Transcriptomic Response of Clonostachys rosea Mycoparasitizing Rhizoctonia solani
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains
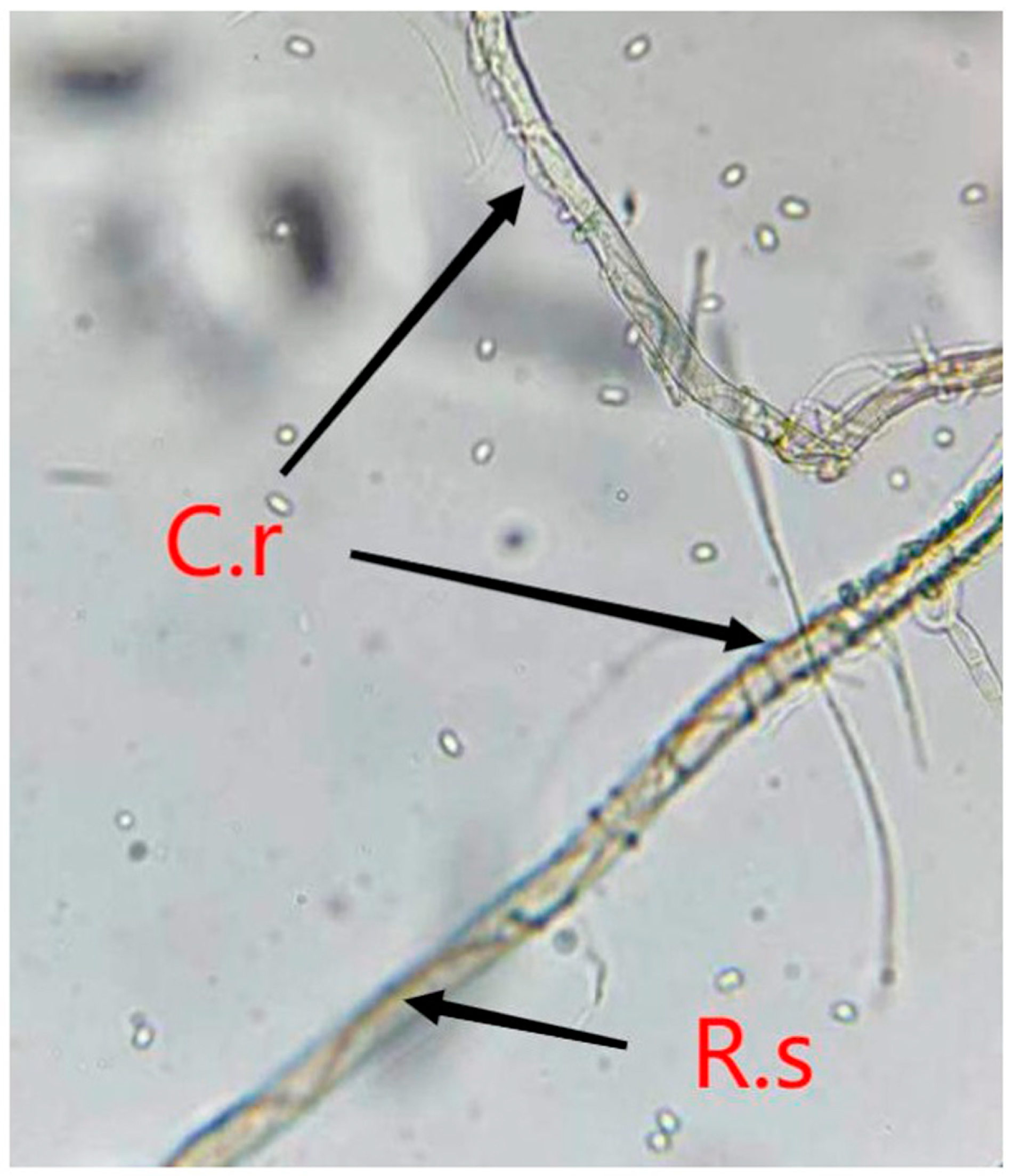
2.2. Sample Collection
2.3. cDNA Library Construction and Transcriptome Sequencing
2.4. Transcriptome Data Processing
2.5. DEG Analysis
2.6. Comparative Transcriptome Analysis
2.7. Quantitative Real-Time PCR Validation
3. Results
3.1. Transcriptome Sequencing and Data Processing
3.2. DEG Analysis
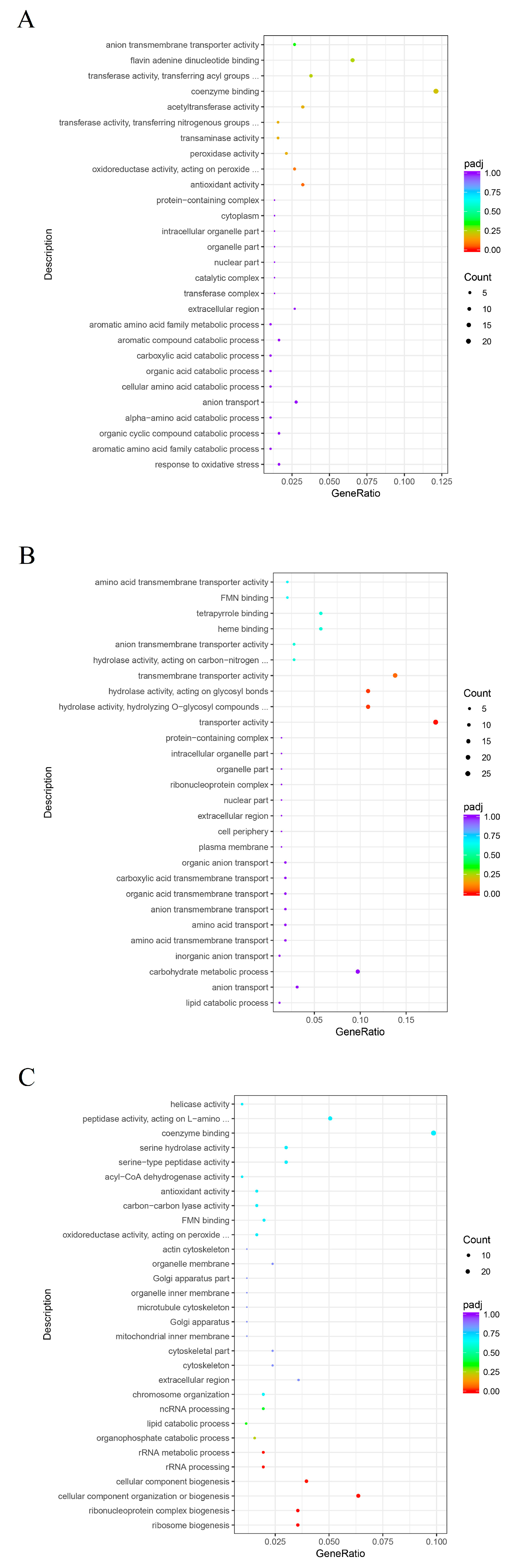
3.3. GO Enrichment Analysis
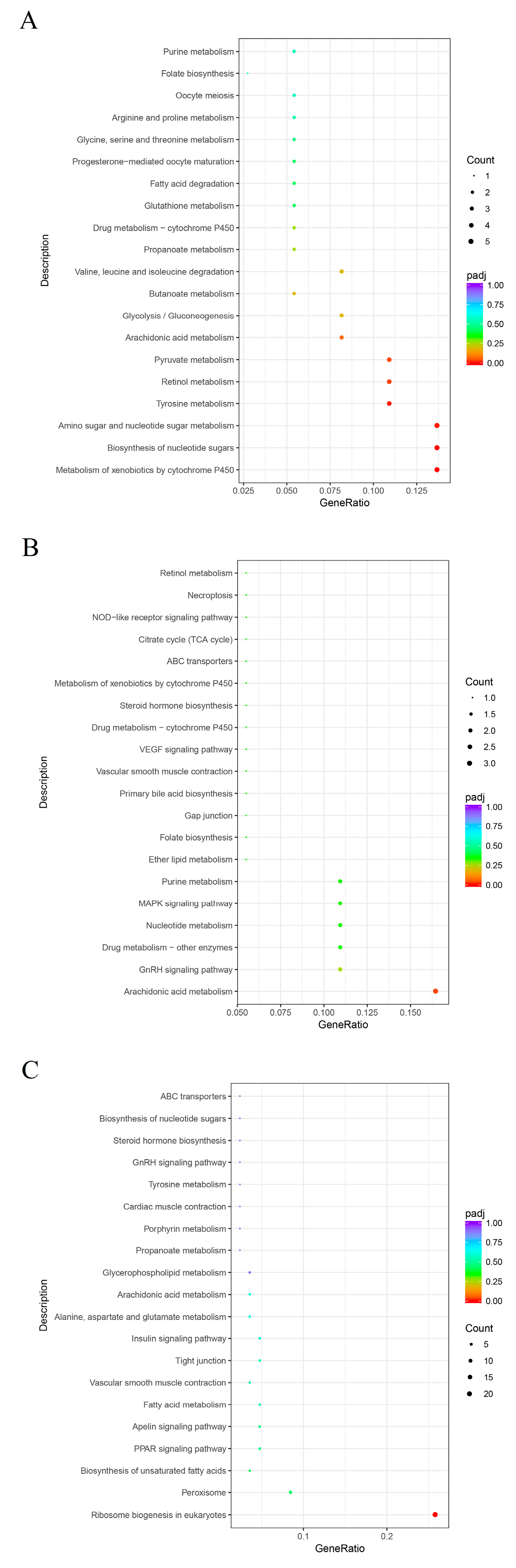
3.4. KEGG Enrichment Analysis
3.5. Screening of Potential Mycoparasitism-Related Genes
3.6. Comparative Transcriptomic Analysis
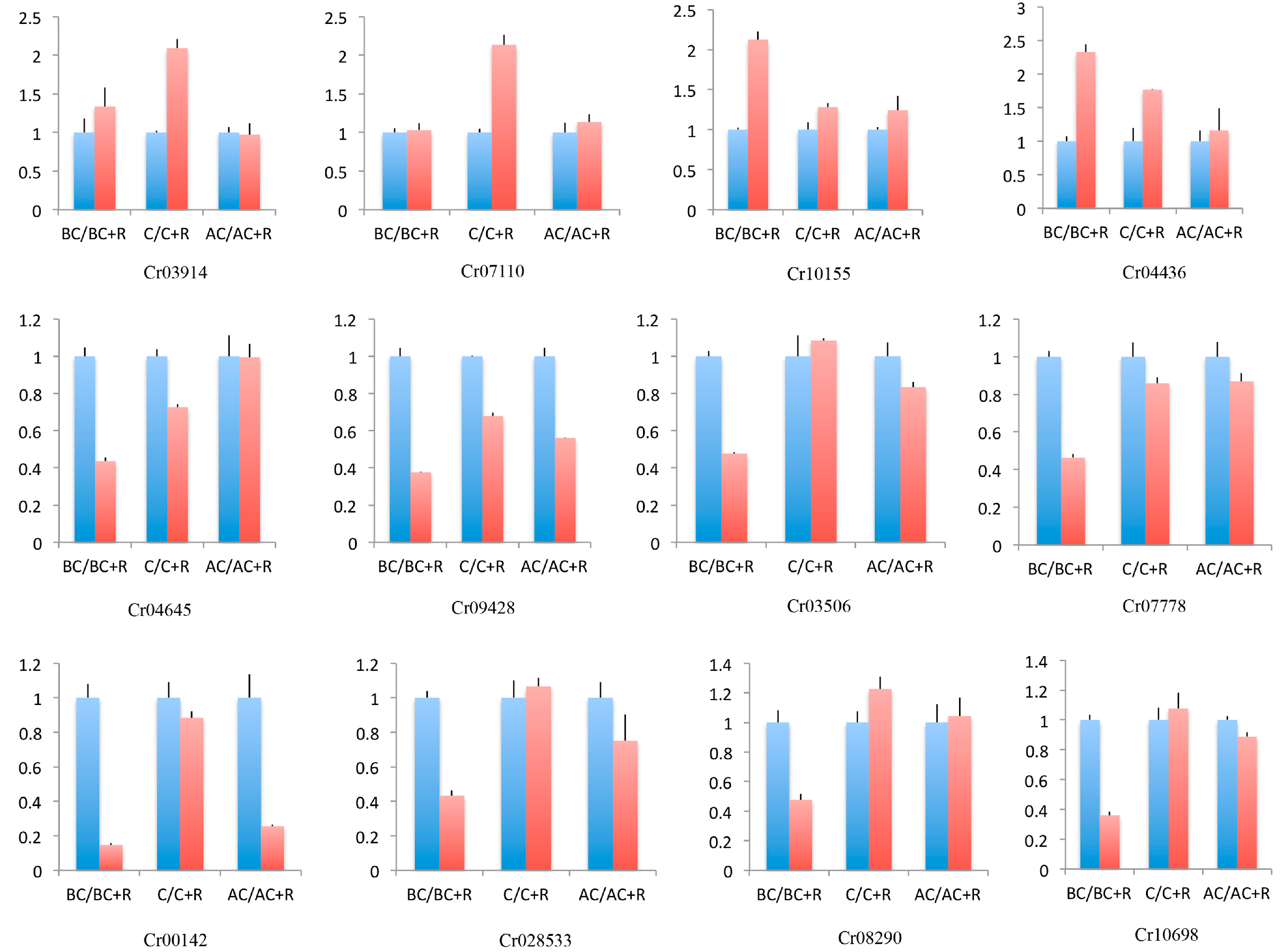
3.7. Quantitative Real-Time PCR Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gonzalez, M.; Pujol, M.; Metraux, J.P.; Gonzalez-Garcia, V.; Bolton, M.D.; Borrás-Hidalgo, O. Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Mol. Plant Pathol. 2011, 12, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Sarkar, M.; Chowdhury, A.; Rakwal, R.; Agrawal, G.K.; Sarkar, A. A comprehensive insight into the biology of Rhizoctonia solani AG1-IA Kühn, the causal organism of the sheath blight disease of rice. J. Plant Pathol. 2022, 104, 79–98. [Google Scholar] [CrossRef]
- Abbas, A.; Khan, S.U.; Khan, W.U.; Saleh, T.A.; Khan, M.H.U.; Ullah, S.; Ali, A.; Ikram, M. Antagonist effects of strains of Bacillus spp. against Rhizoctonia solani for their protection against several plant diseases: Alternatives to chemical pesticides. C R Biol. 2019, 342, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Senapati, M.; Tiwari, A.; Sharma, N.; Chandra, P.; Bashyal, B.M.; Ellur, R.K.; Bhowmick, P.K.; Bollinedi, H.; Vinod, K.K.; Singh, A.K.; et al. Rhizoctonia solani Kühn pathophysiology: Status and prospects of sheath blight disease management in rice. Front Plant Sci. 2022, 13, 881116. [Google Scholar] [CrossRef] [PubMed]
- Sumner, D.R. Sclerotia formation by Rhizoctonia species and their survival. In Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Sneh, B., Jabaji-Hare, S., Neate, S., Dijst, G., Eds.; Kluwer Academic Publications: Dordrecht, NL, USA, 1996; pp. 207–215. [Google Scholar]
