Ena Proteins Respond to PacC-Mediated pH Signaling Pathway and Play a Crucial Role in Patulin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Phylogenetic Relationships and Conserved Domain Analysis
2.3. Mutant Generation and Complementation
2.4. Chromatin Immunoprecipitation Assay
2.5. Phenotypic Analysis
2.6. Reverse Transcription and Quantitative PCR Analysis
2.7. Membrane Potential Assay
2.8. Subcellular Localization of EnaC
2.9. Statistical Analysis
3. Results
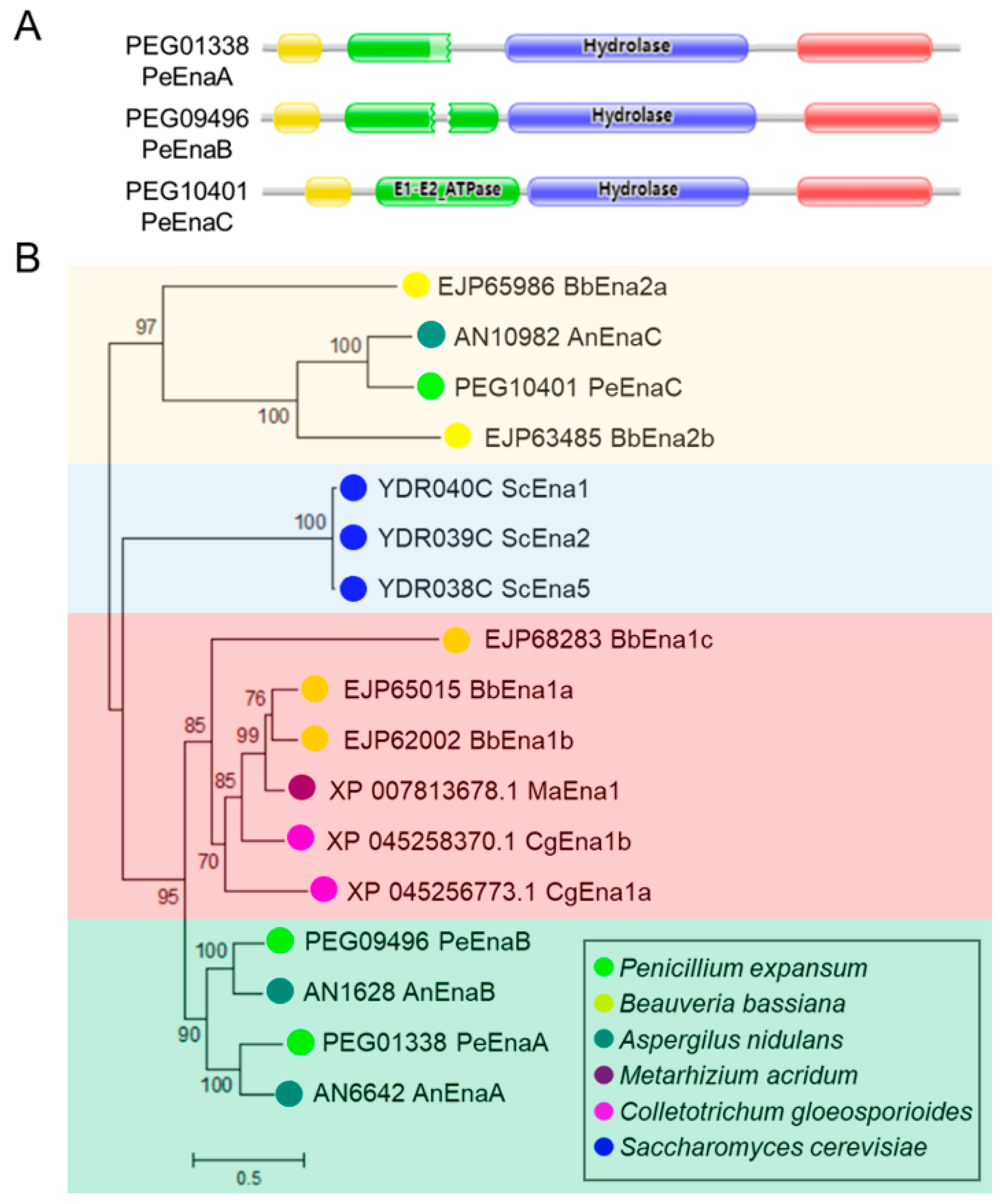
3.1. Sequence Features of Ena ATPases in P. expansum
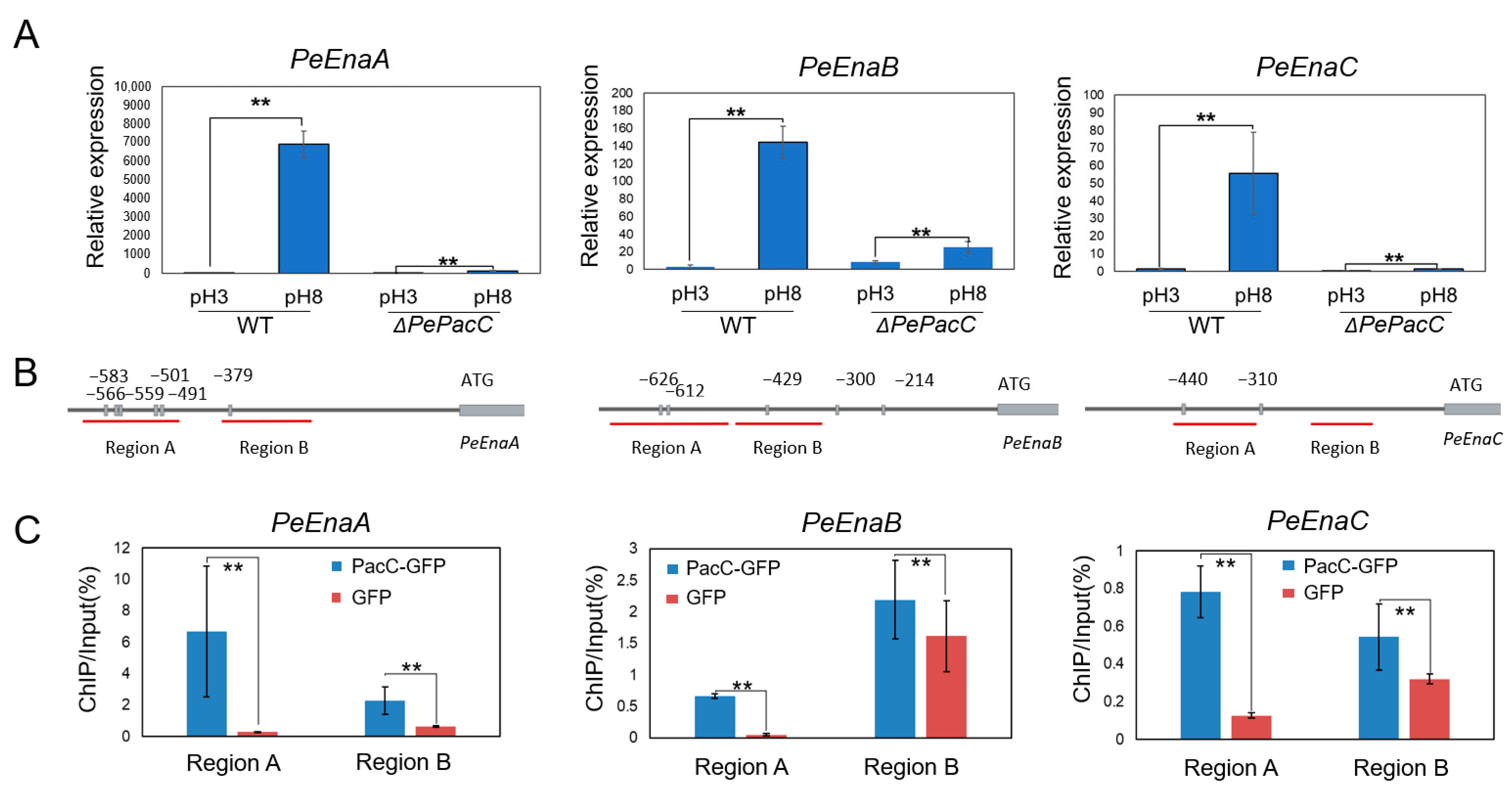
3.2. PePacC Directly Binds to the Promoter Regions of PeEnas and Activates Transcription
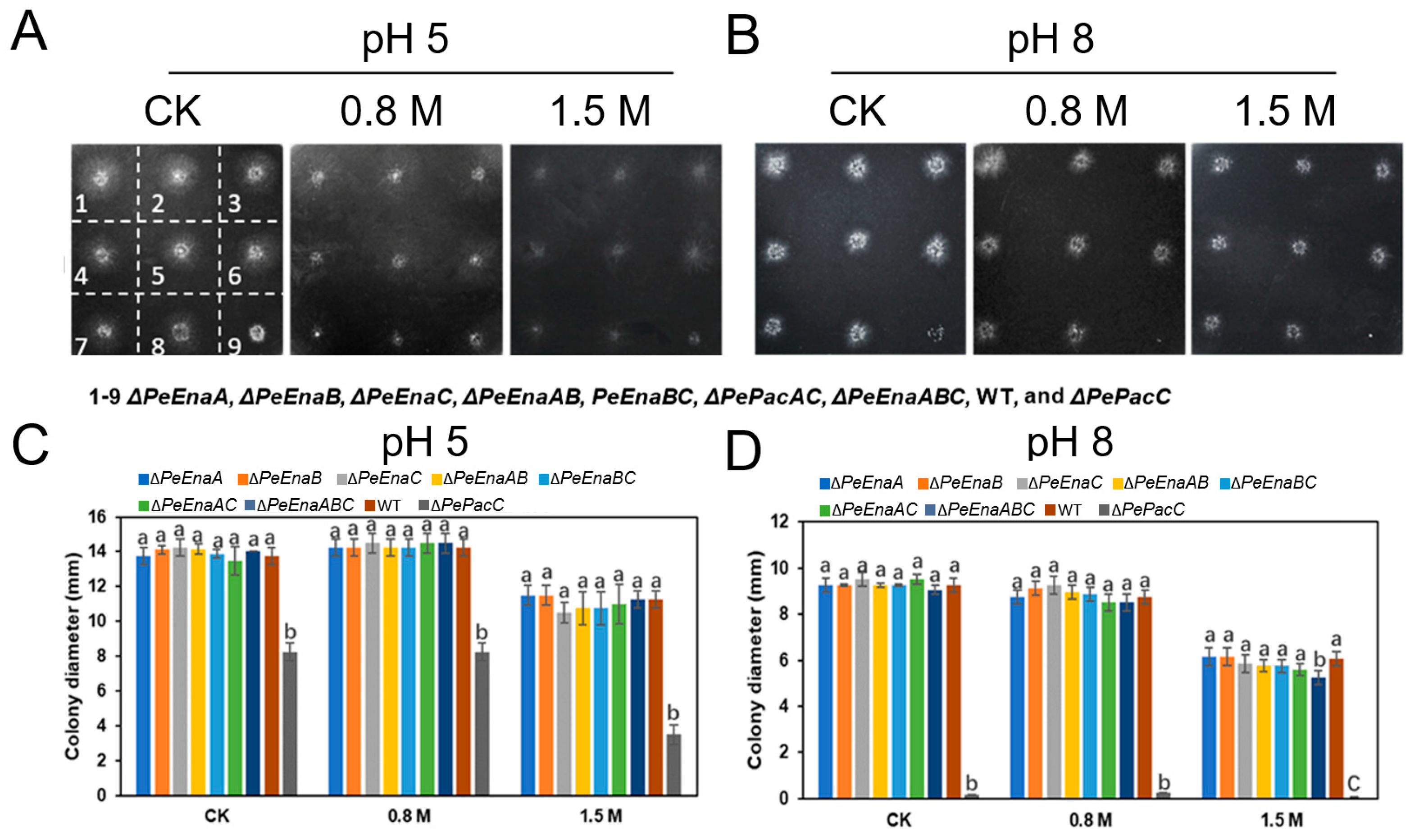
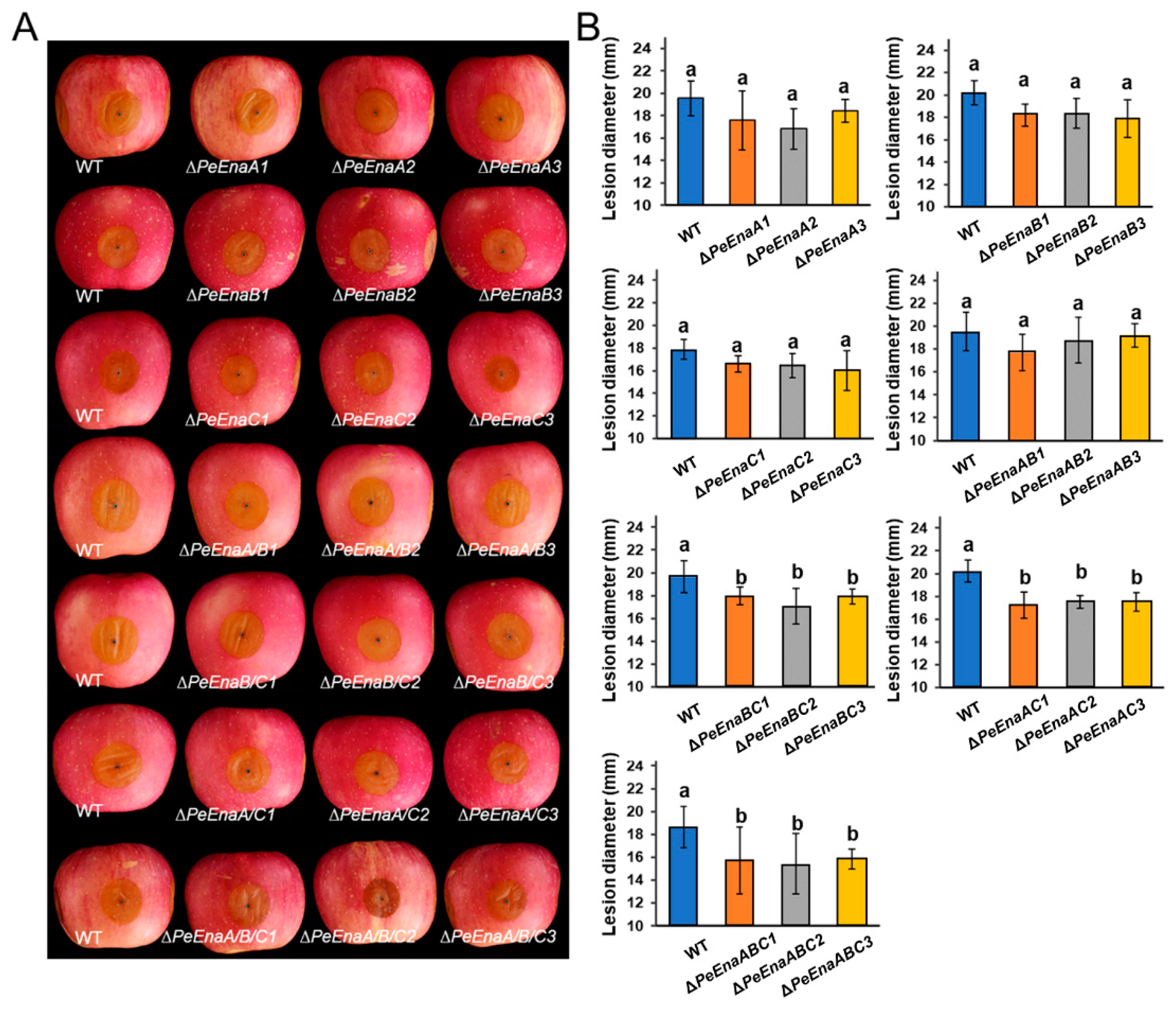
3.3. PeEnas Are Involved in the Growth and Virulence of P. expansum
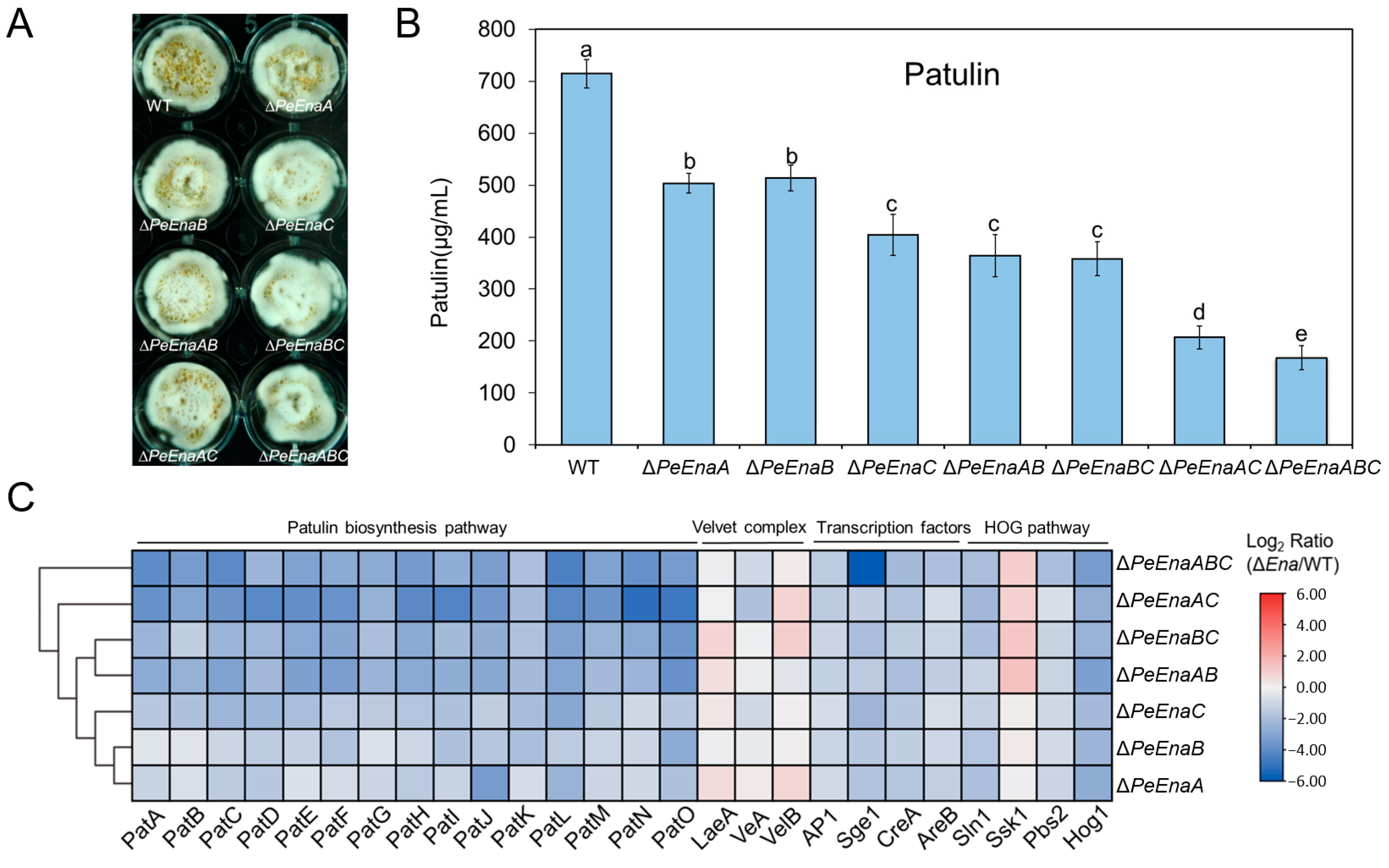
3.4. PeEnas Affect Patulin Biosynthesis in P. expansum
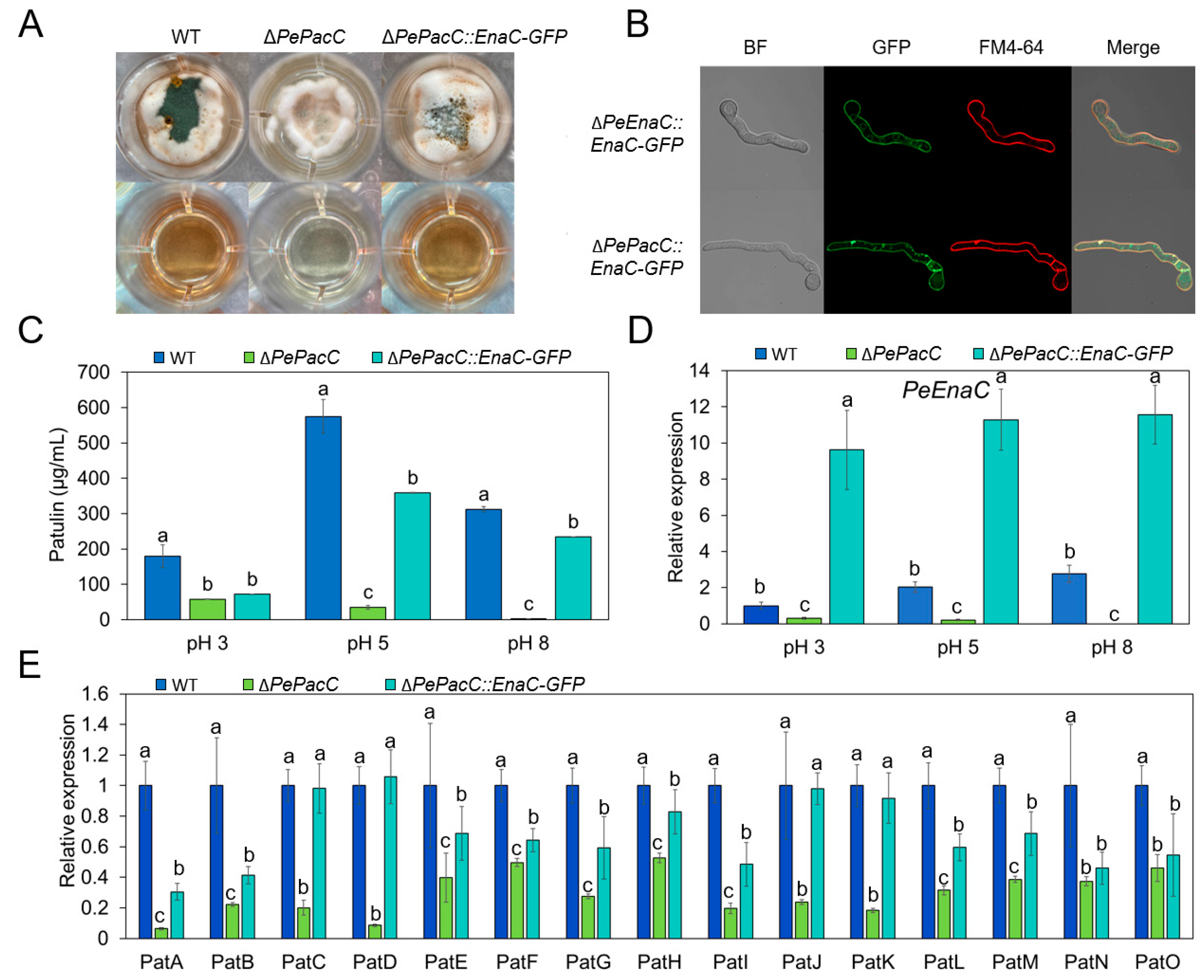
3.5. PeEnaC Rescues the Defective of Patulin Biosynthesis in ΔPePacC
3.6. PeEnas Involve in Maintaining Membrane Potential in P. expansum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, B.; Chen, Y.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular basis and regulation of pathogenicity and patulin biosynthesis in Penicillium expansum. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3416–3438. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, Y.; Zong, Y.; Shang, Y.; Zhang, Z.; Xu, X.; Wang, X.; Long, M.; Tian, S. Dissection of patulin biosynthesis, spatial control and regulation mechanism in Penicillium expansum. Environ. Microbiol. 2019, 21, 1124–1139. [Google Scholar] [CrossRef]
