Abstract
Studies on the population structure and variation of Magnaporthe oryzae in fields are of great significance for the control of rice blast disease. In this study, a total of 462 isolates isolated from different areas of Hunan Province in 2016 and 2018 were analyzed for their population structure and variation tendency. The results showed that from 2016 to 2018, the concentration of fungal races of M. oryzae increased and the diversity decreased; furthermore, 218 isolates in 2016 belonged to ZA, ZB, ZC, ZE, ZF and ZG, with a total of 6 groups and 29 races, in which the dominant-population ZB group accounted for 66.2%; meanwhile, in 2018, 244 isolates were classified into 4 groups and 21 races, including ZA, ZB, ZC and ZG, in which the dominant-population ZB group accounted for 72.54%. In 2018, isolates of ZD, ZE and ZF populations were absent, and the number of total races and isolates of the ZA and ZC groups decreased. Fungal pathogenicity was identified, with 24 monogenic lines (MLs) carrying 24 major R genes. The resistance frequency of R genes to fungal isolates in 2018 decreased significantly, in which except Pikm was 64.5%, the other monogenic lines were less than 50%. Rep-PCR analysis for isolates of Guidong in Hunan also showed that fungal diversity decreased gradually. The influence of R genes on fungal variation was analyzed. The pathogenicity of isolates purified from Xiangwanxian 11 planted with monogenic lines was significantly more enhanced than those without monogenic lines. All the results indicated that in recent years, the fungal abundance in Hunan has decreased while fungal pathogenicity has increased significantly. This study will greatly benefit rice-resistance breeding and the control of rice blast disease in Hunan Province.
1. Introduction
Rice blast caused by Magnaporthe oryzae can lead to a serious loss in rice yield and quality on a worldwide scale. As the most economical and effective measure, the application of resistant varieties has been widely used to control rice blast disease [1]. Rice resistance is based on the interaction between the rice resistance gene (R) and relative fungal avirulence gene (AVR) [2,3]. Pathogens invade plants in various ways, and meanwhile, plants try their best to defend against the invasion of pathogens through related resistant mechanisms [4,5]. The population of M. oryzae in rice fields is richly diverse, and it possesses frequent variations to overcome rice resistance. The continuous wide promotion of one resistant variety can cause the rice blast fungus to evolve into new pathogenic types and can lead to the loss of rice resistance [6]; meanwhile, this will have a great impact on the variation of the population structure of rice blast fungus.
Hunan is at the forefront of rice production in China. However, in Hunan, lots of rice planting districts are located in mountains and hills, and furthermore, the continental subtropical monsoon climate often leads to plentiful rainfall and a high humidity in the nature environment. The climate and geographical environment are suitable for rice blast occurrence and spread and result in the threat of blast disease and a serious loss of rice yield every year (http://www.hnppi.cn/web/hnzbzj/9803/9804/9807/9808/content_335136.html (last accessed on 11 January 2023)) [7]. On the other hand, the abundant ecological rice planting districts and varieties in Hunan may force rice blast fungus to be more diverse than in other provinces [8]. The improvement of the resistance of rice varieties to prevent rice blast disease has been a common measure to assure rice-production security. The consistent monitoring of the fungal population’s structure and pathogenicity constitutes the fundamental research for the control and forecast of rice blast [9,10].
Race classification based on Chinese race differential varieties and pathogenicity assays on monogenic lines (MLs) carrying major R genes have been widely used to analyze the fungal population. With these research methods, it was found that isolates from Jiangsu and Liaoning were similar in terms of both the pathogenicity and genetic structures [11], and Pi9 and Piz5 were the most effective R genes for isolates in Heilongjiang province [12]. Researchers also analyzed the dominant races of rice blast fungus in Fujian and identified the resistant rice varieties through a combination of indoor seedling inoculation identification and molecular marker detection. Furthermore, they guided the promotion and distribution of varieties [13]. The molecular identity and pathogenicity of rice blast fungus in 2012 in Hunan have been characterized, and Pi9, Piz5, Pikh, and Pikm were found to be effective blast resistance genes in Hunan province [8]. These studies have set up a great foundation for the demonstration of the genetic background of rice blast fungus in different provinces of China.
As we know, the nature environment and rice resistance genes are fatal impact factors to fungal variation. In former research, weather conditions have shown obvious discrepancies among three different years in Liuyang district, and analysis on isolates of each year showed that both the fungal population structure and pathogenicity varied [14]. Following the application of abundant new rice varieties and the change of the local climate condition in Hunan, the variation of rice blast fungus in fields is not clear. In order to monitor the genetic variation of M. oryzae and early warnings of blast disease, isolates collected from various regions of Hunan Province in 2016 and 2018 were analyzed. The objectives of this study were to determine (i) the fungal population structure and distribution as well as the variation trend, (ii) the pathogenicity and variation, and (iii) the influence of R genes to fungal variation.
2. Material and Methods
2.1. Fungal Isolates and Rice Varieties
In this study, 218 isolates in 2016 and 244 isolates in 2018 collected from different ecological regions in Hunan Province were analyzed (Table S1). All dried panicle blast samples were sterilized with 70% ethyl alcohol for 10 s and rinsed with distilled water three times, and then immersed in distilled water for 2 h. The damp samples continued to lay on plastic plates with humidity at 27 °C for 24 h. Single spores were isolated under the microscope, then cultured on oatmeal agar, and finally stored on sterilized filter paper at −20 °C [8]. Regarding the name of stored isolates, “HN” represents Hunan, followed by the two digits of the year in which the samples were collected, and the left ones were the serial numbers.