- Chahal, K.S.; Sokhi, S.S.; Rattan, G.S. Investigations on sheath blight of rice in Punjab. Indian Phytopathol. 2003, 56, 22–26. [Google Scholar]
- Kozaka, T. Ecological studies on sheath blight of rice plant caused by Pellicularia sasakii and its chemical control. Chugoku Agric. Res. 1961, 20, 1–13. [Google Scholar]
- Zhang, C.Q.; Liu, Y.H.; Ma, X.Y.; Feng, Z.; Ma, Z.H. Characterization of sensitivity of Rhizoctonia solani, causing rice sheath blight, to mepronil and boscalid. Crop Prot. 2009, 28, 381–386. [Google Scholar] [CrossRef]
- Hussain, M.; Zouhar, M.; Ryšánek, P. Suppression of meloidogyne incognita by the entomopathogenic fungus Lecanicillium muscarium. Plant Dis. 2018, 102, 977–982. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Ji, S.; Wang, Z.; Zhang, H.; Wang, Y.; Liu, Z. Trichoderma asperellum xylanases promote growth and induce resistance in poplar. Microbiol. Res. 2021, 248, 126767. [Google Scholar] [CrossRef]
- Xue, A.G. Biological control of pathogens causing root rot complex in field Pea using Clonostachys rosea strain ACM941. Phytopathology 2003, 93, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Safari Motlagh, M.R.; Jahangiri, B.; Kulus, D.; Tymoszuk, A.; Kaviani, B. Endophytic fungi as potential biocontrol agents against Rhizoctonia solani J.G. Kühn, the causal agent of rice sheath blight disease. Biology 2022, 11, 1282. [Google Scholar] [CrossRef]
- Huang, X.; Chen, L.; Ran, W.; Shen, Q.; Yang, X. Trichoderma harzianum strain SQR-T37 and its bio-organic fertilizer could control Rhizoctonia solani damping-off disease in cucumber seedlings mainly by the mycoparasitism. Appl. Microbiol. Biotechnol. 2011, 91, 741–755. [Google Scholar] [CrossRef]
- Nawrocka, J.; Gromek, A.; Małolepsza, U. Nitric oxide as a beneficial signaling molecule in Trichoderma atroviride TRS25-induced systemic defense responses of cucumber plants against Rhizoctonia solani. Front. Plant Sci. 2019, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Harris, A.R. Biocontrol of Rhizoctonia solani and Pythium ultimum on Capsicum by Trichoderma koningii in potting medium. Microbiol. Res. 1999, 154, 131–135. [Google Scholar] [CrossRef]
