Isolation and Identification of Non-Saccharomyces Yeast Producing 2-Phenylethanol and Study of the Ehrlich Pathway and Shikimate Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Yeast Strains
2.2. Screening of Yeast Strains Producing 2-PE
2.3. Identification of Yeast Strains Producing 2-PE
2.3.1. Observation of Morphology
2.3.2. Physiological Characteristics
2.3.3. Molecular Biological Identification
2.4. Whole-Genome Sequencing, Assembly, and Annotation of Strain R5
2.4.1. Genomic DNA Extraction and Sequencing
2.4.2. Whole-Genome Data Processing and Analysis
2.5. Study of the Synthesis Pathway of 2-PE
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Tolerance of Yeast Strains to 2-PE
3. Results and Discussion
3.1. Isolation and Screening of Yeast Strains Producing 2-PE
3.2. Identification of Morphology, Physiology, and Molecular Biology
3.3. Whole-Genome Sequencing, Assembly, and Annotation of Strain R5
3.4. Identification Synthesis Pathway of 2-PE by Strain R5
3.5. Identification of Potential Genes Involved in 2-PE Synthesis by qRT-PCR
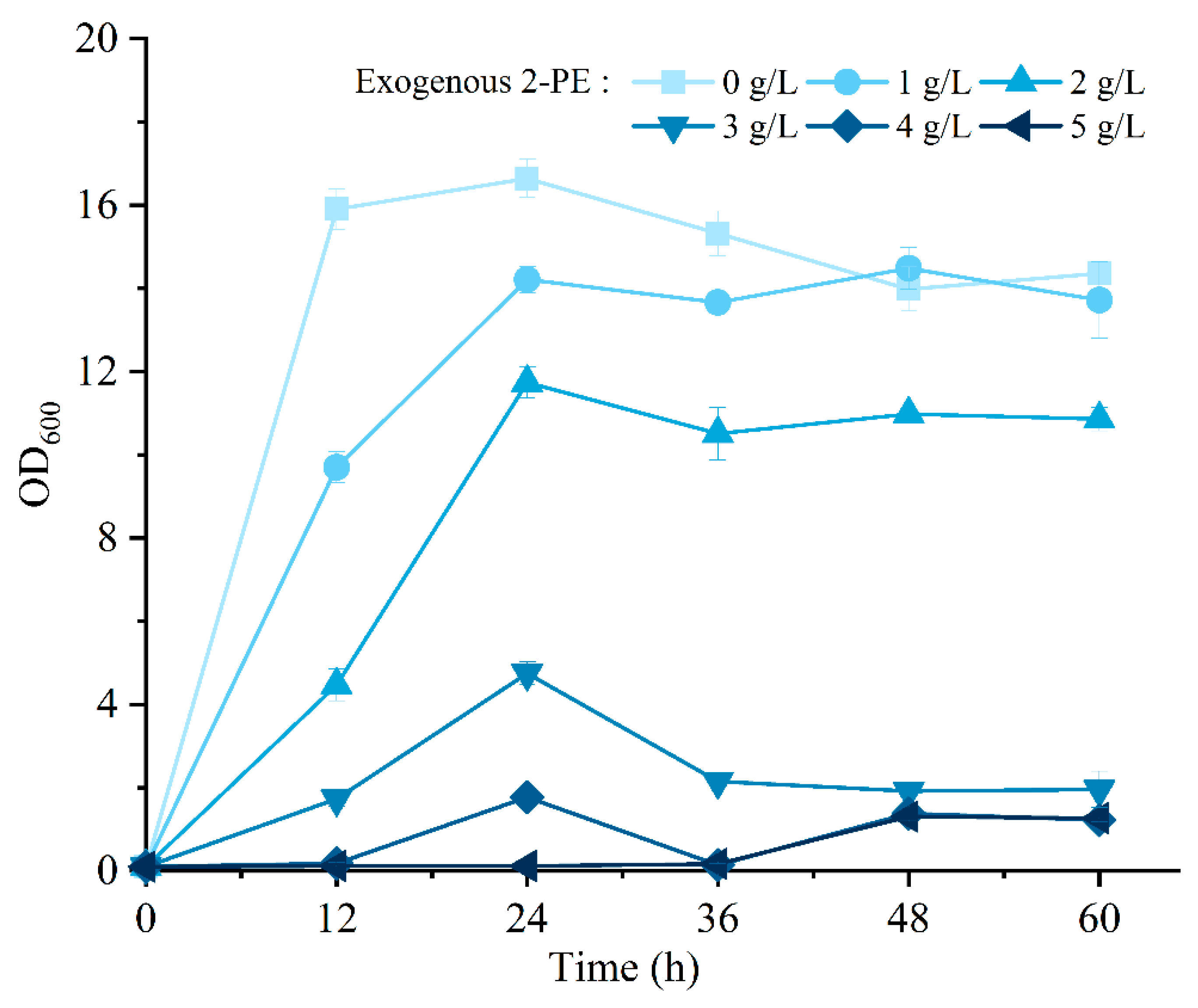
3.6. Effect of 2-PE Stress on the Growth of R5 Strain
4. Conclusions
- (1)
- In this study, a yeast strain was isolated and screened from pear peels that was capable of producing 2-PE. Based on morphology observation, analysis of physiological characteristics, and molecular biological identification, strain R5 was identified as S. bacillaris.
