Recent Advances in the Biosynthesis of Natural Sugar Substitutes in Yeast
Abstract
:1. Introduction
2. Biosynthesis of Oligosaccharides
2.1. 2′-Fucosyllactose (2′-FL)
2.2. Trehalose
3. Biosynthesis of Sugar Alcohols
3.1. Mannitol
3.2. Erythritol
3.3. Threitol
4. Biosynthesis of Glycosides
4.1. Rubusosides
4.2. Glycyrrhizin (GL)
5. Biosynthesis of Rare Monosaccharide
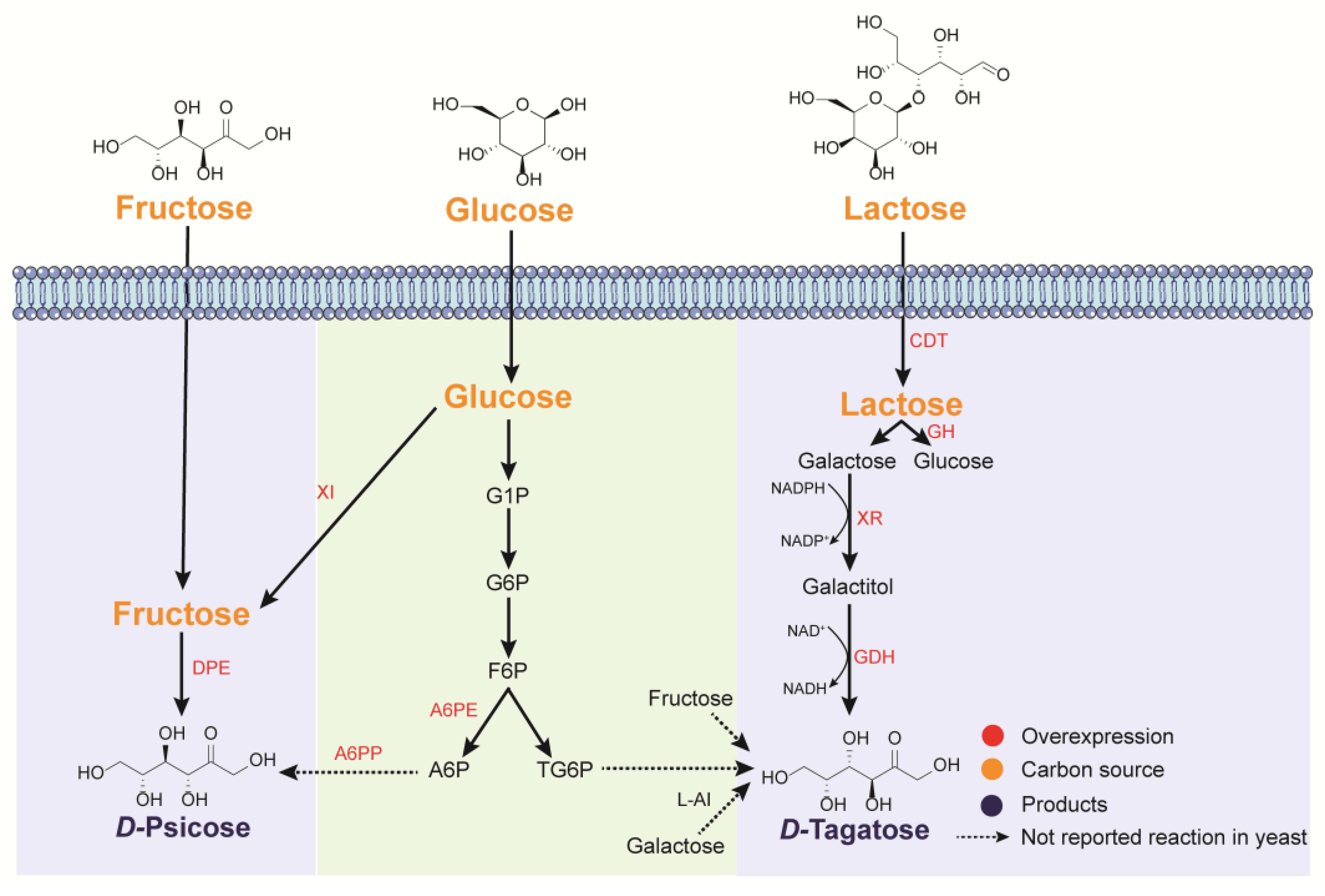
5.1. d-Psicose
5.2. d-Tagatose
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Medina-Remon, A.; Kirwan, R.; Lamuela-Raventos, R.M.; Estruch, R. Dietary patterns and the risk of obesity, type 2 diabetes mellitus, cardiovascular diseases, asthma, and neurodegenerative diseases. Crit. Rev. Food Sci. Nutr. 2018, 58, 262–296. [Google Scholar] [CrossRef]
- Romo-Romo, A.; Aguilar-Salinas, C.A.; Gomez-Diaz, R.A.; Brito-Cordova, G.X.; Gomez-Velasco, D.V.; Lopez-Rocha, M.J.; Almeda-Valdes, P. Non-nutritive sweeteners: Evidence on their association with metabolic diseases and potential effects on glucose metabolism and appetite. Rev. Investig. Clin. 2017, 69, 129–138. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Y.; Liu, Y.; Li, J.; Du, G.; Chen, J.; Lv, X.; Liu, L. Sustainable bioproduction of natural sugar substitutes: Strategies and challenges. Trends Food Sci. Technol. 2022, 129, 512–527. [Google Scholar] [CrossRef]
- World Health Organization. Guideline: Sugars Intake for Adults and Children. 2015. Available online: https://www.ncbi.nlm.nih.gov/books/NBK285537/ (accessed on 2 May 2023).