- Halifu, S.; Deng, X.; Song, X.; Song, R.; Liang, X. Inhibitory Mechanism of Trichoderma virens ZT05 on Rhizoctonia solani. Plants 2020, 9, 912. [Google Scholar] [CrossRef]
- Reithner, B.; Ibarra-Laclette, E.; Mach, R.L.; Herrera-Estrella, A. Identification of mycoparasitism-related genes in Trichoderma atroviride. Appl. Environ. Microbiol. 2011, 77, 4361–4370. [Google Scholar] [CrossRef] [Green Version]
- Atanasova, L.; Le Crom, S.; Gruber, S.; Coulpier, F.; Seidl-Seiboth, V.; Kubicek, C.P.; Druzhinina, I.S. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genom. 2013, 14, 121. [Google Scholar] [CrossRef] [Green Version]
- Keyser, C.A.; Jensen, B.; Meyling, N.V. Dual effects of Metarhizium spp. and Clonostachys rosea against an insect and a seed-borne pathogen in wheat. Pest Manag. Sci. 2016, 72, 517–526. [Google Scholar] [CrossRef]
- Sun, Z.B.; Sun, M.H.; Li, S.D. Identification of mycoparasitism-related genes in Clonostachys rosea 67-1 active against Sclerotinia sclerotiorum. Sci. Rep. 2015, 5, 18169. [Google Scholar] [CrossRef] [Green Version]
- Demissie, Z.A.; Witte, T.; Robinson, K.A.; Sproule, A.; Foote, S.J.; Johnston, A.; Harris, L.J.; Overy, D.P.; Loewen, M.C. Transcriptomic and exometabolomic profiling reveals antagonistic and defensive modes of Clonostachys rosea action against Fusarium graminearum. Mol. Plant Microbe. Interact. 2020, 33, 842–858. [Google Scholar] [CrossRef]
- Nygren, K.; Dubey, M.; Zapparata, A.; Iqbal, M.; Tzelepis, G.D.; Durling, M.B.; Jensen, D.F.; Karlsson, M. The mycoparasitic fungus Clonostachys rosea responds with both common and specific gene expression during interspecific interactions with fungal prey. Evol. Appl. 2018, 11, 931–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lysøe, E.; Dees, M.W.; Brurberg, M.B. A three-way transcriptomic interaction study of a biocontrol agent (Clonostachys rosea), a fungal pathogen (Helminthosporium solani), and a potato host (Solanum tuberosum). Mol. Plant Microbe. Interact. 