The Chinese race differential varieties Tetep, Zhenlong 13, sifeng 43, Dongnong 363, Guandong 51, Hejiang 18 and Lijiangxintuan Heigu (LTH) were used for the classification of races [15]. Rice monogenic lines (MLs) were transferred into a single resistant gene with the background of LTH [16]. With LTH as negative control, 24 rice monogenic lines (MLs), respectively carrying R genes Pik, Piz, Pi11, Piz5, Pikh, Pi7, Pii, Piks, Pi20, Pikp, Pia, Pi3, Pizt, Pit, Pita, Pib, Pish, Pi1, Pi5, Pi9, Pi12, Pi19, Pikm, and Pita2, were used for the pathogenicity assay and resistance analysis of each major blast R gene [16]. In a greenhouse, ten seeds of each rice variety were planted and grown to the three- to four-leaf stage for pathogenicity assays.
2.2. Pathogenicity Assays
For pathogenicity assays, blast isolates were cultured on an oatmeal agar plate for 7 d with dark and white florescent light at 26 °C. Fungal spores were gathered with a 0.25% gelatin solution and filtrated with six layers of cheesecloth. The spore concentration was determined with a hemocytometer and adjusted to 2 × 105/mL for the final inoculation.
During inoculation, a 20 mL spore suspension was sprayed on about 350 rice seedlings at the three- to four-leaf stage. Inoculated plants were transferred into chambers with 95% humidity without light at 26 °C for 24 h and then kept with light for the next six days; then, the disease level was evaluated with a 0-to-5 scale rating system, where 0 to 2 indicated resistance (R) and 3 to 5 indicated susceptibility (S) [17]. Each pathogenicity assay was repeated three times to confirm the disease reactions. The higher disease level was used if ratings disagreed.
The race identity was determined on the basis of disease reactions on the Chinese differential cultivars [15]. A total of 462 isolates were classified into six groups: ZA, ZB, ZC, ZE, ZF, and ZG, in which Z represents Zhongguo (China), and A to G were the abbreviations for each Chinese differential line. In case A was susceptible, the isolate was classified as ZA group; if A was resistant and B was susceptible, then the isolate was classified as ZB group; a similar rule was compiled for the other groups. Pathogenicity assays were repeated once to confirm the disease reactions.
2.3. Fungal DNA Extraction and Rep-PCR Detection
For fungal genomic DNA extraction, the filter paper with purified fungus was cultured on oatmeal agar for 5~7 days, and mycelium blocks with a diameter of 0.5 cm were transferred to 50 mL liquid complete medium (CM) to be cultured in table concentrator at 28 °C and 160 RPM for 3 days in dark. The mycelium collected through a gauze filter was dried with filter paper and ground into powder with liquid nitrogen. The genomic DNA was extracted according to the method provided by the DNA fungus extraction kit (OMEGA Fungal Genomic Extraction Kit).
Rep-PCR primers (POT2-1 and POT2-2) were designed through the end reverse repeat sequence of Pot2 transposon as described by George et al. [18]. The primers were synthesized by Changsha Qingke Biotechnology Co., Ltd., (Changsha, China) Rep-PCR amplification products were detected by electrophoresis with 0.5% Agarose and 0.75% Synergel. Rep-PCR procedure was as follows: 1 cycle of 95 °C for 2.5 min; 3 cycles of 94 °C for 1 min, 62 °C for 1 min, and 65 °C for 10 min; 25 cycles of 94 °C for 30 s, 62 °C for 1 min, and 65 °C for 10 min; and 1 cycle of 65 °C for 15 min. Rep-PCR assay was repeated once to confirm the fungal genome patterns.
2.4. Effect of R Gene on Fungal Population
In order to analyze the effect of the resistance gene on the fungal population, Pi9 and Pizt monogenic lines and susceptible control—Xiangwanxian 11—were planted together in Guidong County, Hunan Province. Three independent experimental plots with a seedling density of 40 rows × 40 columns and 30 cm × 20 cm raw spacing were set up, respectively. A (control group): Xiangwanxian 11 was planted alone; B (Pi9 test group): Xiangwanxian 11 was planted in the middle of a plot with 5 rows × 20 columns surrounded with Pi9 monogenic line; C (Pizt test group): Xiangwanxian 11 was planted in the middle a plot with 5 rows × 20 columns surrounded with Pizt monogenic line. Seeds of rice materials were sowed at the end of May, and blast disease on rice were checked from the end of August. Samples of panicle blast of Xiangwanxian 11 from each plot were collected and purified with a single spore. For isolates from each plot, a race identification and pathogenicity assay on MLs were carried out to analyze the effect of Pi9 and Pizt on the fungal population.
3. Results
3.1. Race Identity
In order to understand the population structure of the isolates, fungal races were classified under seven Chinese differential varieties. The isolates in 2016 were classified into 29 races and 6 groups (ZA, ZB, ZC, ZE, ZF and ZG), in which ZB group was dominant, accounting for 65.6%, and ZB13 was the dominant race, accounting for 39.91%, followed by ZC15 with 10.55% and ZB15 with 9.63%, respectively (Table 1); isolates in 2018 were classified into 21 races and 4 groups (ZA, ZB, ZC and ZG), in which ZB group was dominant, accounting for 72.54%, and ZB13 was the dominant race, accounting for 45.49%, followed by ZA5 with 10.25%. The major variation in these two years was performed on the subdominant race, which changed from ZC15 in 2016 to ZA5 in 2018, and meanwhile, the category numbers of the fungal race and group also decreased from 2016 to 2018.

Table 1.
Race composition of Magnaporthe oryzae in 2016 and 2018.