- Anushkkaran, P. Artificial sweeteners. Int. J. Res. Rev. 2019, 6, 120–128. [Google Scholar]
- Li, K.; Zheng, J.; Yu, L.; Wang, B.; Pan, L. Exploration of the strategy for improving the expression of heterologous sweet protein monellin in Aspergillus niger. J. Fungi 2023, 9, 528. [Google Scholar] [CrossRef]
- Park, Y.C.; Oh, E.J.; Jo, J.H.; Jin, Y.S.; Seo, J.H. Recent advances in biological production of sugar alcohols. Curr. Opin. Biotechnol. 2016, 37, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, K.; Miao, L.; Rong, L.; Zhao, Y.; Li, S.; Ma, L.; Li, J.; Zhang, C.; Xiao, D.; et al. Simultaneous improvement of limonene production and tolerance in Yarrowia lipolytica through tolerance engineering and evolutionary engineering. ACS Synth. Biol. 2021, 10, 884–896. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wu, X.; Ma, H.; Li, J.; Liu, Z.; Guo, X.; Dong, J.; Zou, S.; Luo, Y. High-level production of hydroxytyrosol in engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2022, 11, 3706–3713. [Google Scholar] [CrossRef]
- Liu, H.; Tian, Y.; Zhou, Y.; Kan, Y.; Wu, T.; Xiao, W.; Luo, Y. Multi-modular engineering of Saccharomyces cerevisiae for high-titre production of tyrosol and salidroside. Microb. Biotechnol. 2020, 14, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, X.; Zhang, C.; Zhou, X.; Xu, X.; Han, L.; Lv, X.; Liu, Y.; Liu, S.; Li, J.; et al. De novo biosynthesis of rubusoside and rebaudiosides in engineered yeasts. Nat. Commun. 2022, 13, 3040. [Google Scholar] [CrossRef]
- Guo, X.; Wu, X.; Ma, H.; Liu, H.; Luo, Y. Yeast: A platform for the production of (L)-tyrosine derivatives. Yeast 2023, 40, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rong, L.; Zhao, Y.; Li, S.; Zhang, C.; Xiao, D.; Foo, J.L.; Yu, A. Next-generation metabolic engineering of non-conventional microbial cell factories for carboxylic acid platform chemicals. Biotechnol. Adv. 2020, 43, 107605. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Xu, Y.; Xu, S.; Bilal, M.; Cheng, H. Engineering thermotolerant Yarrowia lipolytica for sustainable biosynthesis of mannitol and fructooligosaccharides. Biochem. Eng. J. 2022, 187, 108604. [Google Scholar] [CrossRef]
- Yang, S.; Pan, X.; Wang, Q.; Lv, Q.; Zhang, X.; Zhang, R.; Rao, Z. Enhancing erythritol production from crude glycerol in a wild-type Yarrowia lipolytica by metabolic engineering. Front. Microbiol. 2022, 13, 1054243. [Google Scholar] [CrossRef]
- Chi, P.; Wang, S.; Ge, X.; Bilal, M.; Fickers, P.; Cheng, H. Efficient D-threitol production by an engineered strain of Yarrowia lipolytica overexpressing xylitol dehydrogenase gene from Scheffersomyces stipitis. Biochem. Eng. J. 2019, 149, 107259. [Google Scholar] [CrossRef]
- Zhu, Y.Y.; Wan, L.; Li, W.; Ni, D.W.; Zhang, W.L.; Yan, X.; Mu, W.M. Recent advances on 2′-fucosyllactose: Physiological properties, applications, and production approaches. Crit. Rev. Food Sci. Nut. 2022, 62, 2083–2092. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Zhang, L.B.; Li, L.J.; Liu, D.W.; Cheng, H.L.; Wang, H.W.; Nawaz, M.Z.; Cheng, H.R.; Deng, Z.X. An alternative approach to synthesizing galactooligosaccharides by cell-surface display of beta-galactosidase on Yarrowia lipolytica. J. Agric. Food Chem. 2016, 64, 3819–3827. [Google Scholar] [CrossRef]
- Kunz, C.; Rudloff, S. Health promoting aspects of milk oligosaccharides. Int. Dairy J. 2006, 16, 1341–1346. [Google Scholar] [CrossRef]
- Du, Z.; Liu, Z.; Tan, Y.; Niu, K.; Guo, W.; Jia, Y.; Fang, X. Lacto-N-biose synthesis via a modular enzymatic cascade with ATP regeneration. iScience 2021, 24, 102236. [Google Scholar] [CrossRef]
- Li, F.; Ma, W.; Liu, L.; Niu, K.; Liu, D.; Yin, W.; Zhang, X.; Han, L.; Fang, X. Reprogramming the Metabolic Network in Kluyveromyces lactis with a transcriptional switch for de novo lacto-N-biose synthesis. J. Agric. Food Chem. 2023, 71, 9031–9039. [Google Scholar] [CrossRef]