2017, 30, 646–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.F.; Ma, G.Z.; Bao, Z.H.; Li, S.D. Field test report about biocontrol fungus Gliocladium roseum WP control rice sheath blight. J. Anhui Agri. Sci. 2013, 41, 13242–13255. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Audic, S.; Claverie, J.M. The significance of digital gene expression profiles. Genome Res. 1997, 7, 986–995. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Dubey, M.; Jensen, D.F.; Karlsson, M. The ABC transporter ABCG29 is involved in H2O2 tolerance and biocontrol traits in the fungus Clonostachys rosea. Mol. Genet. Genom. 2016, 291, 677–686. [Google Scholar] [CrossRef]
- Sun, Z.B.; Wang, Q.; Sun, M.H.; Li, S.D. The heat shock protein 70 gene is involved for colony morphology, sporulation and mycoparasitism of Clonostachys rosea. FEMS Microbiol. Lett. 2019, 366, fnz188. [Google Scholar] [CrossRef]
- Sun, Z.B.; Wang, Q.; Zhang, J.; Jiang, W.Z.; Wang, Q.; Li, S.D.; Ma, G.Z.; Sun, M.H. The transcription factor-encoding gene crtf is involved in Clonostachys chloroleuca mycoparasitism on Sclerotinia sclerotiorum. Microbiol. Res. 2018, 210, 6–11. [Google Scholar] [CrossRef]
- Sun, Z.B.; Sun, M.H.; Zhou, M.; Li, S.D. Transformation of the endochitinase gene Chi67-1 in Clonostachys rosea 67-1 increases its biocontrol activity against Sclerotinia sclerotiorum. AMB Express 2017, 7, 1. [Google Scholar] [CrossRef] [Green Version]
- Fatema, U.; Broberg, A.; Jensen, D.F.; Karlsson, M.; Dubey, M. Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea. Sci. Rep. 2018, 8, 15009. [Google Scholar] [CrossRef] [Green Version]
- Zou, C.G.; Tao, N.; Liu, W.J.; Yang, J.K.; Huang, X.W.; Liu, X.Y.; Tu, H.H.; Gan, Z.W.; Zhang, K.Q. Regulation of subtilisin-like protease prC expression by nematode cuticle in the nematophagous fungus Clonostachys rosea. Environ. Microbiol. 2010, 12, 3243–3252. [Google Scholar] [CrossRef]
- Ruocco, M.; Lanzuise, S.; Vinale, F.; Marra, R.; Turrà, D.; Woo, S.L.; Lorito, M. Identification of a new biocontrol gene in Trichoderma atroviride: The role of an ABC transporter membrane pump in the interaction with different plant-pathogenic fungi. Mol. Plant Microbe. Interact. 2009, 22, 291–301. [Google Scholar] [CrossRef] [Green Version]