- Barad, S.; Sela, N.; Kumar, D.; Kumar-Dubey, A.; Glam-Matana, N.; Sherman, A.; Prusky, D. Fungal and host transcriptome analysis of pH-regulated genes during colonization of apple fruits by Penicillium expansum. BMC Genom. 2016, 17, 330. [Google Scholar] [CrossRef]
- Tannous, J.; Atoui, A.; El Khoury, A.; Francis, Z.; Oswald, I.P.; Puel, O.; Lteif, R. A study on the physicochemical parameters for Penicillium expansum growth and patulin production: Effect of temperature, pH, and water activity. Food Sci. Nutr. 2016, 4, 611–622. [Google Scholar] [CrossRef]
- Penalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N., Jr. Ambient pH gene regulation in fungi: Making connections. Trends Microbiol. 2008, 16, 291–300. [Google Scholar] [CrossRef]
- Diez, E.; Alvaro, J.; Espeso, E.A.; Rainbow, L.; Suarez, T.; Tilburn, J.; Arst, H.N., Jr.; Penalva, M.A. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 2002, 21, 1350–1359. [Google Scholar] [CrossRef]
- Rollins, J.A.; Dickman, M.B. pH signaling in Sclerotinia sclerotiorum: Identification of a pacC/RIM1 homolog. Appl. Environ. Microbiol. 2001, 67, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Chen, Y.; Tian, S. Function of pH-dependent transcription factor PacC in regulating development, pathogenicity, and mycotoxin biosynthesis of phytopathogenic fungi. FEBS J. 2021, 289, 1723–1730. [Google Scholar] [CrossRef]