3.2. Resistance Analysis of MLs
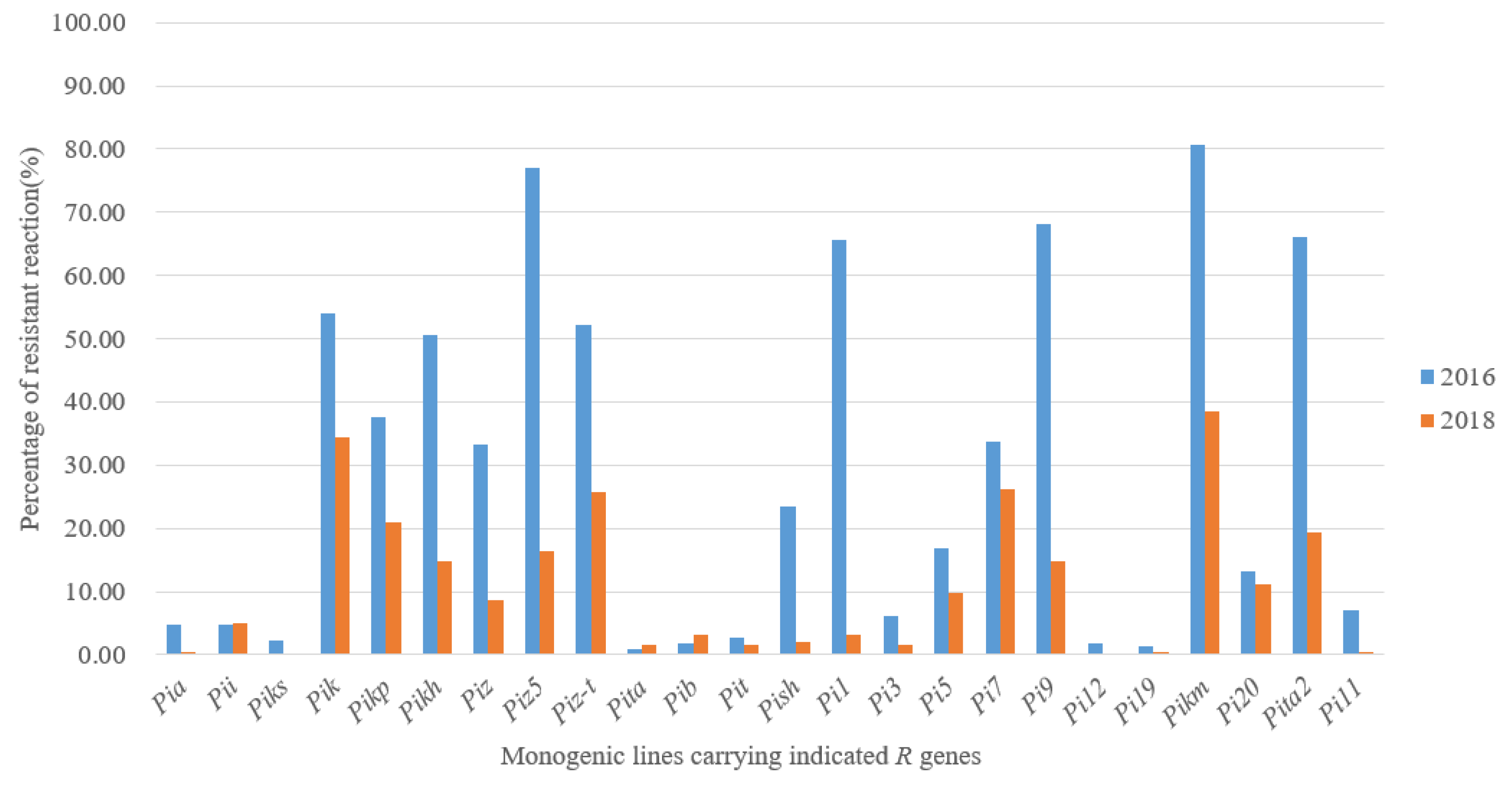
The results of pathogenicity determination on MLs are shown in Table S2. For the isolates in 2016, the resistance frequencies of Pikh, Pizt, Pik, Pi1, Pita2, Pi9, Piz5 and Pikm were over 50%, in which Pikm was the most effective, accounting for 80.75%. However, for isolates in 2018, even though Pikm was still the most effective, accounting for 64.5%, all the other MLs were less than 50%, and also more than half of the MLs were below 10% (Figure 1). The resistance frequency of different MLs varied greatly in recent years, especially the former highly resistant MLs. The resistance frequency of Pi9 dropped from 68.08% in 2016 to only 13.5% in 2018, and also Piz5 and Pizt were similar.
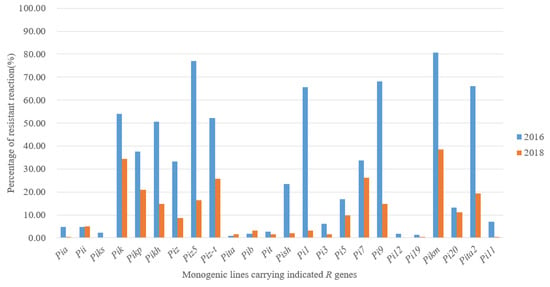
Figure 1.
The resistance frequency of MLs to Magnaporthe oryzae in 2016 and 2018 in Hunan.