- Djonović, S.; Vittone, G.; Mendoza-Herrera, A.; Kenerley, C.M. Enhanced biocontrol activity of Trichoderma virens transformants constitutively coexpressing beta-1,3- and beta-1,6-glucanase genes. Mol. Plant Pathol. 2007, 8, 469–480. [Google Scholar] [CrossRef]
- Pozo, M.J.; Baek, J.M.; García, J.M.; Kenerley, C.M. Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet. Biol. 2004, 41, 336–348. [Google Scholar] [CrossRef]
- Limón, M.C.; Pintor-Toro, J.A.; Benítez, T. Increased antifungal activity of Trichoderma harzianum transformants that overexpress a 33-kDa chitinase. Phytopathology 1999, 89, 254–261. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.B.; Li, S.D.; Ren, Q.; Xu, J.L.; Lu, X.; Sun, M.H. Biology and applications of Clonostachys rosea. J. Appl. Microbiol. 2020, 129, 486–495. [Google Scholar] [CrossRef] [Green Version]
- Pasqualetti, M.; Barghini, P.; Giovannini, V.; Fenice, M. High production of chitinolytic activity in halophilic conditions by a new marine strain of Clonostachys rosea. Molecules 2019, 24, 1880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasqualetti, M.; Gorrasi, S.; Giovannini, V.; Braconcini, M.; Fenice, M. Polyextremophilic chitinolytic activity by a marine strain (IG119) of Clonostachys rosea. Molecules 2022, 27, 688. [Google Scholar] [CrossRef] [PubMed]
- Chatterton, S.; Punja, Z.K. Chitinase and beta-1,3-glucanase enzyme production by the mycoparasite Clonostachys rosea f. catenulata against fungal plant pathogens. Can. J. Microbiol. 2009, 55, 356–367. [Google Scholar] [PubMed]
- Li, J.; Yang, J.K.; Huang, X.W.; Zhang, K.Q. Purification and characterization of an extracellular serine protease from Clonostachys rosea and its potential as a pathogenic factor. Process Biochem. 2006, 41, 925–929. [Google Scholar] [CrossRef]
- Pathania, S.; Lore, J.S.; Kalia, A.; Kaur, A.; Sharma, M.; Mangat, G.S.; Sandhu, J.S. Conversion of sheath blight susceptible indica and japonica rice cultivars into moderately resistant through expression of antifungal β-1,3-glucanase transgene from Trichoderma spp. Transgenic. Res. 2022, 31, 537–551. [Google Scholar] [CrossRef]