- Yu, S.; Liu, J.J.; Yun, E.J.; Kwak, S.; Kim, K.H.; Jin, Y.S. Production of a human milk oligosaccharide 2′-fucosyllactose by metabolically engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2018, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, Y.; Wang, H.; Wan, L.; Zhang, W.; Mu, W. Strategies for enhancing microbial production of 2′-fucosyllactose, the most abundant human milk oligosaccharide. J. Agric. Food Chem. 2022, 70, 11481–11499. [Google Scholar] [CrossRef] [PubMed]
- Albermann, C.; Piepersberg, W.; Wehmeier, U.F. Synthesis of the milk oligosaccharide 2′-fucosyllactose using recombinant bacterial enzymes. Carbohyd. Res. 2001, 334, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.Y.; Mosleh, I.; Abbaspourrad, A. Engineered microbial routes for human milk oligosaccharides synthesis. ACS Synth. Biol. 2021, 10, 923–938. [Google Scholar] [CrossRef]
- Coyne, M.J.; Reinap, B.; Lee, M.M.; Comstock, L.E. Human symbionts use a host-like pathway for surface fucosylation. Science 2005, 307, 1778–1781. [Google Scholar] [CrossRef]
- Liu, J.J.; Kwak, S.; Pathanibul, P.; Lee, J.W.; Yu, S.; Yun, E.J.; Lim, H.; Kim, K.H.; Jin, Y.S. Biosynthesis of a functional human milk oligosaccharide, 2′-fucosyllactose, and l-fucose using engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2018, 7, 2529–2536. [Google Scholar] [CrossRef]
- Hollands, K.; Baron, C.M.; Gibson, K.J.; Kelly, K.J.; Krasley, E.A.; Laffend, L.A.; Lauchli, R.M.; Maggio-Hall, L.A.; Nelson, M.J.; Prasad, J.C.; et al. Engineering two species of yeast as cell factories for 2′-fucosyllactose. Metab. Eng. 2019, 52, 232–242. [Google Scholar] [CrossRef]
- Lee, J.W.; Kwak, S.; Liu, J.J.; Yu, S.; Yun, E.J.; Kim, D.H.; Liu, C.; Kim, K.H.; Jin, Y.S. Enhanced 2′-fucosyllactose production by engineered Saccharomyces cerevisiae using xylose as a co-substrate. Metab. Eng. 2020, 62, 322–329. [Google Scholar] [CrossRef]
- Xu, M.; Meng, X.; Zhang, W.; Shen, Y.; Liu, W. Improved production of 2′-fucosyllactose in engineered Saccharomyces cerevisiae expressing a putative α-1, 2-fucosyltransferase from Bacillus cereus. Microb. Cell Fact. 2021, 20, 165–176. [Google Scholar] [CrossRef]
- Xu, M.; Sun, M.; Meng, X.; Zhang, W.; Shen, Y.; Liu, W. Engineering pheromone-mediated quorum sensing with enhanced response output increases fucosyllactose production in Saccharomyces cerevisiae. ACS Synth. Biol. 2022, 12, 238–248. [Google Scholar] [CrossRef]
- Liu, J.J.; Zhang, G.C.; Kwak, S.; Oh, E.J.; Yun, E.J.; Chomvong, K.; Cate, J.H.D.; Jin, Y.S. Overcoming the thermodynamic equilibrium of an isomerization reaction through oxidoreductive reactions for biotransformation. Nat. Commun. 2019, 10, 1356. [Google Scholar] [CrossRef]
- Peng, B.; Shen, Y.; Li, X.; Chen, X.; Hou, J.; Bao, X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 9–18. [Google Scholar] [CrossRef]
- Li, N.; Wang, H.W.; Li, L.J.; Cheng, H.L.; Liu, D.W.; Cheng, H.R.; Deng, Z.X. Integrated approach to producing high-purity trehalose from maltose by the yeast Yarrowia lipolytica displaying trehalose synthase (TreS) on the cell surface. J. Agric. Food Chem. 2016, 64, 6179–6187. [Google Scholar] [CrossRef]
- Costenoble, R.; Adler, L.; Niklasson, C.; Liden, G. Engineering of the metabolism of Saccharomyces cerevisiae for anaerobic production of mannitol. FEMS Yeast Res. 2003, 3, 17–25. [Google Scholar] [CrossRef]
- Savergave, L.S.; Gadre, R.V.; Vaidya, B.K.; Jogdand, V.V. Two-stage fermentation process for enhanced mannitol production using Candida magnoliae mutant R9. Bioprocess Biosyst. Eng. 2013, 36, 193–203. [Google Scholar] [CrossRef]
- Meng, Q.; Zhang, T.; Wei, W.; Mu, W.; Miao, M. Production of Mannitol from a high concentration of glucose by Candida parapsilosis SK26.001. Appl. Biochem. Biotechnol. 2017, 181, 391–406. [Google Scholar] [CrossRef]
- Lee, J.K.; Oh, D.K.; Song, H.Y.; Kim, I.W. Ca2+ and Cu2+ supplementation increases mannitol production by Candida magnoliae. Biotechnol. Lett. 2007, 29, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Mironczuk, A.M.; Furgala, J.; Rakicka, M.; Rymowicz, W. Enhanced production of erythritol by Yarrowia lipolytica on glycerol in repeated batch cultures. J. Ind. Microbiol. Biotechnol. 2014, 41, 57–64. [Google Scholar] [CrossRef]