- Chen, Y.; Li, B.; Xu, X.; Zhang, Z.; Tian, S. The pH-responsive PacC transcription factor plays pivotal roles in virulence and patulin biosynthesis in Penicillium expansum. Environ. Microbiol. 2018, 20, 4063–4078. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, Z.; Tian, S.; Li, B. Application of -omic technologies in postharvest pathology: Recent advances and perspectives. Curr. Opin. Food Sci. 2022, 45, 100820. [Google Scholar] [CrossRef]
- Zhuo, R.; Li, G.; Peng, H.; Zong, Y.; Wang, X.; Lu, S.; Chen, Y.; Zhang, Z.; Tian, S.; Li, B. Extensive regulation of pH-responsive transcription factor PacC on secondary metabolism contributes to development and virulence of Botrytis cinerea. Postharvest Biol. Technol. 2023, 197, 112219. [Google Scholar] [CrossRef]
- Lamb, T.M.; Mitchell, A.P. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003, 23, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Serra-Cardona, A.; Petrezselyova, S.; Canadell, D.; Ramos, J.; Arino, J. Coregulated expression of the Na+/phosphate Pho89 transporter and Ena1 Na+-ATPase allows their functional coupling under high-pH stress. Mol. Cell. Biol. 2014, 34, 4420–4435. [Google Scholar] [CrossRef]
- Ma, Q.; Jin, K.; Peng, G.; Xia, Y. An ENA ATPase, MaENA1, of Metarhizium acridum influences the Na+-, thermo- and UV-tolerances of conidia and is involved in multiple mechanisms of stress tolerance. Fungal Genet. Biol. 2015, 83, 68–77. [Google Scholar] [CrossRef]
- Dyla, M.; Kjaergaard, M.; Poulsen, H.; Nissen, P. Structure and mechanism of P-Type ATPase ion pumps. Annu. Rev. Biochem. 2020, 89, 583–603. [Google Scholar] [CrossRef]