- Baek, J.M.; Howell, C.R.; Kenerley, C.M. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr. Genet. 1999, 35, 41–50. [Google Scholar] [CrossRef]
- Ferrari, M.; Negri, A.; Romeo, C.; Boccazzi, I.V.; Nodari, R.; Habluetzel, A.; Molteni, G.; Corbett, Y. Adenosine triphosphate-binding cassette transporters are not involved in the detoxification of azadirachta indica extracts In anopheles stephensi Larvae. J. Am. Mosq. Control Assoc. 2018, 34, 311–314. [Google Scholar] [CrossRef]
- Dubey, M.K.; Jensen, D.F.; Karlsson, M. An ATP-binding cassette pleiotropic drug transporter protein is required for xenobiotic tolerance and antagonism in the fungal biocontrol agent Clonostachys rosea. Mol. Plant Microbe. Interact. 2014, 27, 725–732. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Mei, J.; Huang, P.; Tian, Y.; Liang, Y.; Jiang, X.L.; Li, M. Gα3 subunit Thga3 positively regulates conidiation, mycoparasitism, chitinase activity, and hydrophobicity of Trichoderma harzianum. AMB Express 2020, 10, 221. [Google Scholar] [CrossRef]
- Reithner, B.; Brunner, K.; Schuhmacher, R.; Peissl, I.; Seidl, V.; Krska, R.; Zeilinger, S. The G protein alpha subunit Tga1 of Trichoderma atroviride is involved in chitinase formation and differential production of antifungal metabolites. Fungal Genet. Biol. 2005, 42, 749–760. [Google Scholar] [CrossRef]
- Rocha-Ramirez, V.; Omero, C.; Chet, I.; Horwitz, B.A.; Herrera-Estrella, A. Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 2002, 1, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Zeilinger, S.; Reithner, B.; Scala, V.; Peissl, I.; Lorito, M.; Mach, R.L. Signal transduction by Tga3, a novel g protein subunit alpha of Trichoderma atroviride. Appl. Environ. Microbiol. 2005, 71, 1591–1597. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, P.K.; Latha, J.; Hadar, R.; Horwitz, B.A. Role of two G-protein alpha subunits, TgaA and TgaB, in the antagonism of plant pathogens by Trichoderma virens. Appl. Environ. Microbiol. 2004, 70, 542–549. [Google Scholar] [CrossRef] [Green Version]