- (2)
- Subsequently, through the analysis of whole-genome sequencing and comparison with the KEGG database, we found that strain R5 possesses a metabolic pathway producing 2-PE. To further investigate the synthesis pathway of 2-PE, strain R5 was inoculated into three M3 media. When strain R5 was cultured in M3 (Phe) medium, 2-PE was biosynthesized from L-phe via the Ehrlich pathway. When strain R5 was inoculated in M3 (NH4+) medium, 2-PE was produced from glucose through the shikimate pathway. In M3 (Phe) medium, the maximum concentration of 2-PE reached 1.28 g/L, which was 16-fold and 2.29-fold higher than that in M3 (NH4+) and M3 (Phe + NH4+) media, respectively. These results show that the Ehrlich pathway is the main synthetic pathway for producing 2-PE in strain R5.
- (3)
- The qRT-PCR results revealed that the transport of L-phe was inhibited when both NH4+ and Phe were present in the medium. The key gene catalyzing the dehydrogenation of benzaldehyde into 2-PE is ADH5. And genes ADH5, PDC, hisC, and GOT1 are not sensitive to nitrogen metabolism inhibition. These findings provide evidence that strain R5 is capable of biotransforming L-phe into 2-PE.
- (4)
- In summary, all the above results illustrated that the Ehrlich pathway and shikimate pathway synthesize 2-PE in strain R5, mainly for the Ehrlich pathway, which had not been previously investigated. These findings fill the research gap relating to the synthesis pathway of 2-PE in strain R5 and provide a solid basis for future research on the application of non-Saccharomyces yeasts in the wine industry.
5. Accession Number
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lobs, A.K.; Schwartz, C.; Wheeldon, I. Genome and metabolic engineering in non-conventional yeasts: Current advances and applications. Synth. Syst. Biotechnol. 2017, 2, 198–207. [Google Scholar] [CrossRef]
- Mills, D.A.; Johannsen, E.A.; Cocolin, L. Yeast diversity and persistence in botrytis-affected wine fermentations. Appl. Environ. Microbiol. 2002, 68, 4884–4893. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Candida zemplinina sp. nov., an osmotolerant and psychrotolerant yeast that ferments sweet botrytized wines. Int. J. Syst. Evol. Microbiol. 2003, 53, 2079–2083. [Google Scholar] [CrossRef] [PubMed]
- Duarte, F.L.; Pimentel, N.H.; Teixeira, A.; Fonseca, A. Saccharomyces bacillaris is not a synonym of Candida stellata: Reinstatement as Starmerella bacillaris comb. nov. Antonie Van Leeuwenhoek 2012, 102, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Masneuf-Pomarede, I.; Bely, M.; Marullo, P.; Albertin, W. The Genetics of non-conventional wine yeasts: Current knowledge and future challenges. Front. Microbiol. 2015, 6, 1563. [Google Scholar] [CrossRef]