- Palmgren, M.G.; Nissen, P. P-type ATPases. Annu. Rev. Biophys. 2011, 40, 243–266. [Google Scholar] [CrossRef]
- Arino, J.; Ramos, J.; Sychrova, H. Monovalent cation transporters at the plasma membrane in yeasts. Yeast 2019, 36, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Arino, J.; Ramos, J.; Sychrova, H. Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 2010, 74, 95–120. [Google Scholar] [CrossRef] [PubMed]
- Garciadeblas, B.; Rubio, F.; Quintero, F.J.; Banuelos, M.A.; Haro, R.; Rodrigueznavarro, A. Differential expression of two genes encoding isoforms of the ATPase involved in sodium effiux in Saccharomyces cerevisiae. Mol. Genet. Genom. 1993, 236, 363–368. [Google Scholar] [CrossRef]
- Markina-Inarrairaegui, A.; Spielvogel, A.; Etxebeste, O.; Ugalde, U.; Espeso, E.A. Tolerance to alkaline ambient pH in Aspergillus nidulans depends on the activity of ENA proteins. Sci. Rep. 2020, 10, 14325. [Google Scholar] [CrossRef] [PubMed]
- Platara, M.; Ruiz, A.; Serrano, R.; Palomino, A.; Moreno, F.; Arino, J. The transcriptional response of the yeast Na+-ATPase ENA1 gene to alkaline stress involves three main signaling pathways. J. Biol. Chem. 2006, 281, 36632–36642. [Google Scholar] [CrossRef] [PubMed]
- Idnurm, A.; Walton, F.J.; Floyd, A.; Reedy, J.L.; Heitman, J. Identification of ENA1 as a virulence gene of the human pathogenic fungus Cryptococcus neoformans through signature-tagged insertional mutagenesis. Eukaryot. Cell 2009, 8, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Mou, Y.N.; Gao, B.J.; Ren, K.; Tong, S.M.; Ying, S.H.; Feng, M.G. P-type Na+/K+ ATPases essential and nonessential for cellular homeostasis and insect pathogenicity of Beauveria bassiana. Virulence 2020, 11, 1415–1431. