Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances
Abstract
:1. Introduction
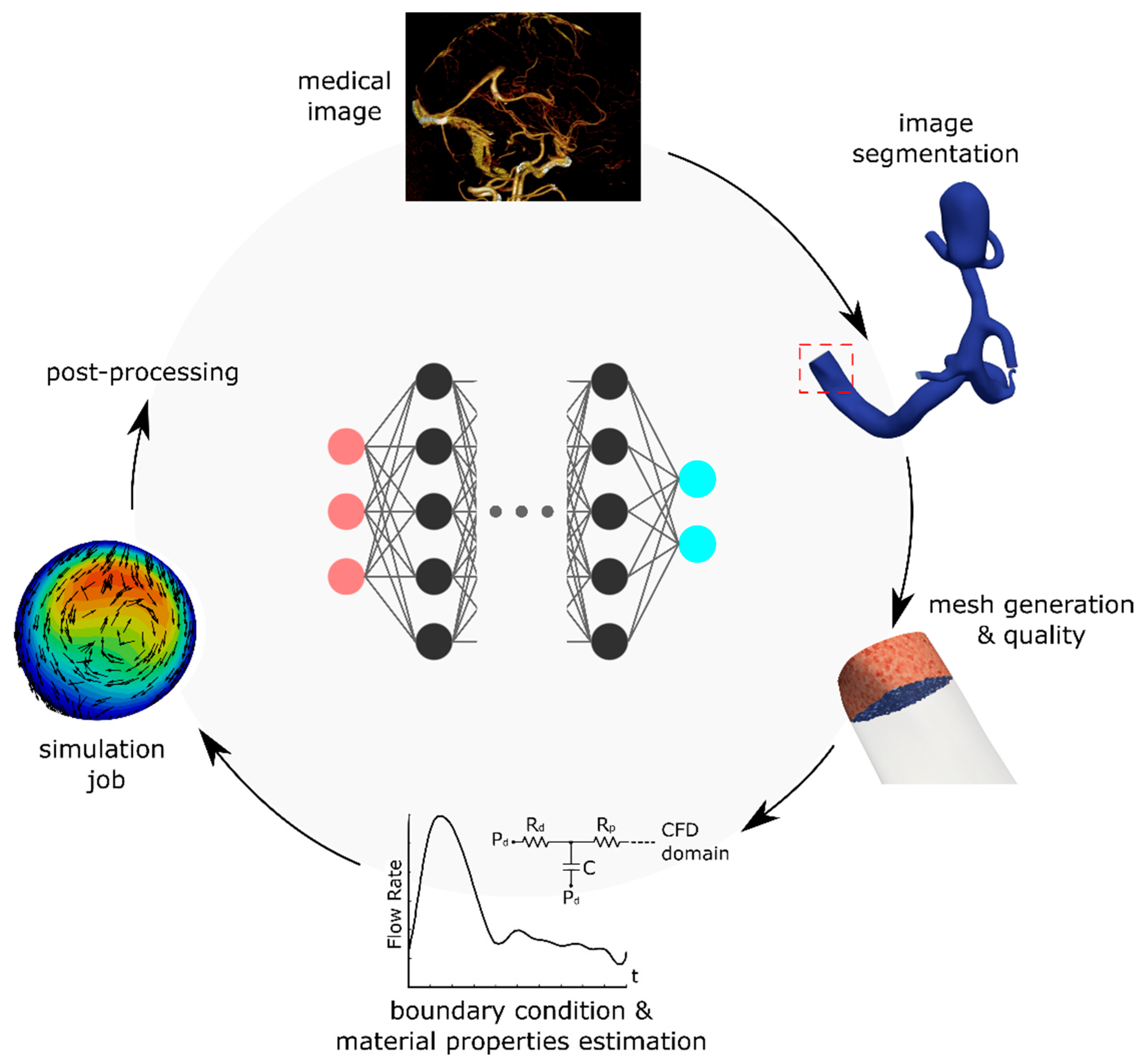
2. Methods
3. Deep Learning for Fluid Mechanics
4. Hemodynamics Applications
4.1. Hemodynamics of Aorta
4.2. Cerebral Hemodynamics
4.3. 4D Flow Magnetic Resonance Imaging
5. Discussion and Future Directions
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reid, L. An Introduction to Biomedical Computational Fluid Dynamics. In Biomedical Visualisation; Springer: Cham, Switzerland, 2021; pp. 205–222. [Google Scholar]
- Can, A.; Du, R. Association of Hemodynamic Factors With Intracranial Aneurysm Formation and Rupture. Neurosurgery 2016, 78, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Ameenuddin, M.; Anand, M. A Mixture Theory Model for Blood Combined With Low-Density Lipoprotein Transport to Predict Early Atherosclerosis Regions in Idealized and Patient-Derived Abdominal Aorta. J. Biomech. Eng. 2020, 142, 101008. [Google Scholar] [CrossRef] [PubMed]
- Shadden, S.C.; Arzani, A. Lagrangian Postprocessing of Computational Hemodynamics. Ann. Biomed. Eng. 2015, 43, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Khalili, F.; Gamage, P.T.; Taebi, A.; Johnson, M.E.; Roberts, R.B.; Mitchell, J. Spectral Decomposition of the Flow and Characterization of the Sound Signals through Stenoses with Different Levels of Severity. Bioengineering 2021, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Khalili, F.; Gamage, P.T.; Taebi, A.; Johnson, M.E.; Roberts, R.B.; Mitchel, J. Spectral Decomposition and Sound Source Localization of Highly Disturbed Flow through a Severe Arterial Stenosis. Bioengineering 2021, 8, 34. [Google Scholar] [CrossRef]
- Gamage, P.P.T.; Khalili, F.; Azad, K.; Mansy, H.A. Modeling Inspiratory Flow in a Porcine Lung Airway. J. Biomech. Eng. 2018, 140, 061003. [Google Scholar] [CrossRef]
- Nowak, N.; Kakade, P.P.; Annapragada, A.V. Computational Fluid Dynamics Simulation of Airflow and Aerosol Deposition in Human Lungs. Ann. Biomed. Eng. 2003, 31, 374–390. [Google Scholar] [CrossRef]
- Xi, J.; April Si, X.; Dong, H.; Zhong, H. Effects of Glottis Motion on Airflow and Energy Expenditure in a Human Upper Airway Model. Eur. J. Mech. B/Fluids 2018, 72, 23–37. [Google Scholar] [CrossRef]
- Tully, B.; Ventikos, Y. Coupling Poroelasticity and CFD for Cerebrospinal Fluid Hydrodynamics. IEEE Trans. Biomed. Eng. 2009, 56, 1644–1651. [Google Scholar] [CrossRef]
- Heidari Pahlavian, S.; Bunck, A.C.; Loth, F.; Shane Tubbs, R.; Yiallourou, T.; Robert Kroeger, J.; Heindel, W.; Martin, B.A. Characterization of the Discrepancies Between Four-Dimensional Phase-Contrast Magnetic Resonance Imaging and In-Silico Simulations of Cerebrospinal Fluid Dynamics. J. Biomech. Eng. 2015, 137, 051002. [Google Scholar] [CrossRef]