- Nadai, C.; Fernandes Lemos, W.J.J.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol activity of Starmerella bacillaris yeast against blue mold disease on apple fruit and its effect on cider fermentation. PLoS ONE 2018, 13, e0204350. [Google Scholar] [CrossRef] [PubMed]
- Englezos, V.; Rantsiou, K.; Torchio, F.; Rolle, L.; Gerbi, V.; Cocolin, L. Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation:Physiological and molecular characterizations. Int. J. Food Microbiol. 2015, 199, 33–40. [Google Scholar] [CrossRef]
- Lemos Junior, W.J.F.; Nadai, C.; Crepalde, L.T.; De Oliveira, V.S.; De Matos, A.D.; Giacomini, A.; Corich, V. Potential use of Starmerella bacillaris as fermentation starter for the production of low-alcohol beverages obtained from unripe grapes. Int. J. Food Microbiol. 2019, 303, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Rantsiou, K.; Englezos, V.; Torchio, F.; Risse, P.A.; Cravero, F.; Gerbi, V.; Rolle, L.; Cocolin, L. Modeling of the fermentation behavior of Starmerella bacillaris. Am. J. Enol. Vitic. 2017, 68, 378–385. [Google Scholar] [CrossRef]
- Bahut, F.; Romanet, R.; Sieczkowski, N.; Schmitt-Kopplin, P.; Nikolantonaki, M.; Gougeon, R.D. Antioxidant activity from inactivated yeast: Expanding knowledge beyond the glutathione-related oxidative stability of wine. Food Chem. 2020, 325, 126941. [Google Scholar] [CrossRef]
- Lemos, W.J.R.; Bovo, B.; Nadai, C.; Crosato, G.; Carlot, M.; Favaron, F.; Giacomini, A.; Corich, V. Biocontrol ability and action mechanism of Starmerella bacillaris (Synonym Candida zemplinina) isolated from wine musts against gray mold disease agent botrytis cinerea on grape and their effects on alcoholic fermentation. Front. Microbiol. 2016, 7, 1249. [Google Scholar] [CrossRef]
- Eeglezos, V.; Giacosa, S.; Rantsiou, K.; Rolle, L.; Cocolin, L. Starmerella bacillaris in winemaking: Opportunities and risks. Curr. Opin. Food Sci. 2017, 17, 30–35. [Google Scholar] [CrossRef]
- Sartor, S.; Toaldo, I.M.; Panceri, C.P.; Caliari, V.; Luna, A.S.; De Gois, J.S.; Bordignon-Luiz, M.T. Changes in organic acids, polyphenolic and elemental composition of rose sparkling wines treated with mannoproteins during over-lees aging. Food Res. Int. 2019, 124, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Xia, H.; Yang, C.; Chen, X. Sensing, uptake and catabolism of L-phenylalanine during 2-phenylethanol biosynthesis via the Ehrlich pathway in Saccharomyces cerevisiae. Front. Microbiol. 2021, 12, 601963. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Ma, J.; Zhu, Y.; Xu, P. Refactoring Ehrlich pathway for high-yield 2-phenylethanol production in Yarrowia lipolytica. ACS Synth. Biol. 2020, 9, 623–633. [Google Scholar] [CrossRef]
- Chreptowicz, K.; Wielechowska, M.; Glowczyk-Zubek, J.; Rybak, E.; Mierzejewska, J. Production of natural 2-phenylethanol: From biotransformation to purified product. Food Bioprod. Process. 2016, 100, 275–281. [Google Scholar] [CrossRef]