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zong, Y.; Du, Z.; Chen, Y.; Zhang, Z.; Qin, G.; Zhao, W.; Tian, S. Genomic characterization reveals insights into patulin biosynthesis and pathogenicity in Penicillium species. Mol. Plant Microbe Interact. 2015, 28, 635–647. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Li, B.; Tian, S. Histone H3K4 methyltransferase PeSet1 regulates colonization, patulin biosynthesis, and stress responses of Penicillium expansum. Microbiol. Spectr. 2023, 11, e0354522. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Wang, Y.; Zhao, X.; Yuan, B.; Zhang, M.; Tan, Z.; Zhang, X.; Chen, Y.; Wu, H.; Luo, Y.; et al. Inhibition of histone acetyltransferase GCN5 by a transcription factor FgPacC controls fungal adaption to host-derived iron stress. Nucleic Acids Res. 2022, 50, 6190–6210. [Google Scholar] [CrossRef]
- Wang, W.; Wang, P.; Li, X.; Wang, Y.; Tian, S.; Qin, G. The transcription factor SlHY5 regulates the ripening of tomato fruit at both the transcriptional and translational levels. Hort. Res. 2021, 8, 83. [Google Scholar] [CrossRef]
- Veerana, M.; Yu, N.N.; Bae, S.J.; Kim, I.; Kim, E.S.; Ketya, W.; Lee, H.Y.; Kim, N.Y.; Park, G. Enhancement of fungal enzyme production by radio-frequency electromagnetic fields. J. Fungi 2022, 8, 1187. [Google Scholar] [CrossRef]