- Qiu, X.; Xu, P.; Zhao, X.; Du, G.; Zhang, J.; Li, J. Combining genetically-encoded biosensors with high throughput strain screening to maximize erythritol production in Yarrowia lipolytica. Metab. Eng. 2020, 60, 66–76. [Google Scholar] [CrossRef]
- Wang, N.; Chi, P.; Zou, Y.; Xu, Y.; Xu, S.; Bilal, M.; Fickers, P.; Cheng, H. Metabolic engineering of Yarrowia lipolytica for thermoresistance and enhanced erythritol productivity. Biotechnol. Biofuels 2020, 13, 176. [Google Scholar] [CrossRef]
- Jeya, M.; Lee, K.-M.; Tiwari, M.K.; Kim, J.-S.; Gunasekaran, P.; Kim, S.-Y.; Kim, I.-W.; Lee, J.-K. Isolation of a novel high erythritol-producing Pseudozyma tsukubaensis and scale-up of erythritol fermentation to industrial level. Appl. Microbiol. Biotechnol. 2009, 83, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Suwanapetch, C.; Vanichsriratana, W. Media optimization for erythritol production by Moniliella sp. BCC25224. Sugar Tech 2022, 25, 257–261. [Google Scholar] [CrossRef]
- Xu, K.; Zhao, Y.J.; Ahmad, N.; Wang, J.N.; Lv, B.; Wang, Y.; Ge, J.; Li, C. O-glycosyltransferases from homo sapiens contributes to the biosynthesis of glycyrrhetic acid 3-O-mono-beta-D-glucuronide and glycyrrhizin in Saccharomyces cerevisiae. Synth. Syst. Biotechnol. 2021, 6, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.Z.; Zhu, X.X.; Zheng, Z.; Mu, D.D.; Jiang, S.T.; Luo, S.Z.; Wu, Y.; Du, M.R. Cell regeneration and cyclic catalysis of engineered Kluyveromyces marxianus of a d-psicose-3-epimerase gene from Agrobacterium tumefaciens for d-allulose production. World J. Microbiol. Biotechnol. 2018, 34, 7. [Google Scholar] [CrossRef] [PubMed]
- Pilauri, V.; Bewley, M.; Diep, C.; Hopper, J. Gal80 dimerization and the yeast GAL gene switch. Genetics 2005, 169, 1903–1914. [Google Scholar] [CrossRef]
- Cai, X.; Seitl, I.; Mu, W.; Zhang, T.; Stressler, T.; Fischer, L.; Jiang, B. Biotechnical production of trehalose through the trehalose synthase pathway: Current status and future prospects. Appl. Environ. Microbiol. 2018, 102, 2965–2976. [Google Scholar] [CrossRef]
- Wang, J.Q.; Ren, X.D.; Wang, R.M.; Su, J.; Wang, F. Structural characteristics and function of a new kind of thermostable trehalose synthase from Thermobaculum terrenum. J. Agric. Food Chem. 2017, 65, 7726–7735. [Google Scholar] [CrossRef]
- Kim, T.K.; Jang, J.H.; Cho, H.Y.; Lee, H.S.; Kim, Y.W. Gene cloning and characterization of a trehalose synthase from Corynebacterium glutamicum ATCC13032. Food Sci. Biotechnol. 2010, 19, 565–569. [Google Scholar] [CrossRef]
- Ferreira, J.C.; Thevelein, J.M.; Hohmann, S.; Paschoalin, V.M.; Trugo, L.C.; Panek, A.D. Trehalose accumulation in mutants of Saccharomyces cerevisiae deleted in the UDPG-dependent trehalose synthase-phosphatase complex. Biochim. Biophys. Acta 1997, 1335, 40–50. [Google Scholar] [CrossRef]
- Hohmann, S.; Bell, W.; Neves, M.J.; Valckx, D.; Thevelein, J.M. Evidence for trehalose-6-phosphate-dependent and -independent mechanisms in the control of sugar influx into yeast glycolysis. Mol. Microbiol. 1996, 20, 981–991. [Google Scholar] [CrossRef]
- Torres, E.M.; Dephoure, N.; Panneerselvam, A.; Tucker, C.M.; Whittaker, C.A.; Gygi, S.P.; Dunham, M.J.; Amon, A. Identification of aneuploidy-tolerating mutations. Cell 2010, 143, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Liang, P.; Cao, M.; Li, J.; Wang, Q.; Dai, Z. Expanding sugar alcohol industry: Microbial production of sugar alcohols and associated chemocatalytic derivatives. Biotechnol. Adv. 2023, 64, 108105. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, C.; Ferreira, C.; Goncalves, L.G.; Turner, D.L.; Leandro, M.J.; Salema-Oom, M.; Santos, H.; Goncalves, P. A new pathway for mannitol metabolism in yeasts suggests a link to the evolution of alcoholic fermentation. Front. Microbiol. 2019, 10, 2510. [Google Scholar] [CrossRef] [PubMed]