- Kelly, W.J.; Muske, K.R. Optimal Operation of High-Pressure Homogenization for Intracellular Product Recovery. Bioprocess Biosyst. Eng. 2004, 27, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Yarmush, G.; Santos, L.; Yarmush, J.; Koundinyan, S.; Saleem, M.; Nativ, N.I.; Yarmush, M.L.; Berthiaume, F.; Maguire, T.J.; Guaghan, C. CFD Assessment of the Effect of Convective Mass Transport on the Intracellular Clearance of Intracellular Triglycerides in Macrosteatotic Hepatocytes. Biomech. Model. Mechanobiol. 2017, 16, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Taebi, A.; Khalili, F. Advances in Noninvasive Diagnosis Based on Body Sounds and Vibrations—A Review. In Biomedical and Biotechnology; American Society of Mechanical Engineers: New York, NY, USA, 2021; Volume 5. [Google Scholar] [CrossRef]
- Khalili, F.; Taebi, A. Advances in Computational Fluid Dynamics Modeling of Cardiac Sounds as a Non-Invasive Diagnosis Method. In Biomedical and Biotechnology; American Society of Mechanical Engineers: New York, NY, USA, 2021; Volume 5. [Google Scholar] [CrossRef]
- Cook, J.; Umar, M.; Khalili, F.; Taebi, A. Body Acoustics for the Non-Invasive Diagnosis of Medical Conditions. Bioengineering 2022, 9, 149. [Google Scholar] [CrossRef] [PubMed]
- Taebi, A.; Solar, B.; Bomar, A.; Sandler, R.; Mansy, H. Recent Advances in Seismocardiography. Vibration 2019, 2, 64–86. [Google Scholar] [CrossRef] [Green Version]
- Meschi, S.S.; Farghadan, A.; Arzani, A. Flow Topology and Targeted Drug Delivery in Cardiovascular Disease. J. Biomech. 2021, 119, 110307. [Google Scholar] [CrossRef]
- Li, B.; Feng, Y. In Silico Study to Enhance Delivery Efficiency of Charged Nanoscale Nasal Spray Aerosols to the Olfactory Region Using External Magnetic Fields. Bioengineering 2022, 9, 40. [Google Scholar] [CrossRef]
- Taebi, A.; Vu, C.T.; Roncali, E. Multiscale Computational Fluid Dynamics Modeling for Personalized Liver Cancer Radioembolization Dosimetry. J. Biomech. Eng. 2021, 143, 011002. [Google Scholar] [CrossRef]
- Roncali, E.; Taebi, A.; Foster, C.; Vu, C.T. Personalized Dosimetry for Liver Cancer Y-90 Radioembolization Using Computational Fluid Dynamics and Monte Carlo Simulation. Ann. Biomed. Eng. 2020, 48, 1499–1510. [Google Scholar] [CrossRef]
- Ghosh, R.P.; Marom, G.; Rotman, O.M.; Slepian, M.J.; Prabhakar, S.; Horner, M.; Bluestein, D. Comparative Fluid–Structure Interaction Analysis of Polymeric Transcatheter and Surgical Aortic Valves’ Hemodynamics and Structural Mechanics. J. Biomech. Eng. 2018, 140, 121002. [Google Scholar] [CrossRef]
- Uchiyama, Y.; Fujimura, S.; Takao, H.; Suzuki, T.; Hayakawa, M.; Ishibashi, T.; Karagiozov, K.; Fukudome, K.; Murayama, Y.; Yamamoto, M. Hemodynamic Investigation of the Effectiveness of a Two Overlapping Flow Diverter Configuration for Cerebral Aneurysm Treatment. Bioengineering 2021, 8, 143. [Google Scholar] [CrossRef]