- Molina-Hernandez, J.B.; Capelli, F.; Laurita, R.; Tappi, S.; Laika, J.; Gioia, L.; Valbonetti, L.; Chaves-López, C. A comparative study on the antifungal efficacy of cold atmospheric plasma at low and high surface density on Aspergillus chevalieri and mechanisms of action. Innov. Food Sci. Emerg. Technol. 2022, 82, 103194. [Google Scholar] [CrossRef]
- Schuster, M.; Steinberg, G. The fungicide dodine primarily inhibits mitochondrial respiration in Ustilago maydis, but also affects plasma membrane integrity and endocytosis, which is not found in Zymoseptoria tritici. Fungal Genet. Biol. 2020, 142, 103414. [Google Scholar] [CrossRef]
- Kane, P.M. Proton transport and pH control in fungi. Adv. Exp. Med. Biol. 2016, 892, 33–68. [Google Scholar] [CrossRef] [PubMed]
- Barda, O.; Maor, U.; Sadhasivam, S.; Bi, Y.; Zakin, V.; Prusky, D.; Sionov, E. The pH-responsive transcription factor PacC governs pathogenicity and ochratoxin A biosynthesis in Aspergillus carbonarius. Front. Microbiol. 2020, 11, 210. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, Q.; Xia, Y.; Jin, K. MaPacC, a pH-responsive transcription factor, negatively regulates thermotolerance and contributes to conidiation and virulence in Metarhizium acridum. Curr. Genet. 2020, 66, 397–408. [Google Scholar] [CrossRef]
- Loss, O.; Bertuzzi, M.; Yan, Y.; Fedorova, N.; McCann, B.L.; Armstrong-James, D.; Espeso, E.A.; Read, N.D.; Nierman, W.C.; Bignell, E.M. Mutual independence of alkaline- and calcium-mediated signalling in Aspergillus fumigatus refutes the existence of a conserved druggable signalling nexus. Mol. Microbiol. 