- Huang, C.; Qian, Y.; Viana, T.; Siegumfeldt, H.; Arneborg, N.; Larsen, N.; Jespersen, L. The quorum-sensing molecule 2-phenylethanol impaired conidial germination, hyphal membrane integrity and growth of Penicillium expansum and Penicillium nordicum. J. Appl. Microbiol. 2020, 129, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Binati, R.L.; Lemos Junior, W.J.F.; Luzzini, G.; Slaghenaufi, D.; Ugliano, M.; Torriani, S. Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: A study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol. 2020, 318, 108470. [Google Scholar] [CrossRef] [PubMed]
- Lorenzini, M.; Simonato, B.; Slaghenaufi, D.; Ugliano, M.; Zapparoli, G. Assessment of yeasts for apple juice fermentation and production of cider volatile compounds. LWT 2019, 99, 224–230. [Google Scholar] [CrossRef]
- Englezos, V.; Rantsiou, K.; Cravero, F.; Torchio, F.; Pollon, M.; Fracassetti, D.; Ortiz-Julien, A.; Gerbi, V.; Rolle, L.; Cocolin, L. Volatile profile of white wines fermented with sequential inoculation of Starmerella bacillaris and Saccharomyces cerevisiae. Food Chem. 2018, 257, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Mo, Q.; Chen, H.; Fan, C.; Zhang, D.; Liu, L.; Fu, B.; Yuan, J. Utilization of a styrene-derived pathway for 2-phenylethanol production in budding yeast. Appl. Microbiol. Biotechnol. 2021, 105, 2333–2340. [Google Scholar] [CrossRef]
- Li, M.; Lang, X.; Moran, C.M.; De, K.S.; Sun, X.; Da, S.N.; Wheeldon, I. CRISPR-mediated multigene integration enables Shikimate pathway refactoring for enhanced 2-phenylethanol biosynthesis in Kluyveromyces marxianus. Biotechnol. Biofuels 2021, 14, 3. [Google Scholar] [CrossRef]
- Chreptowicz, K.; Sternicka, M.K.; Kowalska, P.D.; Mierzejewska, J. Screening of yeasts for the production of 2-phenylethanol (rose aroma) in organic waste-based media. Lett. Appl. Microbiol. 2018, 66, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Lu, X.; Zong, H.; Zhuge, B. Genetic engineering of an industrial yeast Candida glycerinogenes for efficient production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 2020, 104, 10481–10491. [Google Scholar] [CrossRef] [PubMed]
- Mitri, S.; Koubaa, M.; Maroun, R.G.; Rossignol, T.; Nicaud, J.M.; Louka, N. Bioproduction of 2-phenylethanol through yeast fermentation on synthetic media and on agro-industrial waste and by-products: A review. Foods 2022, 11, 109. [Google Scholar] [CrossRef]
- Hossain, M.N.; Afrin, S.; Hunayun, S.; Ahmed, M.M.; Saha, B.K. Identification and growth characterization of a novel strain of Saccharomyces boulardii isolated from soya paste. Front. Nutr. 2020, 7, 27. [Google Scholar] [CrossRef]