- Khalili, F. Fluid Dynamics Modeling and Sound Analysis of a Bileaflet Mechanical Heart Valve. Ph.D. Thesis, University of Central Florida, Orlando, FL, USA, 2018. [Google Scholar]
- Caballero, A.; Mao, W.; McKay, R.; Sun, W. The Impact of Balloon-Expandable Transcatheter Aortic Valve Replacement on Concomitant Mitral Regurgitation: A Comprehensive Computational Analysis. J. R. Soc. Interface 2019, 16, 20190355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamage, P.T.; Dong, P.; Lee, J.; Gharaibeh, Y.; Zimin, V.N.; Dallan, L.A.P.; Bezerra, H.G.; Wilson, D.L.; Gu, L. Hemodynamic Alternations Following Stent Deployment and Post-Dilation in a Heavily Calcified Coronary Artery: In Silico and Ex-Vivo Approaches. Comput. Biol. Med. 2021, 139, 104962. [Google Scholar] [CrossRef] [PubMed]
- Ge, L.; Leo, H.-L.; Sotiropoulos, F.; Yoganathan, A.P. Flow in a Mechanical Bileaflet Heart Valve at Laminar and Near-Peak Systole Flow Rates: CFD Simulations and Experiments. J. Biomech. Eng. 2005, 127, 782–797. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.A.; Steinman, D.A. Image-Based Modeling of Blood Flow and Vessel Wall Dynamics: Applications, Methods and Future Directions. Ann. Biomed. Eng. 2010, 38, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Updegrove, A.; Wilson, N.M.; Maher, G.D.; Shadden, S.C.; Marsden, A.L. A Re-Engineered Software Interface and Workflow for the Open-Source SimVascular Cardiovascular Modeling Package. J. Biomech. Eng. 2018, 140, 024501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
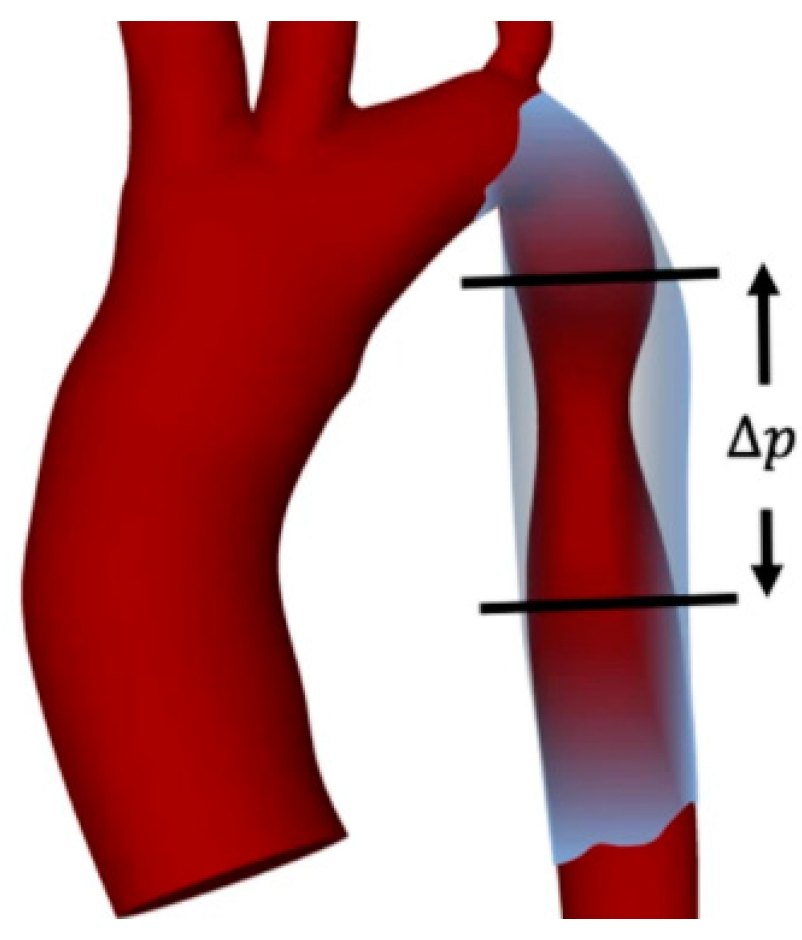
- Feiger, B.; Gounley, J.; Adler, D.; Leopold, J.A.; Draeger, E.W.; Chaudhury, R.; Ryan, J.; Pathangey, G.; Winarta, K.; Frakes, D.; et al. Accelerating Massively Parallel Hemodynamic Models of Coarctation of the Aorta Using Neural Networks. Sci. Rep. 2020, 10, 9508. [Google Scholar] [CrossRef]
- Pfaller, M.R.; Pham, J.; Verma, A.; Wilson, N.M.; Parker, D.W.; Yang, W.; Marsden, A.L. Automated Generation of 0D and 1D Reduced-Order Models of Patient-Specific Blood Flow. arXiv 2021, arXiv:2111.04878. [Google Scholar]
- Westerhof, N.; Lankhaar, J.W.; Westerhof, B.E. The Arterial Windkessel. Med. Biol. Eng. Comput. 2009, 47, 131–141. [Google Scholar] [CrossRef] [Green Version]
- Arzani, A.; Wang, J.-X.; D’Souza, R.M. Uncovering Near-Wall Blood Flow from Sparse Data with Physics-Informed Neural Networks. Phys. Fluids 2021, 33, 071905. [Google Scholar] [CrossRef]
- Kissas, G.; Yang, Y.; Hwuang, E.; Witschey, W.R.; Detre, J.A.; Perdikaris, P. Machine Learning in Cardiovascular Flows Modeling: Predicting Arterial Blood Pressure from Non-Invasive 4D Flow MRI Data Using Physics-Informed Neural Networks. Comput. Methods Appl. Mech. Eng. 2020, 358, 112623. [Google Scholar] [CrossRef]