2017, 106, 861–875. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Park, A.R.; Lim, J.Y.; Lee, Y.W. Fss1 is involved in the regulation of an ENA5 homologue for sodium and lithium tolerance in Fusarium graminearum. Environ. Microbiol. 2015, 17, 2048–2063. [Google Scholar] [CrossRef] [PubMed]
- Perez-Perez, W.D.; Carrasco-Navarro, U.; Garcia-Estrada, C.; Kosalkova, K.; Gutierrez-Ruiz, M.C.; Barrios-Gonzalez, J.; Fierro, F. bZIP transcription factors PcYap1 and PcRsmA link oxidative stress response to secondary metabolism and development in Penicillium chrysogenum. Microb. Cell Fact. 2022, 21, 50. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 2015, 11, 671–677. [Google Scholar] [CrossRef]
- Li, Y.; He, P.; Tian, C.; Wang, Y. CgHog1 controls the adaptation to both sorbitol and fludioxonil in Colletotrichum gloeosporioides. Fungal Genet. Biol. 2020, 135, 103289. [Google Scholar] [CrossRef]
- Chen, Y.; Kistler, H.C.; Ma, Z. Fusarium graminearum trichothecene mycotoxins: Biosynthesis, regulation, and management. Annu. Rev. Phytopathol. 2019, 57, 15–39. [Google Scholar] [CrossRef]
- Reijngoud, J.; Arentshorst, M.; Ruijmbeek, C.; Reid, I.; Alazi, E.D.; Punt, P.J.; Tsang, A.; Ram, A.F.J. Loss of function of the carbon catabolite repressor CreA leads to low but inducer-independent expression from the feruloyl esterase B promoter in Aspergillus niger. Biotechnol. Lett. 2021, 43, 1323–1336. [Google Scholar] [CrossRef]