3.3. Fungal Genetic Diversity with Rep-PCR
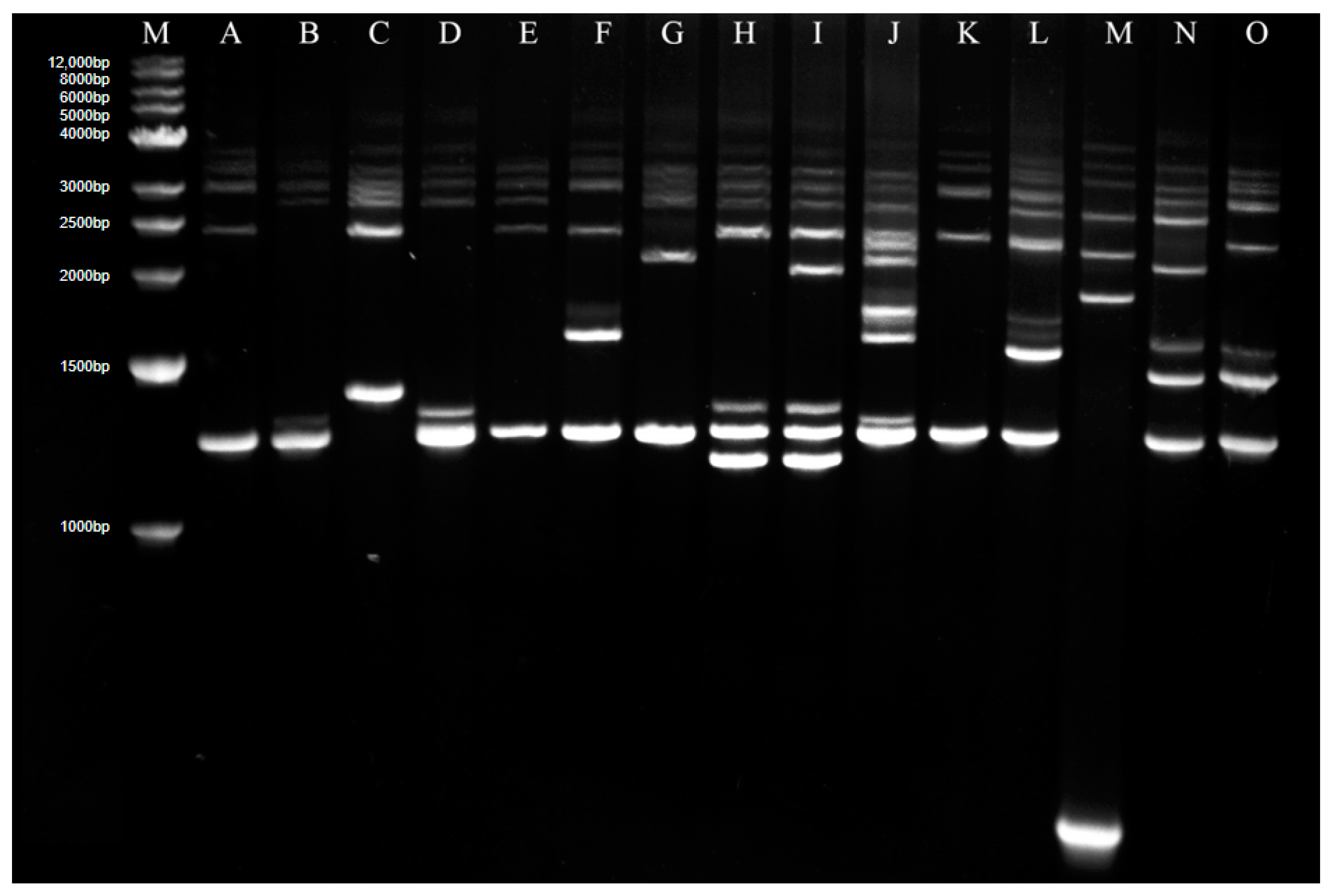
Genome analysis of 27 isolates in 2016 and 73 isolates in 2018 in Guidong County was carried out with Rep-PCR, and 15 Rep-PCR patterns marked as A to O were found in total (Figure 2), in which A type was dominant and accounted for 51%.
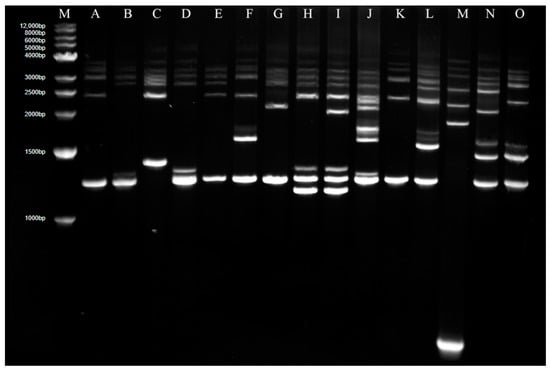
Figure 2.
Rep-PCR analysis for genomes of all isolates in Guidong County. M—Marker; A–O—Different Rep-PCR patterns.
For isolates in 2016, 9 Rep-PCR patterns, A, B, D, E, F, G, K, L and M, were found, in which the frequencies of F, G B, D, A, L and M were 18.52%, 18.52%, 14.8%,14.8%, 11.11%, 7.41% and 7.41%, respectively; both E and K patterns have only one isolate. For isolates in 2018, 8 patterns, A, C, F, H, I, J, N and O, were found, in which pattern A was absolutely dominant, accounting for 65.75%, followed by the J-banding type with 16.44%, while the other patterns were less than 5% (Table 2). The even distribution of different patterns for isolates in 2016 and highly intense distribution for isolates in 2018 suggested an obvious variation in the fungal genome and a decrease of fungal diversity in recent years.

Table 2.
Isolate numbers of different Rep-PCR patterns.
3.4. Variation in Fungal Population around Pizt and Pi9 MLs
Panicle blast samples were collected from susceptible variety Xiangwanxian 11 in different test groups, and each isolate was purified with a single spore. There were 21 isolates in group A (susceptible control group), 23 isolates in group B (Pi9 test group) and 40 isolates in group C (Pizt test group), respectively.