- Brunton, S.L.; Noack, B.R.; Koumoutsakos, P. Machine Learning for Fluid Mechanics. Annu. Rev. Fluid Mech. 2020, 52, 477–508. [Google Scholar] [CrossRef] [Green Version]
- Kochkov, D.; Smith, J.A.; Alieva, A.; Wang, Q.; Brenner, M.P.; Hoyer, S. Machine Learning–Accelerated Computational Fluid Dynamics. Proc. Natl. Acad. Sci. USA 2021, 118, e2101784118. [Google Scholar] [CrossRef] [PubMed]
- Taira, K.; Brunton, S.L.; Dawson, S.T.M.; Rowley, C.W.; Colonius, T.; McKeon, B.J.; Schmidt, O.T.; Gordeyev, S.; Theofilis, V.; Ukeiley, L.S. Modal Analysis of Fluid Flows: An Overview. AIAA J. 2017, 55, 4013–4041. [Google Scholar] [CrossRef] [Green Version]
- Kutz, J.N.; Brunton, S.L.; Brunton, B.W.; Proctor, J.L. Dynamic Mode Decomposition: Data-Driven Modeling of Complex Systems; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2016. [Google Scholar]
- Carvalho, V.; Pinho, D.; Lima, R.A.; Teixeira, J.C.; Teixeira, S. Blood Flow Modeling in Coronary Arteries: A Review. Fluids 2021, 6, 53. [Google Scholar] [CrossRef]
- Kutz, J.N. Deep Learning in Fluid Dynamics. J. Fluid Mech. 2017, 814, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Milano, M.; Koumoutsakos, P. Neural Network Modeling for Near Wall Turbulent Flow. J. Comput. Phys. 2002, 182, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Fukami, K.; Fukagata, K.; Taira, K. Super-Resolution Reconstruction of Turbulent Flows with Machine Learning. J. Fluid Mech. 2019, 870, 106–120. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Franz, E.; Chu, M.; Thuerey, N. TempoGAN: A Temporally Coherent, Volumetric GAN for Super-Resolution Fluid Flow. ACM Trans. Graph. 2018, 37, 1–15. [Google Scholar] [CrossRef]
- Wang, R.; Kashinath, K.; Mustafa, M.; Albert, A.; Yu, R. Towards Physics-Informed Deep Learning for Turbulent Flow Prediction. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, ACM, New York, NY, USA, 6–10 June 2020; pp. 1457–1466. [Google Scholar]
- Jin, X.; Cai, S.; Li, H.; Karniadakis, G.E. NSFnets (Navier-Stokes Flow Nets): Physics-Informed Neural Networks for the Incompressible Navier-Stokes Equations. J. Comput. Phys. 2021, 426, 109951. [Google Scholar] [CrossRef]
- Raissi, M.; Yazdani, A.; Karniadakis, G.E. Hidden Fluid Mechanics: Learning Velocity and Pressure Fields from Flow Visualizations. Science 2020, 367, 1026–1030. [Google Scholar] [CrossRef]
- Yin, M.; Zheng, X.; Humphrey, J.D.; Karniadakis, G.E. Non-Invasive Inference of Thrombus Material Properties with Physics-Informed Neural Networks. Comput. Methods Appl. Mech. Eng. 2021, 375, 113603. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Jagtap, A.D.; Karniadakis, G.E. Physics-Informed Neural Networks for High-Speed Flows. Comput. Methods Appl. Mech. Eng. 2020, 360, 112789. [Google Scholar] [CrossRef]