- Patra, F.; Tomar, S.K.; Arora, S. Technological and functional applications of low-calorie sweeteners from Lactic Acid Bacteria. J. Food Sci. 2009, 74, R16–R23. [Google Scholar] [CrossRef]
- Novotny, M.J.; Reizer, J.; Esch, F.; Saier, M.H. Purification and properties of D-mannitol-1-phosphate dehydrogenase and D-glucitol-6-phosphate dehydrogenase from Escherichia coli. J. Bacteriol. 1984, 159, 986–990. [Google Scholar] [CrossRef]
- Bilal, M.; Xu, S.; Iqbal, H.M.N.; Cheng, H. Yarrowia lipolytica as an emerging biotechnological chassis for functional sugars biosynthesis. Crit. Rev. Food. Sci. Nutr. 2021, 61, 535–552. [Google Scholar] [CrossRef]
- Zhu, H.; Li, J.; Su, Y.; Gu, L.; Chang, C.; Yang, Y. Sugar alcohols as cryoprotectants of egg yolk: Inhibiting crystals and interactions. J. Food Eng. 2023, 342, 111360. [Google Scholar] [CrossRef]
- Vidal, L.; Lebrun, E.; Park, Y.K.; Mottet, G.; Nicaud, J.M. Bidirectional hybrid erythritol-inducible promoter for synthetic biology in Yarrowia lipolytica. Microb. Cell Fact. 2023, 22, 7. [Google Scholar] [CrossRef]
- Mirończuk, A.M.; Dobrowolski, A.; Rakicka, M.; Rywińska, A.; Rymowicz, W. Newly isolated mutant of Yarrowia lipolytica MK1 as a proper host for efficient erythritol biosynthesis from glycerol. Process Biochem. 2015, 50, 61–68. [Google Scholar] [CrossRef]
- Rakicka, M.; Mironczuk, A.M.; Tomaszewska-Hetman, L.; Rywinska, A.; Rymowicz, W. An effective method of continuous production of erythritol from glycerol by Yarrowia lipolytica MK1. Food Technol. Biotech. 2017, 55, 125–130. [Google Scholar] [CrossRef]
- Shahsavarani, H.; Sugiyama, M.; Kaneko, Y.; Chuenchit, B.; Harashima, S. Superior thermotolerance of Saccharomyces cerevisiae for efficient bioethanol fermentation can be achieved by overexpression of RSP5 ubiquitin ligase. Biotechnol. Adv. 2012, 30, 1289–1300. [Google Scholar] [CrossRef]
- Liang, P.; Li, J.; Wang, Q.; Dai, Z. Enhancing the thermotolerance and erythritol production of Yarrowia lipolytica by introducing heat-resistant devices. Front. Bioeng. Biotechnol. 2023, 11, 1108653. [Google Scholar] [CrossRef]
- Carly, F.; Vandermies, M.; Telek, S.; Steels, S.; Thomas, S.; Nicaud, J.M.; Fickers, P. Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metab. Eng. 2017, 42, 19–24. [Google Scholar] [CrossRef]
- Mironczuk, A.M.; Rzechonek, D.A.; Biegalska, A.; Rakicka, M.; Dobrowolski, A. A novel strain of Yarrowia lipolytica as a platform for value-added product synthesis from glycerol. Biotechnol. Biofuels 2016, 9, 180. [Google Scholar] [CrossRef]
- Jagtap, S.S.; Bedekar, A.A.; Singh, V.; Jin, Y.S.; Rao, C.V. Metabolic engineering of the oleaginous yeast Yarrowia lipolytica PO1f for production of erythritol from glycerol. Biotechnol. Biofuels 2021, 14, 188. [Google Scholar] [CrossRef]
- Köpf-Maier, P.; Saß, G. Antitumor activity of treosulfan against human breast carcinomas. Cancer Chemoth. Pharm. 1992, 31, 103–110. [Google Scholar] [CrossRef]
- Miller, L.K.; Smith, J.S. Production of threitol and sorbitol by an adult insect: Association with freezing tolerance. Nature 1975, 258, 519–520. [Google Scholar] [CrossRef]
- Rizzi, M.; Harwart, K.; Erlemann, P.; Bui-Thanh, N.-A.; Dellweg, H. Purification and properties of the NAD+-xylitol-dehydrogenase from the yeast Pichia stipitis. J. Ferment. Bioeng. 1989, 67, 20–24. [Google Scholar] [CrossRef]
- Richman, A.; Swanson, A.; Humphrey, T.; Chapman, R.; McGarvey, B.; Pocs, R.; Brandle, J. Functional genomics uncovers three glucosyltransferases involved in the synthesis of the major sweet glucosides of Stevia rebaudiana. Plant J. 2005, 41, 56–67. [Google Scholar] [CrossRef]
- Li, Y.; Luo, H.M.; Sun, C.; Song, J.Y.; Sun, Y.Z.; Wu, Q.; Wang, N.; Yao, H.; Steinmetz, A.; Chen, S.L. EST analysis reveals putative genes involved in glycyrrhizin biosynthesis. BMC Genom. 2010, 11, 268. [Google Scholar] [CrossRef]