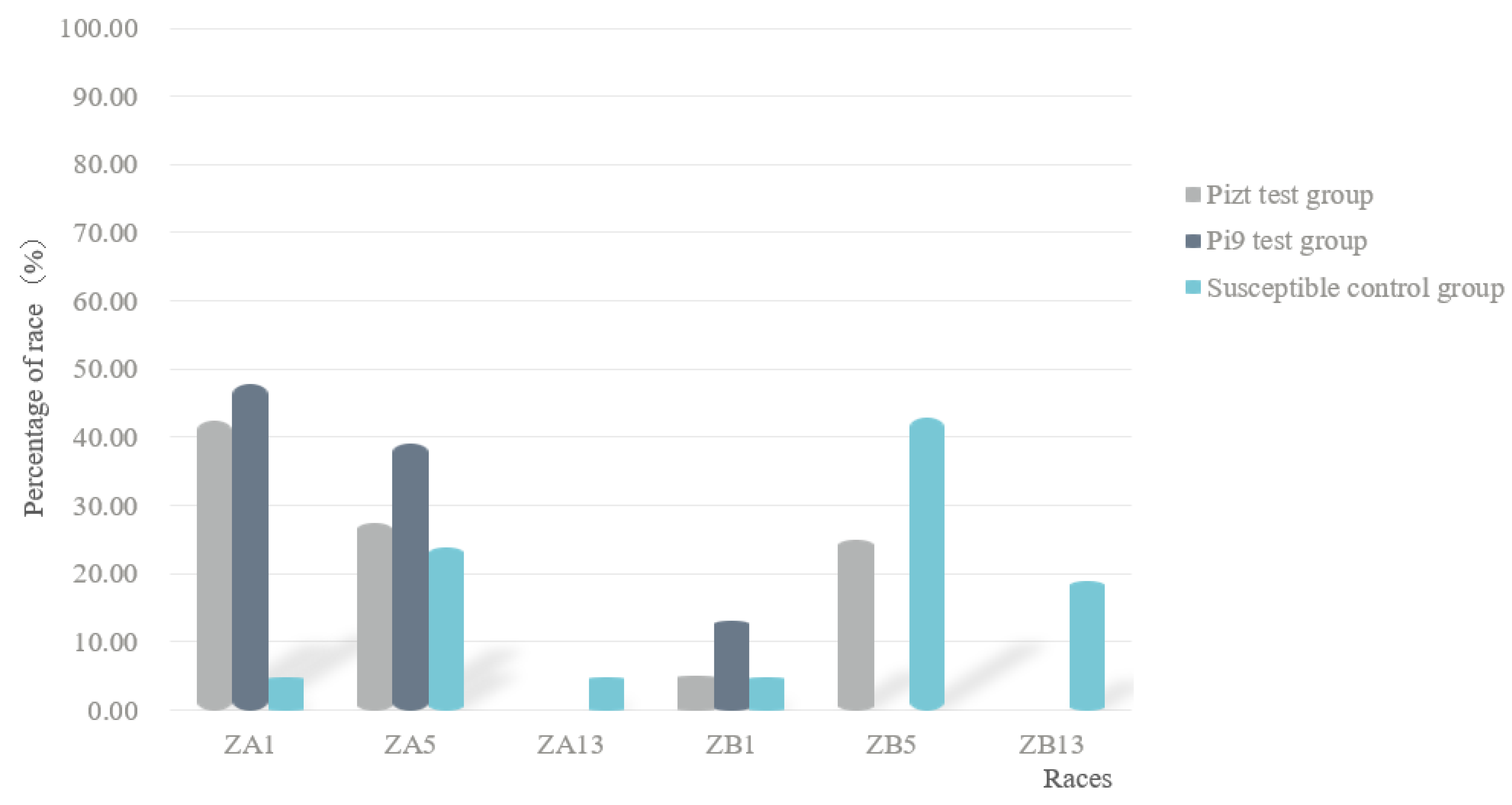
Race classification results showed that in group A, six races were found, in which ZB5 was dominant, accounting for 42.86%; in group B, three races were found, in which ZA1 was dominant, accounting for 47.83%; in group C, four races were found, in which ZA1 was dominant, accounting for 42.5% (Figure 3). These results showed that under the condition of the existence of MLs, the dominant race changed from ZB5 to ZA1, and the race number also decreased.
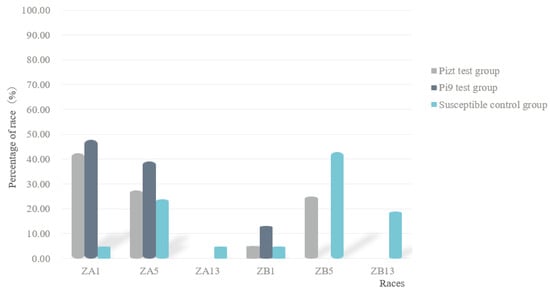
Figure 3.
Fungal race structures of control group and of Pizt and Pi9 test groups.
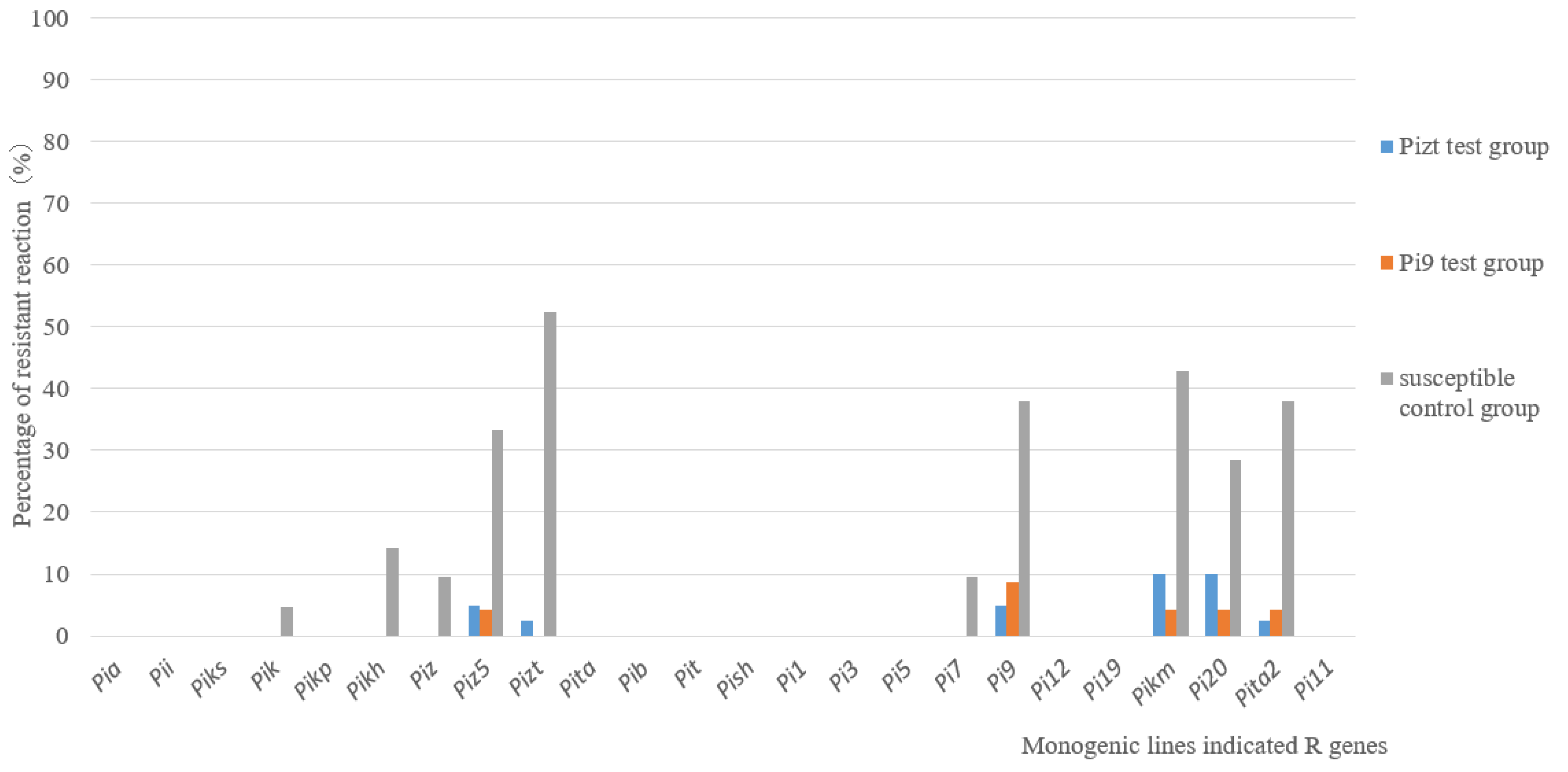
The pathogenicity assay on 24 MLs showed that there were also significant differences between the control group and test groups. For control group A, the resistance frequencies of MLs with Piz5, Pizt, Pi9, Pikm, Pi20 and Pita2 were 33.3%, 52.3%, 38%, 42.8%, 28.5% and 38%, respectively; however, for test groups B and C, the resistance frequency of all MLs was no more than 10% (Figure 4). The results suggested that the virulence of isolates from the test group was much stronger than that from the control group.
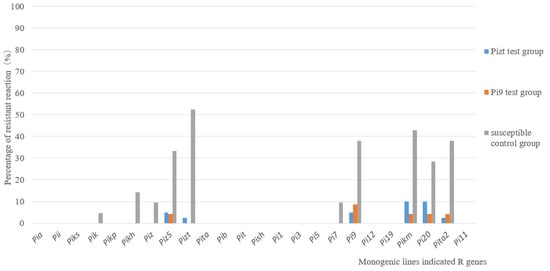
Figure 4.
Pathogenicity assays on MLs for isolates from control group and from Pizt and Pi9 test groups.
4. Discussion
In general, the interaction between M. oryzae and rice in nature maintains a dynamic equilibrium. In order to break through the defense of rice resistance genes, rice blast fungus will undergo different kinds of mutations, such as gene deletion, nucleotide substitution and transposon insertion [19,20]. Consistent monitoring of the rice blast fungus population in the field can clarify the distribution of prevalent isolates in different ecological areas and contribute to early warning, and it is also important for the rational application of rice varieties in order to control rice blast disease [21]. In this study, we identified the variation of the population structure and pathogenicity of rice blast fungus in Hunan Province in recent years and the effects of R gene on fungal variation. These results suggested that rice blast fungus went through significant changes in pathogenicity and genetic identity, and it will lead to a huge risk for rice production safety in the future.