- Cai, S.; Mao, Z.; Wang, Z.; Yin, M.; Karniadakis, G.E. Physics-Informed Neural Networks (PINNs) for Fluid Mechanics: A Review. Acta Mech. Sin. 2022, 37, 1727–1738. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Kong, F.; Martin, C.; Pham, T.; Wang, Q.; Duncan, J.; Sun, W. Machine Learning-Based 3-D Geometry Reconstruction and Modeling of Aortic Valve Deformation Using 3-D Computed Tomography Images. Int. J. Numer. Method. Biomed. Eng. 2017, 33, e2827. [Google Scholar] [CrossRef] [Green Version]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Pouch, A.M.; Wang, H.; Takabe, M.; Jackson, B.M.; Gorman, J.H.; Gorman, R.C.; Yushkevich, P.A.; Sehgal, C.M. Fully Automatic Segmentation of the Mitral Leaflets in 3D Transesophageal Echocardiographic Images Using Multi-Atlas Joint Label Fusion and Deformable Medial Modeling. Med. Image Anal. 2014, 18, 118–129. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Taebi, A.; Roudsari, B.; Vu, C.; Cherry, S.; Roncali, E. Hepatic Arterial Tree Segmentation: Towards Patient-Specific Dosimetry for Liver Cancer Radioembolization. J. Nucl. Med. 2019, 60 (Suppl. 1), 122. [Google Scholar]
- Huang, K.; Krügener, M.; Brown, A.; Menhorn, F.; Bungartz, H.-J.; Hartmann, D. Machine Learning-Based Optimal Mesh Generation in Computational Fluid Dynamics. arXiv 2021, arXiv:2102.12923. [Google Scholar]
- Zhang, Z.; Jimack, P.K.; Wang, H. MeshingNet3D: Efficient Generation of Adapted Tetrahedral Meshes for Computational Mechanics. Adv. Eng. Softw. 2021, 157–158, 103021. [Google Scholar] [CrossRef]
- Chen, X.; Liu, J.; Gong, C.; Li, S.; Pang, Y.; Chen, B. MVE-Net: An Automatic 3-D Structured Mesh Validity Evaluation Framework Using Deep Neural Networks. Comput. Des. 2021, 141, 103104. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, X.; Chi, L.; Liu, J.; Gong, C. A Mesh Quality Discrimination Method Based on Convolutional Neural Network. In Proceedings of the 2020 IEEE International Conference on Artificial Intelligence and Computer Applications, Dalian, China, 27–29 June 2020; pp. 481–486. [Google Scholar]
- Chen, X.; Liu, J.; Pang, Y.; Chen, J.; Chi, L.; Gong, C. Developing a New Mesh Quality Evaluation Method Based on Convolutional Neural Network. Eng. Appl. Comput. Fluid Mech. 2020, 14, 391–400. [Google Scholar] [CrossRef]
- Liang, L.; Mao, W.; Sun, W. A Feasibility Study of Deep Learning for Predicting Hemodynamics of Human Thoracic Aorta. J. Biomech. 2020, 99, 109544. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liang, L.; Sun, W. Estimation of in Vivo Constitutive Parameters of the Aortic Wall Using a Machine Learning Approach. Comput. Methods Appl. Mech. Eng. 2019, 347, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Cilla, M.; Pérez-Rey, I.; Martínez, M.A.; Peña, E.; Martínez, J. On the Use of Machine Learning Techniques for the Mechanical Characterization of Soft Biological Tissues. Int. J. Numer. Method. Biomed. Eng. 2018, 34, e3121. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; Fan, Z.; Baek, S.; Lu, J. Machine Learning-Aided Exploration of Relationship between Strength and Elastic Properties in Ascending Thoracic Aneurysm. Int. J. Numer. Method. Biomed. Eng. 2018, 34, e2977. [Google Scholar] [CrossRef]