- Petretto, G.L.; Mercenaro, L.; Urgeghe, P.P.; Fadda, C.; Valentoni, A.; Del Caro, A. Grape and wine composition in Vitis vinifera L. cv. Cannonau explored by GC-MS and sensory analysis. Foods 2021, 10, 101. [Google Scholar] [CrossRef]
- Cao, W.; Shu, N.; Wen, J.; Yang, Y.; Jin, Y.; Lu, W. Characterization of the key aroma volatile compounds in nine different grape varieties wine by headspace gas chromatography-ion mobility spectrometry (HS-GC-IMS), odor activity values (OAV) and sensory analysis. Foods 2022, 11, 2767. [Google Scholar] [CrossRef] [PubMed]
- Salas-Millan, J.A.; Aznar, A.; Conesa, E.; Conesa-Bueno, A.; Aguayo, E. Fruit wine obtained from melon by-products: Physico-chemical and sensory analysis, and characterization of key aromas by GC-MS. Foods 2022, 11, 3619. [Google Scholar] [CrossRef] [PubMed]
- Rong, Z.; Xiao, C.; Ying, X.; Maobin, C.; Yu, Z.; Qin, L.; Da, Z.; Shangling, F. Research on the application of liquid-liquid extraction-gas chromatography-mass spectrometry (LLE-GC-MS) and headspace-gas chromatography-ion mobility spectrometry (HS-GC-IMS) in distinguishing the Baiyunbian aged liquors. Int. J. Food Eng. 2021, 17, 2. [Google Scholar] [CrossRef]
- Salgado, V.; Fonseca, C.; Lopes, D.S.T.; Roseiro, J.C.; Eusebio, A. Isolation and identification of Magnusiomyces capitatus as a lipase-producing yeast from olive mill wastewater. Waste Biomass Valorization 2019, 11, 3207–3221. [Google Scholar] [CrossRef]
- Hongru, J. Isolation and Identification of Wine-Related Yeast Associated to Different Varieties; Northwest A & F University: Xianyang, China, 2008. [Google Scholar]
- Kim, S.H.; Chon, J.W.; Jeong, H.W.; Song, K.Y.; Kim, D.H.; Bae, D.; Kim, H.; Seo, K.H. Identification and phylogenetic analysis of enterococcus isolates using MALDI-TOF MS and VITEK 2. AMB Express. 2023, 13, 21. [Google Scholar] [CrossRef]
- Wagh, V.S.; Said, M.S.; Bennale, J.S.; Dastager, S.G. Isolation and structural characterization of exopolysaccharide from marine Bacillus sp. and its optimization by microbioreactor. Carbohydr. Polym. 2022, 285, 119241. [Google Scholar] [CrossRef]
- Qu, Y.; Jiang, J.; Yang, X.; Tang, C. Protocol for titrating gene expression levels in budding yeast. Star Protoc. 2020, 1, 100082. [Google Scholar] [CrossRef]
- Wang, L.; Xu, A.; Zhou, P.; Zhao, M.; Xu, C.; Wang, Y.; Wang, K.; Wang, F.; Miao, Y.; Zhao, W.; et al. Rapid detection of Candida tropicalis in clinical samples from different sources using RPA-LFS. Front. Cell Infect. Microbiol. 2022, 12, 898186. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Li, J.; Chen, Y. Characterization of tolerance and multi-enzyme activities in non-Saccharomyces yeasts isolated from vidal blanc icewine fermentation. J. Food Biochem. 2019, 43, e13027. [Google Scholar] [CrossRef]