The population structure of M. oryzae varied in different ecological regions and showed a dynamic development in different years. Fungal races and population analysis based on the Chinese differential system has been widely used [22,23]. In Hunan, 182 isolates in 2012 were found to belong to 6 groups and 28 races, and the dominant race had changed from ZG1 to ZB13 [8]. The consistent analysis on isolates in 2016 and 2018 showed that the dominant race and group was still the ZB13 and ZB groups, but the fungal population composition varied; particularly in 2018 it showed a significant increase of ZA group and decrease of ZC group, and the numbers of fungal group categories also diminished. The whole fungal population in 2018 got more concentrated, and the diversity was reduced. Meanwhile, the genetic background of the fungal population was also analyzed. The genetic polymorphism of M.oryzae is an important indicator for fungal population diversity. Different molecular marker methods, such as the restriction fragment length polymorphism (RFLP) method [24], randomly amplified polymorphic DNA (RAPD) technology, simple repeat sequence (SSR) labeling [25], Rep-PCR technology [17], and amplified fragment length polymorphism (AFLP), were widely used. In this study, Rep-PCR analysis was performed on isolates from Guidong County in 2016 and 2018. In 2016, nine types of Rep-PCR patterns were found and showed a uniform distribution, in which the highest proportion was 18.52%; in 2018, eight types were found, in which pattern A was dominant, accounting for 65.75%, and the other patterns were less than 17%. The isolates with the same genetic background got more concentrated from 2016 to 2018, and this was consistent with the conclusion on variations of race and population structure in this study. The change in fungal pathogenicity performance was direct evidence of fungal variation. For isolates in 2012, the resistance frequency of Pi9, Piz5, Pikh and Pikm was over 85% [8]. Even though most of these four major genes kept a relatively high resistance in 2016, in 2018, only the resistance frequency of Pikm exceeded 60%, and the other three R genes showed a precipitous decline. These results suggested that rice blast fungi in Hunan have undergone a distinct alteration and also provided a fatal warning for rice breeders.