- Tannous, J.; Kumar, D.; Sela, N.; Sionov, E.; Prusky, D.; Keller, N.P. Fungal attack and host defence pathways unveiled in near-avirulent interactions of Penicillium expansum creA mutants on apples. Mol. Plant Pathol. 2018, 19, 2635–2650. [Google Scholar] [CrossRef] [PubMed]
- Gurdaswani, V.; Ghag, S.B.; Ganapathi, T.R. FocSge1 in Fusarium oxysporum f. sp. cubense race 1 is essential for full virulence. BMC Microbiol. 2020, 20, 255. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, K.C.; Cary, J.W.; Montalbano, B.G. Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Acta Biochim. Biophys. Sin. 1999, 1444, 412–417. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuo, R.; Chen, Y.; Xing, M.; Zhang, Z.; Tian, S.; Li, B. Ena Proteins Respond to PacC-Mediated pH Signaling Pathway and Play a Crucial Role in Patulin Biosynthesis. J. Fungi 2023, 9, 806. https://doi.org/10.3390/jof9080806
Zhuo R, Chen Y, Xing M, Zhang Z, Tian S, Li B. Ena Proteins Respond to PacC-Mediated pH Signaling Pathway and Play a Crucial Role in Patulin Biosynthesis. Journal of Fungi. 2023; 9(8):806. https://doi.org/10.3390/jof9080806
Chicago/Turabian StyleZhuo, Ruiling, Yong Chen, Mengyang Xing, Zhanquan Zhang, Shiping Tian, and Boqiang Li. 2023. "Ena Proteins Respond to PacC-Mediated pH Signaling Pathway and Play a Crucial Role in Patulin Biosynthesis" Journal of Fungi 9, no. 8: 806. https://doi.org/10.3390/jof9080806
APA StyleZhuo, R., Chen, Y., Xing, M., Zhang, Z., Tian, S., & Li, B. (2023). Ena Proteins Respond to PacC-Mediated pH Signaling Pathway and Play a Crucial Role in Patulin Biosynthesis. Journal of Fungi, 9(8), 806. https://doi.org/10.3390/jof9080806