- Itkin, M.; Davidovich-Rikanati, R.; Cohen, S.; Portnoy, V.; Doron-Faigenboim, A.; Oren, E.; Freilich, S.; Tzuri, G.; Baranes, N.; Shen, S.; et al. The biosynthetic pathway of the nonsugar, high-intensity sweetener mogroside V from Siraitia grosvenorii. Proc. Natl. Acad. Sci. USA 2016, 113, E7619–E7628. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Wang, Y.; Chen, L.; Yan, M.; Chen, K.; Xu, L.; Ouyang, P. Production of rebaudioside A from stevioside catalyzed by the engineered Saccharomyces cerevisiae. Appl. Biochem. Biotechnol. 2016, 178, 1586–1598. [Google Scholar] [CrossRef]
- Chen, L.; Cai, R.; Weng, J.; Li, Y.; Jia, H.; Chen, K.; Yan, M.; Ouyang, P. Production of rebaudioside D from stevioside using a UGTSL2 Asn358Phe mutant in a multi-enzyme system. Microb. Biotechnol. 2020, 13, 974–983. [Google Scholar] [CrossRef]
- Urban, J.D.; Carakostas, M.C.; Taylor, S.L. Steviol glycoside safety: Are highly purified steviol glycoside sweeteners food allergens? Food Chem. Toxicol. 2015, 75, 71–78. [Google Scholar] [CrossRef]
- Olsson, K.; Carlsen, S.; Semmler, A.; Simon, E.; Mikkelsen, M.D.; Moller, B.L. Microbial production of next-generation stevia sweeteners. Microb. Cell Fact. 2016, 15, 207. [Google Scholar] [CrossRef]
- Shu, W.; Zheng, H.; Fu, X.; Zhen, J.; Tan, M.; Xu, J.; Zhao, X.; Yang, S.; Song, H.; Ma, Y. Enhanced Heterologous Production of glycosyltransferase UGT76G1 by co-expression of endogenous prpD and malK in Escherichia coli and Its transglycosylation application in production of rebaudioside. Int. J. Mol. Sci. 2020, 21, 5752. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, J.; Ma, S.; Huang, T.; Ma, Y.; Liu, W.; Liu, W.; Liu, Z.; Song, H. Heterologous expression of EUGT11 from Oryza sativa in Pichia pastoris for highly efficient one-pot production of rebaudioside D from rebaudioside A. Int. J. Biol. Macromol. 2020, 163, 1669–1676. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, X.; Chen, L.; Mustapha, A.T.; Yu, X.; Zhou, C.; Okonkwo, C.E. Natural and low-caloric rebaudioside A as a substitute for dietary sugars: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2023, 22, 615–642. [Google Scholar] [CrossRef]
- Mizutani, K.; Kuramoto, T.; Tamura, Y.; Ohtake, N.; Doi, S.; Nakaura, M.; Tanaka, O. Sweetness of glycyrrhetic acid 3-O-beta-D-monoglucuronide and the related glycosides. Biosci. Biotechnol. Biochem. 1994, 58, 554–555. [Google Scholar] [CrossRef]
- Zhao, Y.; Lv, B.; Feng, X.; Li, C. Perspective on Biotransformation and de novo biosynthesis of licorice constituents. J. Agric. Food Chem. 2017, 65, 11147–11156. [Google Scholar] [CrossRef]
- Kirby, J.; Romanini, D.W.; Paradise, E.M.; Keasling, J.D. Engineering triterpene production in Saccharomyces cerevisiae-beta-amyrin synthase from Artemisia annua. FEBS J. 2008, 275, 1852–1859. [Google Scholar] [CrossRef]
- Seki, H.; Sawai, S.; Ohyama, K.; Mizutani, M.; Ohnishi, T.; Sudo, H.; Fukushima, E.O.; Akashi, T.; Aoki, T.; Saito, K.; et al. Triterpene functional genomics in licorice for identification of CYP72A154 involved in the biosynthesis of glycyrrhizin. Plant Cell 2011, 23, 4112–4123. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.L.; Bai, P.; Liu, T.; Li, D.S.; Zhang, X.M.; Lu, W.Y.; Yuan, Y.J. Optimization of a cytochrome P450 oxidation system for enhancing protopanaxadiol production in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2016, 113, 1787–1795. [Google Scholar] [CrossRef]
- Chen, K.; Hu, Z.M.; Song, W.; Wang, Z.L.; He, J.B.; Shi, X.M.; Cui, Q.H.; Qiao, X.; Ye, M. Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in Glycyrrhiza uralensis. ACS Synth. Biol. 2019, 8, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Chen, K.; Hu, Z.-m.; Li, K.; Song, W.; Yu, L.-y.; Leung, C.-H.; Ma, D.-L.; Qiao, X.; Ye, M. UGT73F17, a new glycosyltransferase from Glycyrrhiza uralensis, catalyzes the regiospecific glycosylation of pentacyclic triterpenoids. Chem. Commun. 2018, 54, 8594–8597. [Google Scholar] [CrossRef]
- Nomura, Y.; Seki, H.; Suzuki, T.; Ohyama, K.; Mizutani, M.; Kaku, T.; Tamura, K.; Ono, E.; Horikawa, M.; Sudo, H.; et al. Functional specialization of UDP-glycosyltransferase 73P12 in licorice to produce a sweet triterpenoid saponin, glycyrrhizin. Plant J. 2019, 99, 1127–1143. [Google Scholar] [CrossRef]