The natural environment and resistant (R) genes in rice varieties with a large planting scale are the important factors affecting the population variation of M. oryzae [14,26]. There was no significant change for climates in Guidong County between 2016 and 2018. Therefore, R genes will be the core effect factor. In order to confirm the effect of R gene, isolates from a susceptible variety (LTH) with and without Pizt or Pi9-ML planted in high blast risk regions in Guidong were analyzed, respectively. The isolates from LTH alone performed with more diversity than those from LTH surrounded with R gene MLs. These results proved that R genes could affect the variation of the fungal population. Furthermore, the resistance frequencies of Piz5, Pizt, Pi9, Pikm, Pi20 and Pita2 to isolates from LTH alone ranged from 28.5% to 52.3%, but for isolates from the Pi9 or Pizt test groups, they were no higher than 10%. This phenomenon is not only found in rice blast disease but also in wheat stripe rust [27].
Following the attention that has been focused on the damage from rice blast fungus, blast resistance has been the critical index for the authorization of a new rice variety. Hence, many resistant varieties have been promoted to farmers. Huazhan containing the major resistant gene Pi2 proved to be an excellent resistant parent resource for hybrid rice breeding [28]. In recent years, the distribution data of main rice varieties in Hunan has shown that Huazhan series rice varieties accounted for more than 12% of middle and late rice from 2013 to 2017 [29], and the approved varieties of the Huazhan series also showed a gradual increase. As we know, the natural environment and resistant varieties were the major factors to induce fungal variation. In recent years, there was no drastic climate change in Hunan. Hence, the present situation in rice production may explain the variation in blast fungus. It is known that the promotion and planting of diversified rice varieties can help prevent and control the occurrence of rice blast [30]. Different control measures should be considered in response to fugal variation. In any case, we must pay great attention to the risk of the large-scale planting of varieties related to Huazhan, and we must also address the variation in blast fungus and breed new resistant varieties in advance.
5. Conclusions
As we all know, monitoring the structure and variation trend of rice blast fungus flora in rice planting areas is critical work for rice breeding. In the present study, we analyzed the fungal population structure and variation tendency of 462 isolates from different districts of Hunan Province in 2016 and 2018, showed the fungal variation through race population, genetic background, pathogenicity, and identified the influence of R genes on the fungal population. The results showed that from 2016 to 2018, rice blast fungus got more concentrated and its diversity decreased; additionally, the resistance frequency of R genes to fungal isolates decreased significantly in 2018, and fungus showed a stronger pathogenicity. In addition, Rep-PCR analysis of isolates from Guidong district in Hunan also showed that fungal diversity gradually decreased. Analysis on the influence of R genes on fungal variation showed that under the influence of MLs with R gene, the fungal dominant race varied and race number was reduced and also that fungal pathogenicity got stronger. This research is tightly related to the actual rice production situation, provides critical guidance for future rice breeding and contributes to the early warning of the potential risk of blast disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof9070776/s1.
Author Contributions
Conceptualization, Z.P., J.X. (Junjie Xing), Y.F. and H.D.; methodology, Z.P., F.W., J.X. (Junjie Xing), Y.F. and Q.L.; formal analysis, Y.L., Z.Z., L.Y., X.-L.C. and J.X. (Jingbo Xu); investigation, Q.L., Z.P., Y.F. and H.D.; writing—original draft preparation, Z.P., J.X. (Junjie Xing), Y.F. and H.D.; writing—review and editing, Z.P., J.X. (Junjie Xing), J.X. (Jingbo Xu) and H.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32071937), Natural Science Foundation of Hunan Province (2021JJ30016).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Silué, D.; Notteghem, J.L.; Tharreau, D. Evidence of a Gene-for-Gene Relationship in the Oryza sativa-Magnaporthe grisea Pathosystem. Phytopathology 1992, 82, 577–580. [Google Scholar] [CrossRef]
- Cesari, S.; Thilliez, G.; Ribot, C.; Chalvon, V.; Michel, C.; Jauneau, A.; Rivas, S.; Alaux, L.; Kanzaki, H.; Okuyama, Y.; et al. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 2013, 25, 1463–1481. [Google Scholar] [CrossRef]
- Jia, Y.; McAdams, S.A.; Bryan, G.T.; Hershey, H.P.; Valent, B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000, 19, 4004–4014. [Google Scholar] [CrossRef]
- Osés-Ruiz, M.; Cruz-Mireles, N.; Martin-Urdiroz, M. Appressorium-mediated plant infection by Magnaporthe oryzae is regulated by a Pmk1-dependent hierarchical transcriptional network. Nat. Microbiol. 2021, 6, 1383–1397. [Google Scholar] [CrossRef]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Xiao, D.F.; Zhang, P.S.; Wang, L.; Huang, S.W. Research Progress on Populations and Physiological Race Distribution of Rice Blast Pathogen(Magnaporthe grisea) in China. Chin. J. Rice Sci. 2013, 27, 312–320. [Google Scholar]