- Tong, Y.; Wu, H.; Liu, Z.; Wang, Z.; Huang, B. G-Protein Subunit Gαi in mitochondria, MrGPA1, affects conidiation, stress resistance, and virulence of entomopathogenic fungus Metarhizium robertsii. Front. Microbiol. 2020, 11, 1251. [Google Scholar] [CrossRef]
- Zou, C.G.; Tu, H.H.; Liu, X.Y.; Tao, N.; Zhang, K.Q. PacC in the nematophagous fungus Clonostachys rosea controls virulence to nematodes. Environ. Microbiol. 2010, 12, 1868–1877. [Google Scholar] [CrossRef]
- Trushina, N.; Levin, M.; Mukherjee, P.K.; Horwitz, B.A. PacC and pH-dependent transcriptome of the mycotrophic fungus Trichoderma virens. BMC Genom. 2013, 14, 138. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Han, Y.C.; Yang, L.; Wu, M.D.; Zhang, J.; Cheng, J.S.; Wang, M.Y.; Jiang, D.H.; Chen, W.D.; Li, G.Q. CmpacC regulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungus Coniothyrium minitans. Environ. Microbiol. 2015, 17, 4711–4729. [Google Scholar]
- Zhang, M.G.; Wei, Q.L.; Xia, Y.X.; Jin, K. MaPacC, a pH-responsive transcription factor, negatively regulates thermotolerance and contributes to conidiation and virulence in Metarhizium acridum. Curr. Genet. 2020, 66, 397–408. [Google Scholar] [CrossRef]
- Song, Z.Y.; Yin, Y.P.; Lin, Y.L.; Du, F.; Ren, G.W.; Wang, Z.K. The bZIP transcriptional factor activator protein-1 regulates Metarhizium rileyi morphology and mediates microsclerotia formation. Appl. Microbiol. Biotechnol. 2018, 102, 4577–4588. [Google Scholar] [CrossRef]
- Zhao, X.; Yang, X.; Lu, Z.; Wang, H.; He, Z.; Zhou, G.; Luo, Z.; Zhang, Y. MADS-box transcription factor Mcm1 controls cell cycle, fungal development, cell integrity and virulence in the filamentous insect pathogenic fungus Beauveria bassiana. Environ. Microbiol. 2019, 21, 3392–3416. [Google Scholar] [CrossRef]
- Peng, Y.J.; Wang, J.J.; Lin, H.Y.; Ding, J.L.; Feng, M.G.; Ying, S.H. HapX, an indispensable bZIP transcription factor for iron acquisition, regulates infection initiation by orchestrating conidial oleic acid homeostasis and cytomembrane functionality in mycopathogen Beauveria bassiana. mSystems 2020, 5, e00695-20. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, L.; Knox, B.P.; Kubicek, C.P.; Druzhinina, I.S.; Baker, S.E. The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukarot. Cell 2013, 12, 1499–1508. [Google Scholar]
