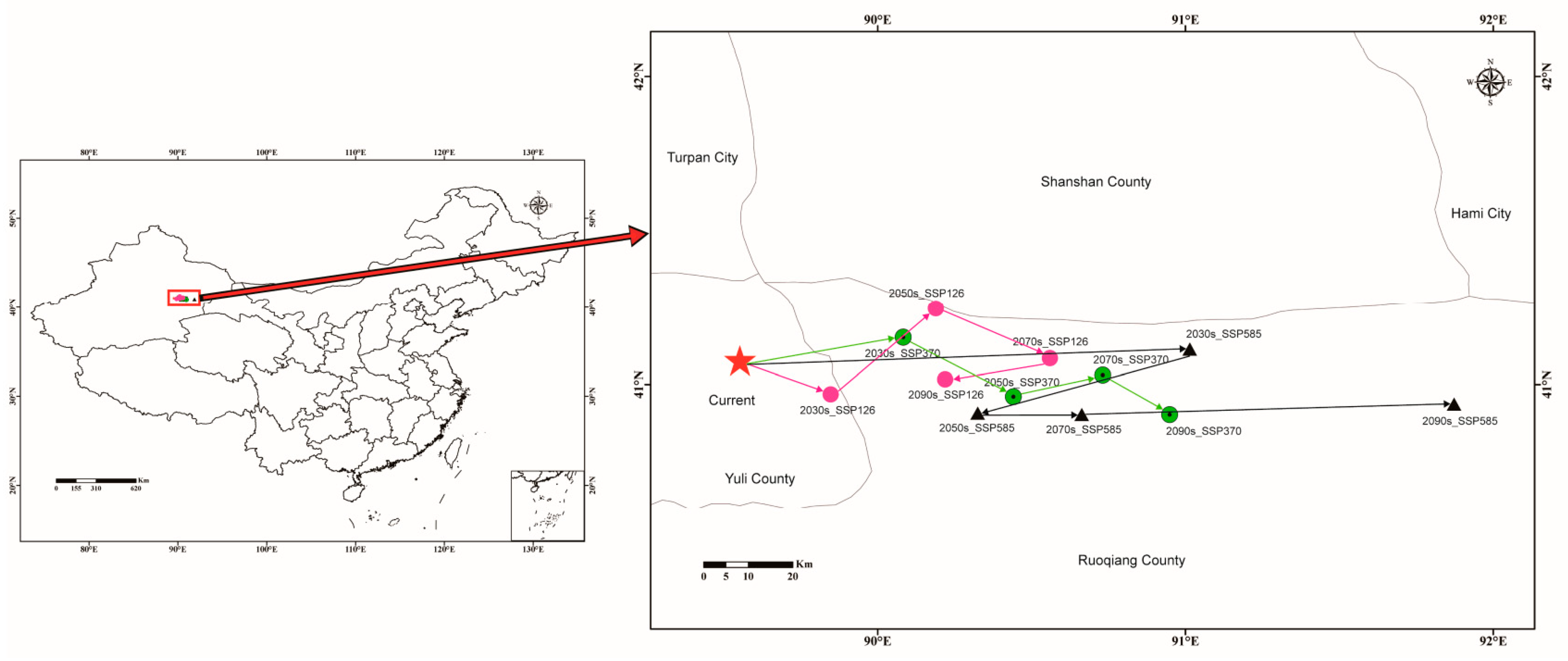
| Climate Scenario | Period | Centroid Coordinates | Direction | Migration Distance (between Two Adjacent Decades)/km | |
|---|---|---|---|---|---|
| Longitude /° E | Latitude /° N | ||||
| Current | 1970–2000 | 89.552582 | 41.076586 | ||
| SSP126 | 2021–2040/2030s | 89.847418 | 40.967573 | Southeast | 3.03 |
| 2041–2060/2050s | 90.188925 | 41.246166 | Northeast | 63.5 | |
| 2061–2080/2070s | 90.559061 | 41.08451 | Northeast | 97.1 | |
| 2081–2100/2090s | 90.218878 | 41.016473 | Northeast | 64.5 | |
| SSP370 | 2021–2040/2030s | 90.082805 | 41.152546 | Northeast | 51.6 |
| 2041–2060/2050s | 90.440685 | 40.959847 | Southeast | 86.4 | |
| 2061–2080/2070s | 90.73097 | 41.03039 | Northeast | 113.8 | |
| 2081–2100/2090s | 90.948488 | 40.901504 | Northeast | 135.7 | |
| SSP585 | 2021–2040/2030s | 91.013858 | 41.113851 | Northeast | 141 |
| 2041–2060/2050s | 90.324757 | 40.906896 | Northeast | 76.3 | |
| 2061–2080/2070s | 90.662871 | 40.90278 | Southeast | 108.4 | |
| 2081–2100/2090s | 91.872354 | 40.936859 | Southeast | 224.2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, C.; Hao, H.; Wang, Z.; Sha, S.; Zhang, Y.; Wang, Q.; Kang, Z.; Huang, L.; Wang, L.; Feng, H. Prediction of Suitable Habitat Distribution of Cryptosphaeria pullmanensis in the World and China under Climate Change. J. Fungi 2023, 9, 739. https://doi.org/10.3390/jof9070739
Yan C, Hao H, Wang Z, Sha S, Zhang Y, Wang Q, Kang Z, Huang L, Wang L, Feng H. Prediction of Suitable Habitat Distribution of Cryptosphaeria pullmanensis in the World and China under Climate Change. Journal of Fungi. 2023; 9(7):739. https://doi.org/10.3390/jof9070739
Chicago/Turabian StyleYan, Chengcai, Haiting Hao, Zhe Wang, Shuaishuai Sha, Yiwen Zhang, Qingpeng Wang, Zhensheng Kang, Lili Huang, Lan Wang, and Hongzu Feng. 2023. "Prediction of Suitable Habitat Distribution of Cryptosphaeria pullmanensis in the World and China under Climate Change" Journal of Fungi 9, no. 7: 739. https://doi.org/10.3390/jof9070739