- Chen, Z.F.; Jin, M.N.; Liu, J.C.H.; Li, Z.X.; Qiu, S.J.; Zhou, X.Y.; Tang, X.Y. Development of a HRM-based Functional Marker for Rice Blast Resistance Gene Pi2. Mol. Plant Breed 2017, 15, 938–943. [Google Scholar]
- Xing, J.; Jia, Y.; Peng, Z.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Zhang, Z.; Deng, H. Characterization of Molecular Identity and Pathogenicity of Rice Blast Fungus in Hunan Province of China. Plant Dis. 2017, 101, 557–561. [Google Scholar] [CrossRef]
- Peng, Z.; Li, L.; Wu, S.; Chen, X.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Sun, P.; Deng, H.; et al. Frequencies and Variations of Magnaporthe oryzae Avirulence Genes in Hunan Province, China. Plant Dis. 2021, 105, 3829–3834. [Google Scholar] [CrossRef]
- Kankanala, P.; Czymmek, K.; Valent, B. Roles for Rice Membrane Dynamics and Plasmodesmata during Biotrophic Invasion by the Blast Fungus. Plant Cell 2007, 19, 706–724. [Google Scholar] [CrossRef]
- Shi, M.; Liu, Z.; Chen, Y.; Tian, A.; Yao, L.; Ren, C.; Miao, Q.; Yu, H. Population diversity of Magnaporthe oryzae of Jiangsu and Liaoning japonica rice area. Acta Phytopathol. Sin. 2015, 45, 158–166. [Google Scholar]
- Ma, J.; Zhang, G.; Xin, A.; Zhang, L.; Deng, L.; Wang, Y.; Wang, Y.; Ren, Y.; Gong, X.; Ge, X.; et al. Comparison of Pathogenicity of Pyricularia oryzae under Different Genetic Backgrounds. Acta Agron. Sin. 2015, 41, 1791–1801. [Google Scholar] [CrossRef]
- Ruan, H.C.; Shi, N.N.; Du, Y.X.; Gan, L.; Yang, X.J.; Dai, Y.L.; Chen, F.R. Analysis on Resistance of Pi Genes to Predominant Races of Mangnaporthe oryzae in Fujian Province, China. Chin. J. Rice Sci. 2017, 31, 105–110. [Google Scholar]
- Peng, Z.; Liu, Y.; He, Q.; Tan, Y.; Pei, Y.; Deng, H.; Xing, J. Population diversity and variation of the rice blast fungus under different climate conditions. Eur. J. Plant Pathol. 2019, 155, 881–889. [Google Scholar] [CrossRef]
- National Joint Test Group on physiological races of Magnaporthe grisea. Research on physiological races of rice blast fungus in China. Acta Phytopathol. Sin. 1980, 10, 71–82. [Google Scholar]
- Tsunematsu, H.; Yanoria, M.J.; Ebron, L.; Hayashi, N.; Ando, I.; Kato, H.; Imbe, T.; Khush, G. Development of Monogenic Lines of Rice for Blast Resistance. Breed Sci. 2000, 50, 229–234. [Google Scholar] [CrossRef]
- Xing, J.; Jia, Y.; Correll, J.C.; Lee, F.N.; Cartwright, R.; Cao, M.; Yuan, L. Analysis of Genetic and Molecular Identity Among Field Isolates of the Rice Blast Fungus with an International Differential System, Rep-PCR, and DNA Sequencing. Plant Dis. 2013, 97, 491–495. [Google Scholar] [CrossRef]
- George, M.L.; Nelson, R.J.; Zeigler, R.S.; Leung, H. Rapid Population Analysis of Magnaporthe grisea by Using rep-PCR and Endogenous Repetitive DNA Sequences. Phytopathology 1998, 88, 223–229. [Google Scholar] [CrossRef]
- Valent, B.; Khang, C.H. Recent advances in rice blast effector research. Curr. Opin. Plant. Biol. 2010, 13, 434–441. [Google Scholar] [CrossRef]
- Dai, Y.; Jia, Y.; Correll, J.; Wang, X.; Wang, Y. Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe Oryzae. Fungal Genet. Biol. 2010, 47, 973–980. [Google Scholar] [CrossRef]
- Yang, Y.; Chu, J.; He, P.F.; Wu, Y.X.; Yi, J.H.; He, Y.Q. Dynamic Change of Avirulence AvrPi9 of Magnaporthe oryzae in Two Rice Blast Nurseries. Chin. Agric. Sci. Bull. 2016, 32, 195–199. [Google Scholar]
- Liu, W.; Wei, S.H.; Zhu, L.j.; Wang, H.N.; Zhang, Z.R.; Li, X.Y. Identification of Physiological Races and Avirulence Genes of Pyricularia oryzae in Liaoning Province. Southwest China J. Agric. Sci. 2020, 33, 1969–1976. [Google Scholar]
- Deng, Y.; Tian, D.G.; Su, Y.; Zhang, J.W.; Wu, J.W. Physiological Races of Magnaporthe grisea and Disease-Resistant Rice in Fujian. Fujian J. Agric. Sci. 2020, 35, 1101–1110. [Google Scholar]
- Farman, M.L.; Kim, Y.S. Telomere hypervariability in Magnaporthe Oryzae. Mol. Plant Pathol. 2005, 6, 287–298. [Google Scholar] [CrossRef]
- Xie, J.J.; Jiang, H.; Mao, X.Q.; Chai, R.Y.; Qiu, H.P.; Zhang, Z.; Wang, Y.L.; Wang, J.Y.; Du, X.F.; Sun, G.C. Analysis of genetic diversity of Magnaporthe oryzae isolates from Zhejiang Province based on SSR markers. Acta Agric. Zhejiangensis 2015, 27, 1781–1788. [Google Scholar]
- Khang, C.H.; Park, S.Y.; Lee, Y.H.; Valent, B.; Kang, S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant Microb. Interact. 2008, 21, 658–670. [Google Scholar] [CrossRef]
- Zhang, Y.; Chao, K.X.; Gao, X.; Liu, Z.G.; Yao, W.Y.; Li, Q.; Wang, B.T. Genetic analysis and SSR markers of wheat stripe rust resistance gene YrElm derived from Elymus mollis (Trin.) Hara. Acta Phytopathol. Sin. 2014, 44, 641–650. [Google Scholar]
- Chen, S.; Su, J.; Hua, L.X.; Wang, W.J.; Wang, C.Y.; Yang, J.Y.; Zeng, L.X.; Zhu, X.Y. Genetic analysis and gene identification of restorer line Huazhan against rice blast. Acta Phytopathol. Sin. 2015, 45, 598–605. [Google Scholar]
- Deng, W.; Tan, J.Y.; Liu, B.; Zhu, Y.L.; Wang, J. Study on Distribution of Main Rice Varieties in Hunan. Hunan Agric. Sci. 2019, 402, 85–88. [Google Scholar]
- Zhu, Y.; Chen, H.; Fan, J.; Wang, Y.; Li, Y.; Chen, J.; Fan, J.; Yang, S.; Hu, L.; Leung, H.; et al. Genetic diversity and disease control in rice. Nature 2000, 406, 718–722. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).