- Liang, L.; Liu, M.; Martin, C.; Sun, W. A Deep Learning Approach to Estimate Stress Distribution: A Fast and Accurate Surrogate of Finite-Element Analysis. J. R. Soc. Interface 2018, 15, 20170844. [Google Scholar] [CrossRef] [Green Version]
- Itu, L.; Rapaka, S.; Passerini, T.; Georgescu, B.; Schwemmer, C.; Schoebinger, M.; Flohr, T.; Sharma, P.; Comaniciu, D. A Machine-Learning Approach for Computation of Fractional Flow Reserve from Coronary Computed Tomography. J. Appl. Physiol. 2016, 121, 42–52. [Google Scholar] [CrossRef] [Green Version]
- Sklet, V. Exploring the Capabilities of Machine Learning (ML) for 1D Blood Flow: Application to Coronary Flow. Master’s Thesis, Norwegian University of Science and Technology, Trondheim, Norway, 2018. [Google Scholar]
- Sarabian, M.; Babaee, H.; Laksari, K. Physics-Informed Neural Networks for Improving Cerebral Hemodynamics Predictions. arXiv 2021, arXiv:2108.11498. [Google Scholar]
- Tran, D.M.; Nguyen, M.T.; Lee, S.-W. Machine Learning Based Evaluation of Functional Index for Coronary Lesion Severity. In Proceedings of the 2nd International Conference on Machine Learning and Soft Computing, Phu Quoc Island, Vietnam, 2–4 February 2018; ACM Press: New York, NY, USA, 2018; pp. 1–4. [Google Scholar]
- Li, G.; Song, X.; Wang, H.; Liu, S.; Ji, J.; Guo, Y.; Qiao, A.; Liu, Y.; Wang, X. Prediction of Cerebral Aneurysm Hemodynamics With Porous-Medium Models of Flow-Diverting Stents via Deep Learning. Front. Physiol. 2021, 12, 1513. [Google Scholar] [CrossRef]
- Gharleghi, R.; Samarasinghe, G.; Sowmya, A.; Beier, S. Deep Learning for Time Averaged Wall Shear Stress Prediction in Left Main Coronary Bifurcations. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1–4. [Google Scholar]
- Charles, R.Q.; Su, H.; Kaichun, M.; Guibas, L.J. PointNet: Deep Learning on Point Sets for 3D Classification and Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 77–85. [Google Scholar]
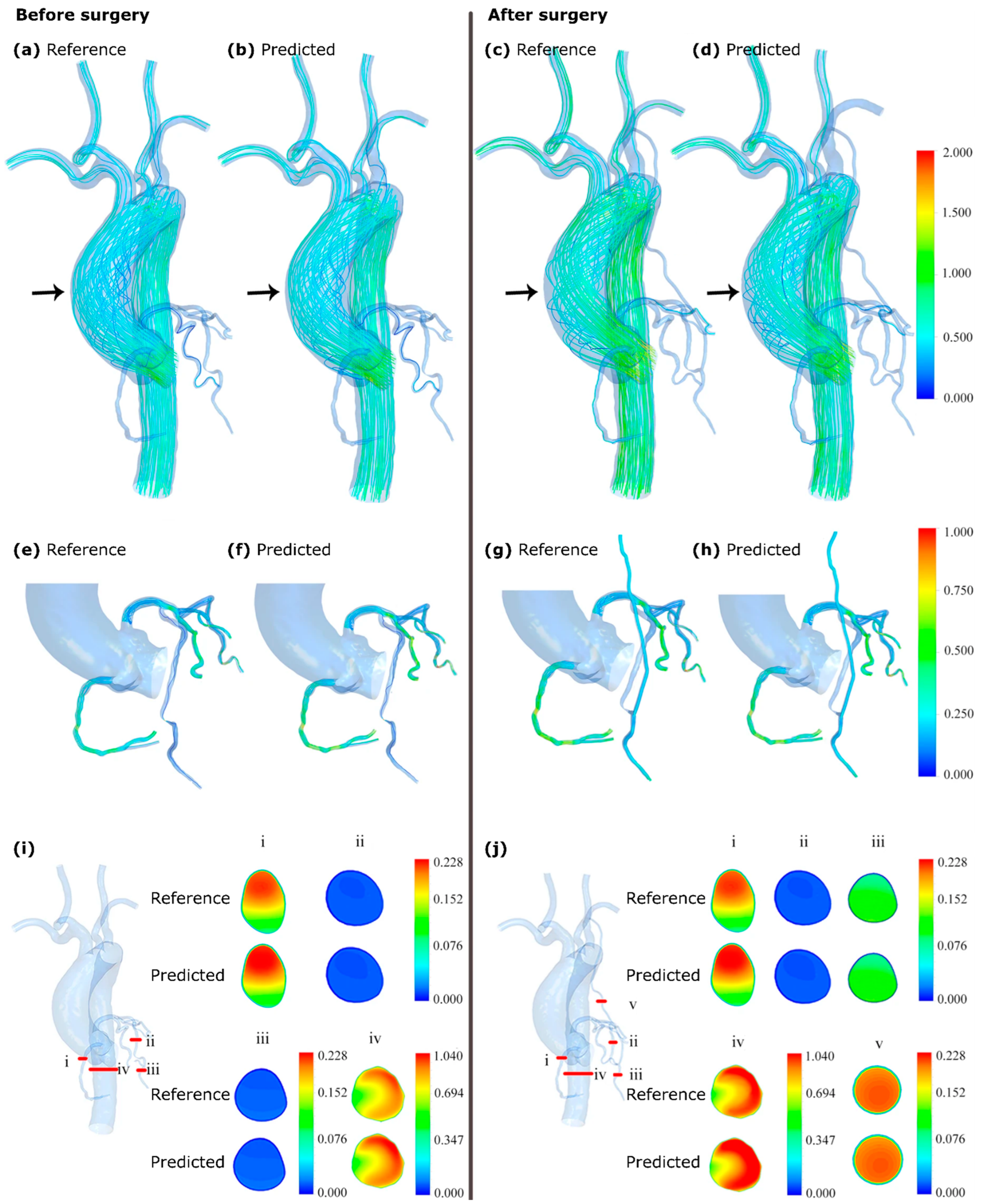
- Li, G.; Wang, H.; Zhang, M.; Tupin, S.; Qiao, A.; Liu, Y.; Ohta, M.; Anzai, H. Prediction of 3D Cardiovascular Hemodynamics before and after Coronary Artery Bypass Surgery via Deep Learning. Commun. Biol. 2021, 4, 99. [Google Scholar] [CrossRef]
- Randles, A.P.; Kale, V.; Hammond, J.; Gropp, W.; Kaxiras, E. Performance Analysis of the Lattice Boltzmann Model Beyond Navier-Stokes. In Proceedings of the IEEE 27th International Symposium on Parallel and Distributed Processing, Cambridge, MA, USA, 20–24 May 2013; pp. 1063–1074. [Google Scholar]