- Raymond Eder, M.L.; Rosa, A.L. Genetic, physiological, and industrial aspects of the fructophilic non-Saccharomyces yeast species, Starmerella bacillaris. Fermentation 2021, 7, 87. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest-HPC: Fast selection of best-fit models of protein evolution. In Proceedings of the Euro-Par 2010 Parallel Processing Workshops: HeteroPar, HPCC, HiBB, CoreGrid, UCHPC, HPCF, PROPER, CCPI, VHPC, Ischia, Italy, 31 August–3 September 2010; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6586, pp. 177–184. [Google Scholar] [CrossRef]
- Ma, W.; Yu, J.; Zhang, X.; Guo, S.; Zhang, F.; Jin, W.; Dong, J.; Jia, S.; Zhong, C.; Xue, J. Whole-genome sequencing exploitation analysis of non-Saccharomyces yeast Nakazawaea ishiwadae GDMCC 60786 and its physiological characterizations. Food Biosci. 2021, 41, 100982. [Google Scholar] [CrossRef]
- Mctaggart, L.R.; Cabrea, A.; Cronin, K.; Kus, J.V. Antifungal susceptibility of clinical yeast isolates from a large Canadian reference laboratory and application of whole-genome sequence analysis to elucidate mechanisms of acquired resistance. Antimicrob. Agents Chemother. 2020, 64, 10–1128. [Google Scholar] [CrossRef]
- Mierzejewska, J.; Tymoszewska, A.; Chreptowicz, K.; Krol, K. Mating of 2 laboratory Saccharomyces cerevisiae strains resulted in enhanced production of 2-phenylethanol by biotransformation of L-phenylalanine. J. Mol. Microbiol. Biotechnol. 2017, 27, 81–90. [Google Scholar] [CrossRef]
- Deed, R.C.; Hou, R.; Kinzurik, M.I.; Gardner, R.C.; Fedrizzi, B. The role of yeast ARO8, ARO9 and ARO10 genes in the biosynthesis of 3-(methylthio)-1-propanol from L-methionine during fermentation in synthetic grape medium. FEMS Yeast Res. 2019, 19, foy109. [Google Scholar] [CrossRef]
- Dai, J.; Li, K.; Song, N.; Yao, W.; Xia, H.; Yang, Q.; Zhang, X.; Li, X.; Wang, Z.; Yao, L.; et al. Zygosaccharomyces rouxii, an aromatic yeast isolated from chili sauce, is able to biosynthesize 2-phenylethanol via the Shikimate or Ehrlich pathways. Front. Microbiol. 2020, 11, 597454. [Google Scholar] [CrossRef]
- Yan, W.; Zhang, X.; Qian, X.; Zhou, J.; Dong, W.; Ma, J.; Zhang, W.; Xin, F.; Jiang, M. Comprehensive investigations of 2-phenylethanol production by high 2-phenylethanol tolerating Meyerozyma sp. strain YLG18. Enzyme Microb. Technol. 2020, 140, 109629. [Google Scholar] [CrossRef]
- Lloyd, D. Book Reviews. J. Eukaryot. Microbiol. 2000, 47, 598. [Google Scholar] [CrossRef]
- Goncalves, P.; Goncalves, C.; Brito, P.H.; Sampaio, J.P. The Wickerhamiella/Starmerella clade-A treasure trove for the study of the evolution of yeast metabolism. Yeast 2020, 37, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Lemos Junior, W.J.F.; Treu, L.; Nadai, C.; Da Silva Duarte, V.; Campanaro, S.; Fabrega-Prats, M.; Giacomini, A.; Corcih, V. Genomic insights into the glutathione metabolism of the non-conventional wine yeast Starmerella bacillaris. OENO One 2021, 55, 4347. [Google Scholar] [CrossRef]