- Zhu, M.; Wang, C.; Sun, W.; Zhou, A.; Wang, Y.; Zhang, G.; Zhou, X.; Huo, Y.; Li, C. Boosting 11-oxo-beta-amyrin and glycyrrhetinic acid synthesis in Saccharomyces cerevisiae via pairing novel oxidation and reduction system from legume plants. Metab. Eng. 2018, 45, 43–50. [Google Scholar] [CrossRef]
- Chahed, A.; Nesler, A.; Aziz, A.; Barka, E.A.; Pertot, I.; Perazzolli, M. A review of knowledge on the mechanisms of action of the rare sugar d-tagatose against phytopathogenic oomycetes. Plant Pathol. 2021, 70, 1979–1986. [Google Scholar] [CrossRef]
- Jiang, S.; Xiao, W.; Zhu, X.; Yang, P.; Zheng, Z.; Lu, S.; Jiang, S.; Zhang, G.; Liu, J. Review on d-allulose: In vivo metabolism, catalytic mechanism, engineering strain construction, bio-production technology. Front. Bioeng. Biotechnol. 2020, 8, 26. [Google Scholar] [CrossRef]
- Li, Y.J.; Shi, T.; Han, P.P.; You, C. Thermodynamics-driven production of value-added d-allulose from inexpensive dtarch by an in vitro enzymatic synthetic biosystem. ACS Catal. 2021, 11, 5088–5099. [Google Scholar] [CrossRef]
- Seo, M.J.; Kwon, E.R.; Kim, S.J.; Choi, M.S.; Kim, Y.S.; Park, C.S. D-allulose production from d-fructose by putative dolichol phosphate mannose synthase from bacillus sp. with potential d-allulose 3-epimrase activity. Biotechnol. Bioproc. E 2021, 26, 976–984. [Google Scholar] [CrossRef]
- Park, C.S.; Kim, T.; Hong, S.H.; Shin, K.C.; Kim, K.R.; Oh, D.K. D-allulose production from d-fructose by permeabilized recombinant cells of Corynebacterium glutamicum cells expressing d-allulose 3-epimerase flavonifractor plautii. PLoS ONE 2016, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Signori, L.; Passolunghi, S.; Ruohonen, L.; Porro, D.; Branduardi, P. Effect of oxygenation and temperature on glucose-xylose fermentation in Kluyveromyces marxianus CBS712 strain. Microb. Cell Fact. 2014, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Y.; Duan, S.; Liu, J.; Yuan, P.; Nakanishi, H.; Gao, X.D. Bioconversion of d-glucose to d-psicose with immobilized d-xylose isomerase and d-psicose 3-epimerase on Saccharomyces cerevisiae spores. J. Ind. Microbiol. Biotechnol. 2015, 42, 1117–1128. [Google Scholar] [CrossRef]
- Torres, P.; Batista-Viera, F. Production of d-tagatose and d-fructose from whey by co-immobilized enzymatic system. Mol. Catal. 2019, 463, 99–109. [Google Scholar] [CrossRef]
- Ravikumar, Y.; Ponpandian, L.N.; Zhang, G.; Yun, J.; Qi, X. Harnessing l-arabinose isomerase for biological production of d-tagatose: Recent advances and its applications. Trends Food Sci. Technol. 2021, 107, 16–30. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, R.; Wang, Q.; Hu, M.; Li, Z.; Xu, M.; Yang, T.; Zhang, R.; Rao, Z. Production of d-tagatose by whole-cell conversion of recombinant Bacillus subtilis in the absence of antibiotics. Biology 2021, 10, 1343. [Google Scholar] [CrossRef]
- Wanarska, M.; Kur, J. A method for the production of d-tagatose using a recombinant Pichia pastoris strain secreting beta-d-galactosidase from Arthrobacter chlorophenolicus and a recombinant l-arabinose isomerase from Arthrobacter sp. 22c. Microb. Cell Fact. 2012, 11, 113. [Google Scholar] [CrossRef]
- Kim, H.J.; Kang, S.Y.; Park, J.J.; Kim, P. Novel activity of UDP-galactose-4-epimerase for free monosaccharide and activity improvement by active site-saturation mutagenesis. Appl. Biochem. Biotechnol. 2011, 163, 444–451. [Google Scholar] [CrossRef]
- Beerens, K.; Soetaert, W.; Desmet, T. UDP-hexose 4-epimerases: A view on structure, mechanism and substrate specificity. Carbohydr. Res. 2015, 414, 8–14. [Google Scholar] [CrossRef]
- Shin, K.-C.; Lee, T.-E.; Seo, M.-J.; Kim, D.W.; Kang, L.-W.; Oh, D.-K. Development of tagaturonate 3-epimerase into tagatose 4-epimerase with a biocatalytic route from fructose to tagatose. ACS Catal. 2020, 10, 12212–12222. [Google Scholar] [CrossRef]