- Yevtushenko, P.; Goubergrits, L.; Gundelwein, L.; Setio, A.; Heimann, T.; Ramm, H.; Lamecker, H.; Kuehne, T.; Meyer, A.; Schafstedde, M. Deep Learning Based Centerline-Aggregated Aortic Hemodynamics: An Efficient Alternative to Numerical Modelling of Hemodynamics. IEEE J. Biomed. Heal. Inform. 2021, 26, 1815–1825. [Google Scholar] [CrossRef] [PubMed]
- Morales, X.; Mill, J.; Juhl, K.A.; Olivares, A.; Jimenez-Perez, G.; Paulsen, R.R.; Camara, O. Deep Learning Surrogate of Computational Fluid Dynamics for Thrombus Formation Risk in the Left Atrial Appendage. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2020; pp. 157–166. [Google Scholar]
- Taebi, A.; Vu, C.T.; Roncali, E. Estimation of Yttrium-90 Distribution in Liver Radioembolization Using Computational Fluid Dynamics and Deep Neural Networks. In Proceedings of the 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 4974–4977. [Google Scholar]
- Taebi, A.; Vu, C.T.; Roncali, E. Prediction of Blood Flow Distribution in Liver Radioembolization Using Convolutional Neural Networks. In Biomedical and Biotechnology; American Society of Mechanical Engineers: New York, NY, USA, 2020; Volume 5. [Google Scholar] [CrossRef]
- Logg, A.; Mardal, K.-A.; Wells, G. (Eds.) Automated Solution of Differential Equations by the Finite Element Method; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
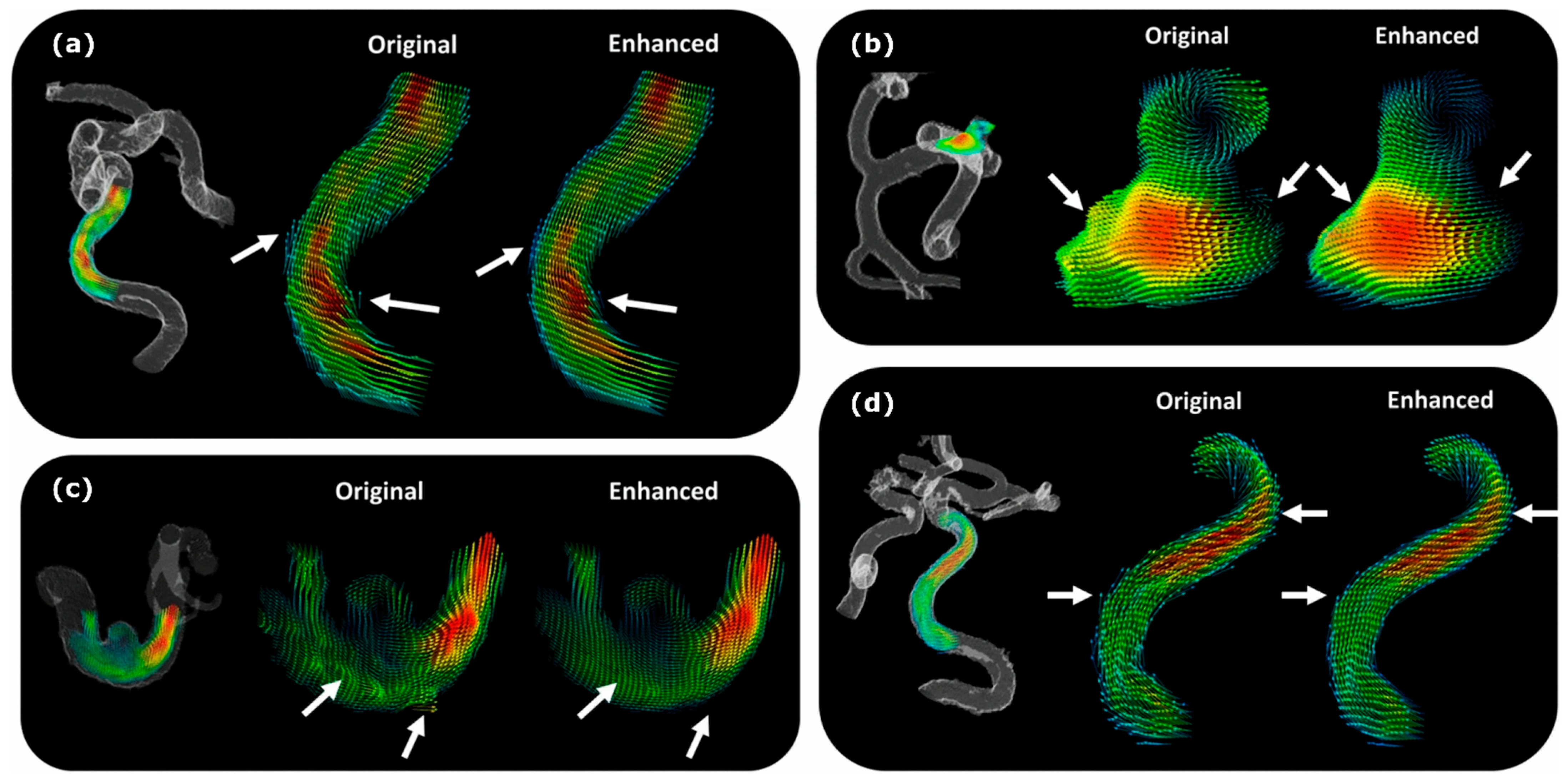
- Rutkowski, D.R.; Roldán-Alzate, A.; Johnson, K.M. Enhancement of Cerebrovascular 4D Flow MRI Velocity Fields Using Machine Learning and Computational Fluid Dynamics Simulation Data. Sci. Rep. 2021, 11, 10240. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Liu, M.; Martin, C.; Elefteriades, J.A.; Sun, W. A Machine Learning Approach to Investigate the Relationship between Shape Features and Numerically Predicted Risk of Ascending Aortic Aneurysm. Biomech. Model. Mechanobiol. 2017, 16, 1519–1533. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, S.; Huang, J.; Metaxas, D.; Axel, L. Sparse Deformable Models with Application to Cardiac Motion Analysis. In International Conference on Information Processing in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2013; pp. 208–219. [Google Scholar]
- Love, B.A.; Fischer, G.W.; Stelzer, P.; Fuster, V. Aortic Coarctation in the Adult. In Pediatric and Congenital Cardiology, Cardiac Surgery and Intensive Care; Springer: London, UK, 2014; pp. 2521–2549. [Google Scholar]
- Warnes, C.A.; Williams, R.G.; Bashore, T.M.; Child, J.S.; Connolly, H.M.; Dearani, J.A.; del Nido, P.; Fasules, J.W.; Graham, T.P.; Hijazi, Z.M.; et al. ACC/AHA 2008 Guidelines for the Management of Adults With Congenital Heart Disease. J. Am. Coll. Cardiol. 2008, 52, e143–e263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markl, M.; Brendecke, S.M.; Simon, J.; Barker, A.J.; Weiller, C.; Harloff, A. Co-Registration of the Distribution of Wall Shear Stress and 140 Complex Plaques of the Aorta. Magn. Reson. Imaging 2013, 31, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhang, J.; Shih, J.; Lopez-Bertoni, F.; Hagaman, J.R.; Maeda, N.; Friedman, M.H. Differences in Aortic Arch Geometry, Hemodynamics, and Plaque Patterns Between C57BL/6 and 129/SvEv Mice. J. Biomech. Eng. 2009, 131, 121005. [Google Scholar] [CrossRef]