- Toopaang, W.; Phonghanpot, S.; Punya, J.; Panyasiri, C.; Klamchao, K.; Wasuwan, R.; Srisuksam, C.; Sangsrakru, D.; Sonthirod, C.; Tangphatsornruang, S.; et al. Targeted disruption of the polyketide synthase gene pks15 affects virulence against insects and phagocytic survival in the fungus Beauveria bassiana. Fungal Biol. 2017, 121, 664–675. [Google Scholar] [CrossRef] [PubMed]
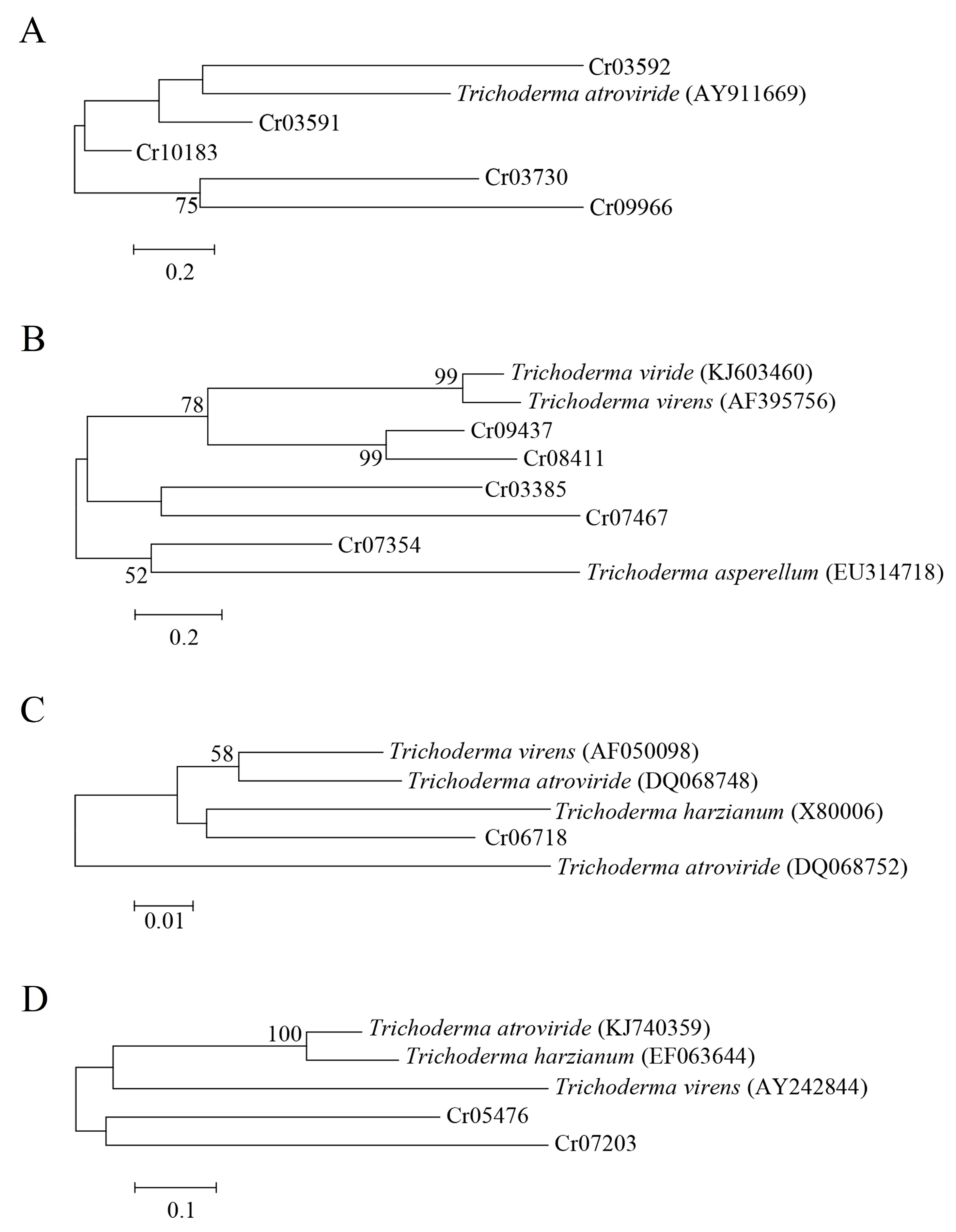
| Gene Annotation | Gene Name | Isolate | Accession No. |
|---|---|---|---|
| ABC transporter | Taabc2 | T. atroviride | AY911669 |
| β-1,3-glucanase | TvBng2 | T. virens | AF395756 |
| Exo-β-1,3-glucanase | tag83 | T. asperellum | EU314718 |
| β-1,3-glucanase | - | Trichoderma spp. | KJ603460 |
| Subtilisin-like serine protease | TghSS42 | T. ghanense | KJ740359 |
| Subtilisin-like protease | SS10 | T. harzianum | EF063644 |
| Serine protease | tvsp1 | T. virens | AY242844 |
| Chitinase | chit42 | T. virens | AF050098 |
| Chitinase | chit33 | T. harzianum | X80006 |
| Chitinase | chi18-10 | T. atroviride | DQ068748 |
| Chitinase | chi18-13 | T. atroviride | DQ068752 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.-B.; Yu, S.-F.; Sun, M.-H.; Li, S.-D.; Hu, Y.-F.; Song, H.-J. Transcriptomic Response of Clonostachys rosea Mycoparasitizing Rhizoctonia solani. J. Fungi 2023, 9, 818. https://doi.org/10.3390/jof9080818
Sun Z-B, Yu S-F, Sun M-H, Li S-D, Hu Y-F, Song H-J. Transcriptomic Response of Clonostachys rosea Mycoparasitizing Rhizoctonia solani. Journal of Fungi. 2023; 9(8):818. https://doi.org/10.3390/jof9080818
Chicago/Turabian StyleSun, Zhan-Bin, Shu-Fan Yu, Man-Hong Sun, Shi-Dong Li, Ya-Feng Hu, and Han-Jian Song. 2023. "Transcriptomic Response of Clonostachys rosea Mycoparasitizing Rhizoctonia solani" Journal of Fungi 9, no. 8: 818. https://doi.org/10.3390/jof9080818
APA StyleSun, Z.-B., Yu, S.-F., Sun, M.-H., Li, S.-D., Hu, Y.-F., & Song, H.-J. (2023). Transcriptomic Response of Clonostachys rosea Mycoparasitizing Rhizoctonia solani. Journal of Fungi, 9(8), 818. https://doi.org/10.3390/jof9080818