- Dai, Y.; Li, C.; Zheng, L.; Jiang, B.; Zhang, T.; Chen, J. Enhanced biosynthesis of d-tagatose from maltodextrin through modular pathway engineering of recombinant Escherichia coli. Biochem. Eng. J. 2022, 178, 108303. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Li, G.; Savolainen, O.; Chen, Y.; Nielsen, J. De novo biosynthesis of bioactive isoflavonoids by engineered yeast cell factories. Nat. Commun. 2021, 12, 6085. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Gao, C.; Ye, C.; Guo, L.; Liu, J.; Chen, X.; Song, W.; Wu, J.; Liu, L. Systems engineering of Escherichia coli for high-level shikimate production. Metab. Eng. 2023, 75, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gainza, P.; Wehrle, S.; Van Hall-Beauvais, A.; Marchand, A.; Scheck, A.; Harteveld, Z.; Buckley, S.; Ni, D.; Tan, S.; Sverrisson, F.; et al. De novo design of protein interactions with learned surface fingerprints. Nature 2023, 617, 176–184. [Google Scholar] [CrossRef]



| Types | Products | Hosts | Substrate | Fermentation Condition | Titers (g/L) | Yield (g/g) | Productivity (g/L/h) | Reference |
|---|---|---|---|---|---|---|---|---|
| Oligosaccharides | 2′-Fucosyllactose | S. cerevisiae | Fucose and Lactose | Shake Flask | 0.503 | NA 1 | NA | [22] |
| S. cerevisiae | Xylose and Lactose | Fed-Batch Bioreactor | 25.5 | NA | 0.35 | [29] | ||
| S. cerevisiae | Glucose and Lactose | 5 L Fed-Batch Bioreactor | 26.63 | NA | 0.28 | [30] | ||
| S. cerevisiae | Glucose and Lactose | 5 L Fed-Batch Bioreactor | 32.05 | NA | 0.67 | [31] | ||
| Trehalose | Y. lipolytica | Maltose | 3 L Batch Bioreactor | 219 | 0.73 | 4.5 | [34] | |
| Sugar alcohols | Mannitol | S. cerevisiae | Glucose | Shake Flask | NA | 0.12 | NA | [35] |
| C. magnoliae | Glucose | 2.5 L Fed-Batch Bioreactor | 240 | 0.81 | 4 | [36] | ||
| C. parapsilosis | Glucose | 30 L Fed-Batch Bioreactor | 97.1 | 0.34 | 0.81 | [37] | ||
| C. magnoliae | Glucose | 10 L Fed-Batch Bioreactor | 223 | 0.89 | 1.72 | [38] | ||
| Y. lipolytica | Glucose | 5 L Fed-Batch Bioreactor | 98.2 | 0.33 | 1.1 | [14] | ||
| Erythritol | Y. lipolytica | Glycerol | 5 L Fed-batch Bioreactor | 220 | 0.43 | 0.54 | [39] | |
| Y. lipolytica | Glucose | Bench-Top Reactors | 148 | NA | NA | [40] | ||
| Sugar alcohols | Erythritol | Y. lipolytica | Glucose | 30 m3 Fed-Batch Bioreactor | 196 | 0.65 | 2.51 | [41] |
| Y. lipolytica | Crude glycerol | 5 L Fed-Batch Bioreactor | 150 | 0.62 | 1.25 | [15] | ||
| P. tsukubaensis | Glucose | 5 L Fed-Batch Bioreactor | 245 | 0.61 | 2.86 | [42] | ||
| Moniliella | Glucose and soybean flour | 10 L Fed-batch Bioreactor | 86.6 | 0.47 | 0.40 | [43] | ||
| Threitol | Y. lipolytica | Glucose | Shake Flask | 112 | 0.37 | NA | [16] | |
| Glycosides | Rubusoside | S. cerevisiae | Glucose | 15 L Fed-Batch Bioreactor | 1.37 | NA | NA | [11] |
| Reb A | S. cerevisiae | Glucose | 15 L Fed-Batch Bioreactor | 21.5 mg/L | NA | NA | [11] | |
| Reb D | S. cerevisiae | Glucose | 15 L Fed-Batch Bioreactor | 44.2 mg/L | NA | NA | [11] | |
| Reb M | S. cerevisiae | Glucose | 15 L Fed-Batch Bioreactor | 67.0 mg/L | NA | NA | [11] | |
| GL | S. cerevisiae | Glucose | Shake Flask | 5.98 mg/L | NA | NA | [44] | |
| Rare monosaccharide | d-Psicose | K. marxianus | Fructose | Whole-cell Reaction | 190 | 0.253 | 15.83 | [45] |
| d-Tagatose | S. cerevisiae | Lactose | 2 L Fed-Batch Bioreactor | 37.69 | 0.526 | 0.1126 | [32] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, H.; Liu, H.; Luo, Y. Recent Advances in the Biosynthesis of Natural Sugar Substitutes in Yeast. J. Fungi 2023, 9, 907. https://doi.org/10.3390/jof9090907
Li J, Li H, Liu H, Luo Y. Recent Advances in the Biosynthesis of Natural Sugar Substitutes in Yeast. Journal of Fungi. 2023; 9(9):907. https://doi.org/10.3390/jof9090907
Chicago/Turabian StyleLi, Jian, Honghao Li, Huayi Liu, and Yunzi Luo. 2023. "Recent Advances in the Biosynthesis of Natural Sugar Substitutes in Yeast" Journal of Fungi 9, no. 9: 907. https://doi.org/10.3390/jof9090907
APA StyleLi, J., Li, H., Liu, H., & Luo, Y. (2023). Recent Advances in the Biosynthesis of Natural Sugar Substitutes in Yeast. Journal of Fungi, 9(9), 907. https://doi.org/10.3390/jof9090907