- Etschmann, M.M.; Bluemke, W.; Ssee, D.; Schrader, J. Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 2002, 59, 1–8. [Google Scholar] [CrossRef]
- Etschmann, M.; Sell, D.; Schrader, J. Screening of yeasts for the production of the aroma compound 2-phenylethanol in a molasses-based medium. Biotechnol. Lett. 2003, 25, 531–536. [Google Scholar] [CrossRef]
- Yin, S.; Zhou, H.; Xiao, X.; Lang, T.; Liang, J.; Wang, C. Improving 2-phenylethanol production via Ehrlich pathway using genetic engineered Saccharomyces cerevisiae strains. Curr. Microbiol. 2015, 70, 762–767. [Google Scholar] [CrossRef]
- Romano, P.; Braschi, G.; Siesto, G.; Patrignani, F.; Lanciotti, R. Role of yeasts on the sensory component of wines. Foods 2022, 11, 1921. [Google Scholar] [CrossRef]
- Jin, D.; Gu, B.; Xiong, D.; Huang, G.; Huang, X.; Liu, L.; Xiao, J. A transcriptomic analysis of Saccharomyces cerevisiae under the stress of 2-phenylethanol. Curr. Microbiol. 2018, 75, 1068–1076. [Google Scholar] [CrossRef]
- Zhu, L.; Xu, S.; Li, Y.; Shi, G. Improvement of 2-phenylethanol production in Saccharomyces cerevisiae by evolutionary and rational metabolic engineering. PLoS ONE 2021, 16, e0258180. [Google Scholar] [CrossRef]
- Hua, D.; Xu, P. Recent advances in biotechnological production of 2-phenylethanol. Biotechnol. Adv. 2011, 29, 654–660. [Google Scholar] [CrossRef]
- De Lima, L.A.; Diniz, R.H.S.; De Queiroz, M.V.; Fietto, L.G.; Da Silveira, W.B. Screening of yeasts isolated from Brazilian environments for the 2-phenylethanol (2-PE) production. Biotechnol. Bioprocess Eng. 2018, 23, 326–332. [Google Scholar] [CrossRef]
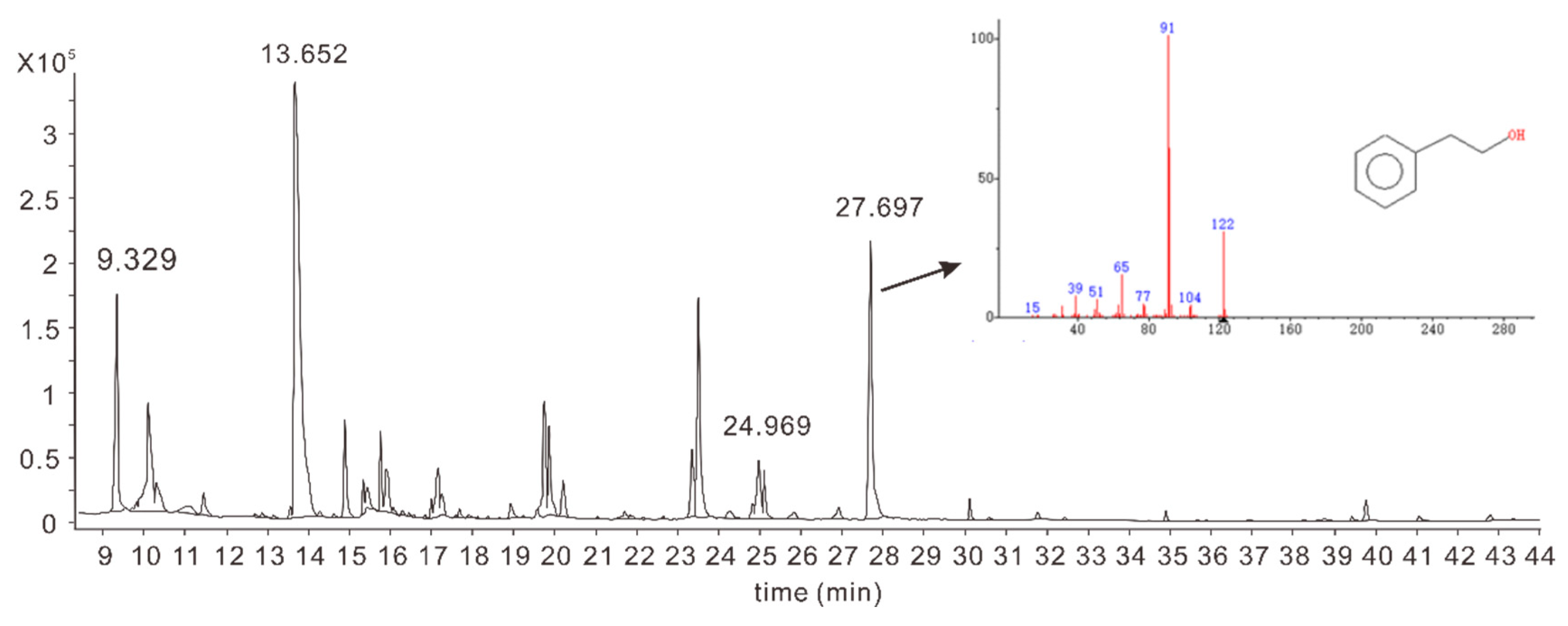
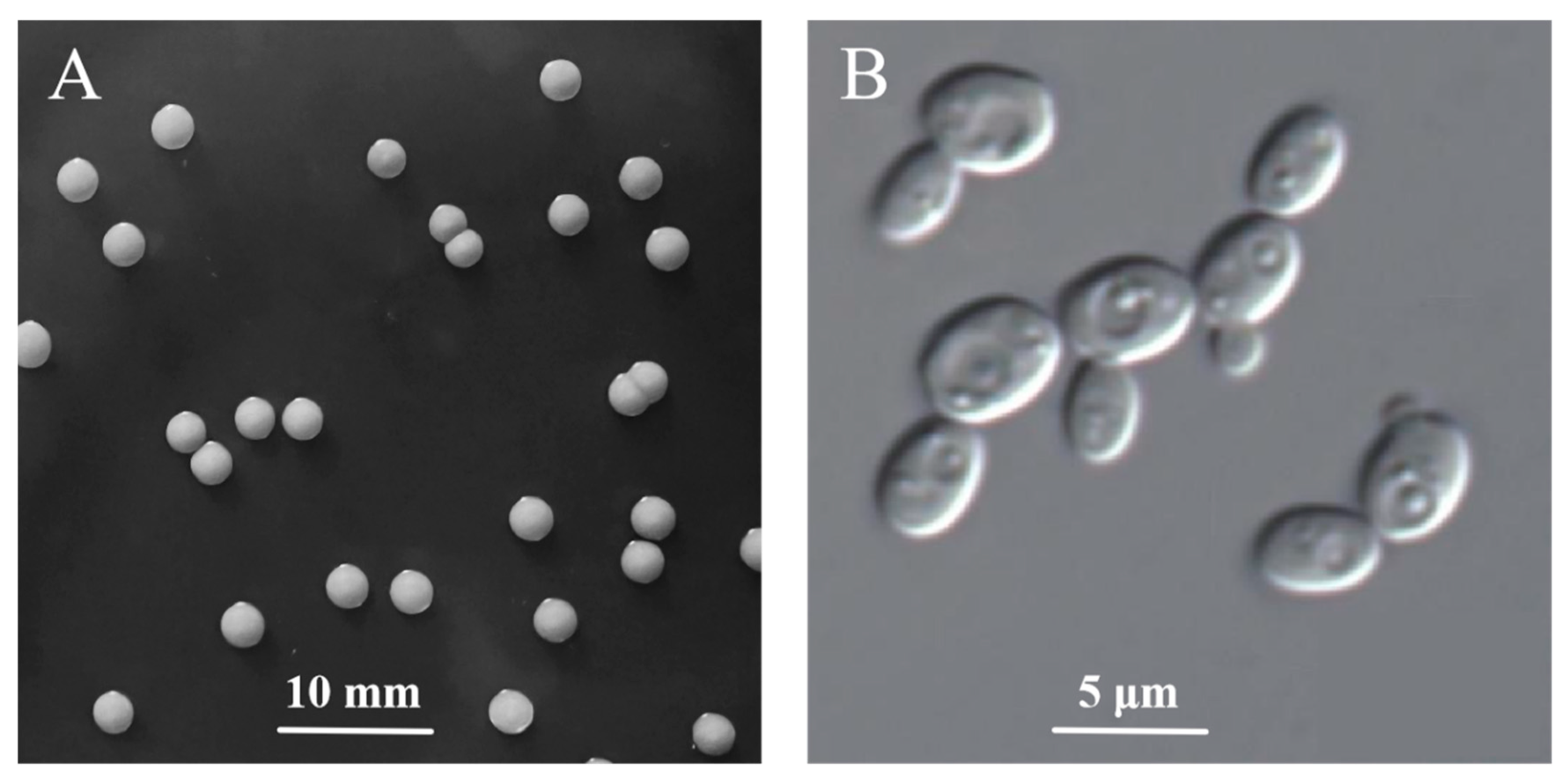

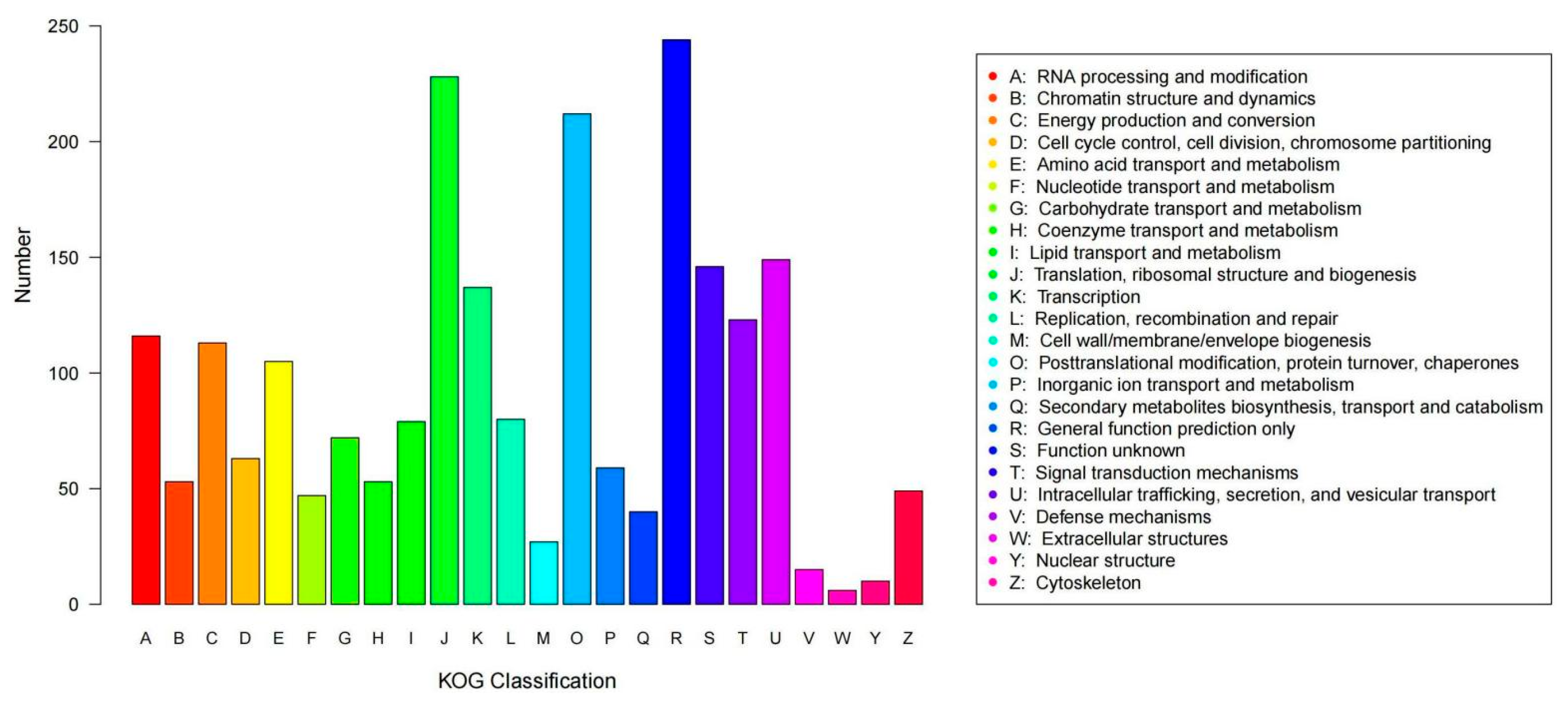

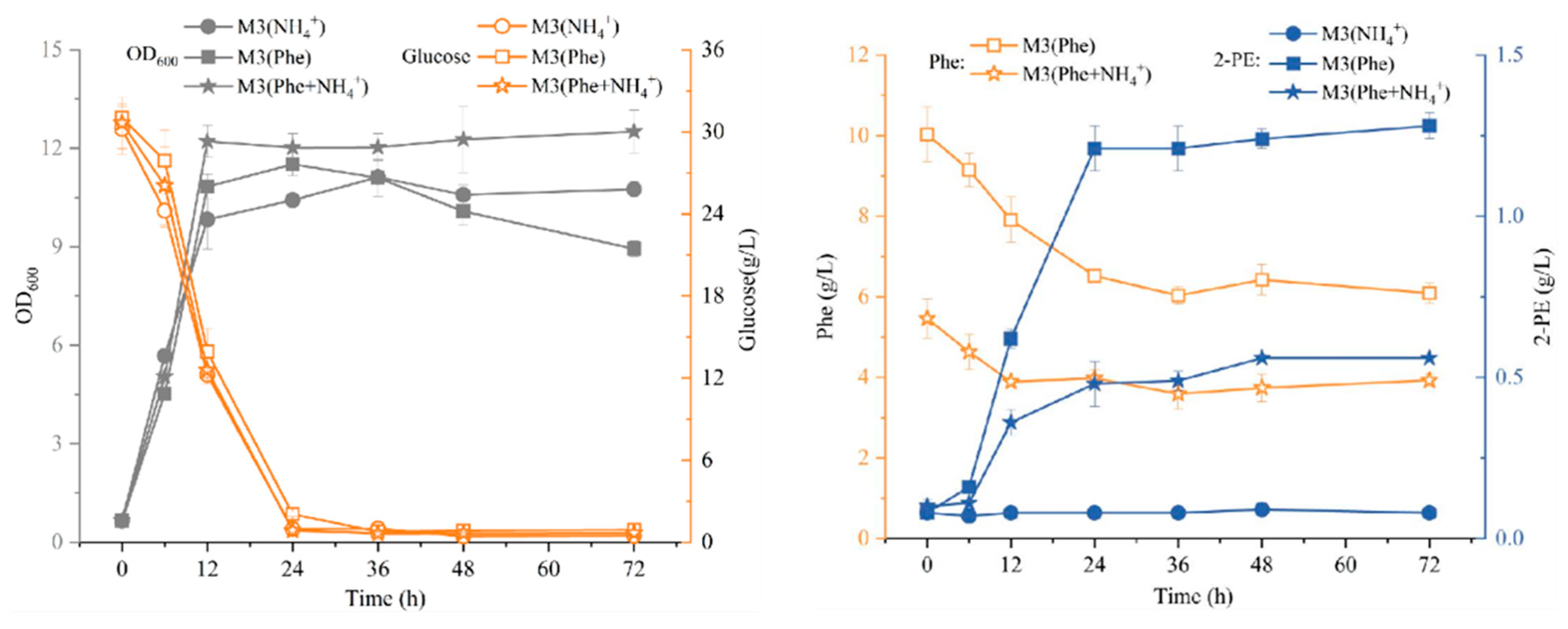

| Glucose (g/L) | Sucrose (g/L) | YNB (g/L) | L-phe (g/L) | (NH4)2SO4 (g/L) | MgSO4·7H2O (g/L) | |
|---|---|---|---|---|---|---|
| M3 (Phe) | 30 | 8 | 1.7 | 9 | - | 0.5 |
| M3 (Phe + NH4+) | 30 | 8 | 1.7 | 4.5 | 2.25 | 0.5 |
| M3 (NH4+) | 30 | 8 | 1.7 | - | 4.5 | 0.5 |
| Primers | Sequences |
|---|---|
| ENO F | 5′-TGCTATTGACGCTGCTGGTTA-3′ |
| ENO R | 5′-GCCTTGGACTTGTCGGAGTTA-3′ |
| YAT F | 5′-GGCGAGGTAGCAGTTAGATTC-3′ |
| YAT R | 5′-TGCGGCGGTCAATTCCAA-3′ |
| GOT1 F | 5′-TCACCAGTCAACTCGCTTACC-3′ |
| GOT1 R | 5′-CAATTTACCACGCAGGGCTTT-3′ |
| hisC F | 5′-GCACCTTCAATGGCACCACTA-3′ |
| hisC R | 5′-GTCAGCAGTAAGAACGGAGA-3′ |
| PDC F | 5′-GGTGTCGTTGATGAGGTTGAGA-3′ |
| PDC R | 5′-GGAAGCGTAGAAGGCGTGAA-3′ |
| ADH5 F | 5′-TGGCTCAACCTATCAAGTGTCA-3′ |
| ADH5 R | 5′-ACCAAGAACAGAAGGCGTGAA-3′ |
| Items | Results | Items | Results |
|---|---|---|---|
| D-pine trisaccharides assimilate | − | L-lysine arylamase | − |
| L-malic acid assimilate | + | D-sorbitol assimilate | + |
| 2-Keto-gluconate assimilate | − | L-rhamnose assimilate | − |
| Glucuronic acid assimilate | + | Xylitol | − |
| Erythritol assimilate | − | D-sorbitol assimilate | + |
| Glycerol assimilate | − | Sucrose assimilate | + |
| D-brown sugar assimilate | − | Urease | − |
| β-N-acetyl glucosaminidase | + | α-glucosidase | − |
| Myric acid assimilate | + | Tyrosine arylamase | − |
| Amygdalin assimilate | − | D-trehalose assimilate | + |
| α-galactose assimilate | + | Nitrate assimilate | − |
| Gentiobiose assimilate | + | L-arabinose assimilate | + |
| D-glucose assimilate | + | Lactose assimilate | + |
| D-galacturaldehyde assimilate | + | Aesculin hydrolysis | + |
| Methyl glucoside assimilate | − | L-Glutamate assimilate | + |
| D-cellobiose assimilate | + | Xylose assimilate | − |
| γ-glutamyl transferase | − | DL-lactate assimilate | − |
| D-maltose assimilate | + | Acetate assimilate | − |
| D-raffinose assimilate | − | Citrate assimilate | − |
| PNP-n-acetyl-BD-galactosidase | − | Arginine | + |
| D-mannose assimilate | + | L-proline assimilate | − |
| D-melibiose assimilate | − | Leucine arylamase | − |
| N-Acetyl-Glucosamine assimilate | − | D-gluconate assimilate | − |
| Assembly Feature | R5 |
|---|---|
| Assembled sequence (bp) | 9,474,513 |
| No. of scaffolds | 250 |
| Sequence depth | 204.03 |
| Maximum contig length (bp) | 4,175,501 |
| N50 length (bp) | 3,806,193 |
| N90 length (bp) | 1,100,691 |
| GC content in genome (%) | 39.62 |
| Strain | S. bacillaris R5 | S. bacillaris PAS13 | S. cerevisiae S288c |
|---|---|---|---|
| Ploidy | n | N | n |
| Genome size (Mb) | 9.47 | 9.4 | 12.3 |
| GC content in genome (%) | 39.62 | 39.45 | 38.3 |
| Total number of CDS | 3991 | 4321 | 5769 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; Song, Q.; Xia, H.; Song, N.; Yang, Q.; Zhang, X.; Yao, L.; Yang, S.; Dai, J.; Chen, X. Isolation and Identification of Non-Saccharomyces Yeast Producing 2-Phenylethanol and Study of the Ehrlich Pathway and Shikimate Pathway. J. Fungi 2023, 9, 878. https://doi.org/10.3390/jof9090878
Zhou R, Song Q, Xia H, Song N, Yang Q, Zhang X, Yao L, Yang S, Dai J, Chen X. Isolation and Identification of Non-Saccharomyces Yeast Producing 2-Phenylethanol and Study of the Ehrlich Pathway and Shikimate Pathway. Journal of Fungi. 2023; 9(9):878. https://doi.org/10.3390/jof9090878
Chicago/Turabian StyleZhou, Rong, Qingyi Song, Huili Xia, Na Song, Qiao Yang, Xiaoling Zhang, Lan Yao, Shihui Yang, Jun Dai, and Xiong Chen. 2023. "Isolation and Identification of Non-Saccharomyces Yeast Producing 2-Phenylethanol and Study of the Ehrlich Pathway and Shikimate Pathway" Journal of Fungi 9, no. 9: 878. https://doi.org/10.3390/jof9090878