- Miles, K.A.; Lee, T.-Y.; Goh, V.; Klotz, E.; Cuenod, C.; Bisdas, S.; Groves, A.M.; Hayball, M.P.; Alonzi, R.; Brunner, T. Current Status and Guidelines for the Assessment of Tumour Vascular Support with Dynamic Contrast-Enhanced Computed Tomography. Eur. Radiol. 2012, 22, 1430–1441. [Google Scholar] [CrossRef]
- Markl, M.; Frydrychowicz, A.; Kozerke, S.; Hope, M.; Wieben, O. 4D Flow MRI. J. Magn. Reson. Imaging 2012, 36, 1015–1036. [Google Scholar] [CrossRef]
- Plein, S.; Bloomer, T.N.; Ridgway, J.P.; Jones, T.R.; Bainbridge, G.J.; Sivananthan, M.U. Steady-State Free Precession Magnetic Resonance Imaging of the Heart: Comparison with Segmented k-Space Gradient-Echo Imaging. J. Magn. Reson. Imaging 2001, 14, 230–236. [Google Scholar] [CrossRef]
- Ferdian, E.; Suinesiaputra, A.; Dubowitz, D.J.; Zhao, D.; Wang, A.; Cowan, B.; Young, A.A. 4DFlowNet: Super-Resolution 4D Flow MRI Using Deep Learning and Computational Fluid Dynamics. Front. Phys. 2020, 8, 138. [Google Scholar] [CrossRef]
- Miyanawala, T.P.; Jaiman, R.K. An Efficient Deep Learning Technique for the Navier-Stokes Equations: Application to Unsteady Wake Flow Dynamics. arXiv 2017, arXiv:1710.09099. [Google Scholar]
- Wang, Z.; Xiao, D.; Fang, F.; Govindan, R.; Pain, C.C.; Guo, Y. Model Identification of Reduced Order Fluid Dynamics Systems Using Deep Learning. Int. J. Numer. Methods Fluids 2018, 86, 255–268. [Google Scholar] [CrossRef] [Green Version]
- Lye, K.O.; Mishra, S.; Ray, D. Deep Learning Observables in Computational Fluid Dynamics. J. Comput. Phys. 2020, 410, 109339. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; You, D. Prediction of Laminar Vortex Shedding over a Cylinder Using Deep Learning. arXiv 2017, arXiv:1712.07854. [Google Scholar]
- Lee, S.; You, D. Data-Driven Prediction of Unsteady Flow over a Circular Cylinder Using Deep Learning. J. Fluid Mech. 2019, 879, 217–254. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; He, Y.; Wu, J.; Cui, C.; Chen, M.; Sun, B. A Method of Parameter Estimation for Cardiovascular Hemodynamics Based on Deep Learning and Its Application to Personalize a Reduced-order Model. Int. J. Numer. Method. Biomed. Eng. 2022, 38, e3533. [Google Scholar] [CrossRef]
- Guo, X.; Li, W.; Iorio, F. Convolutional Neural Networks for Steady Flow Approximation. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM Press: New York, NY, USA, 2016; pp. 481–490. [Google Scholar]
- Ku, D.N. Blood Flow in Arteries. Annu. Rev. Fluid Mech. 1997, 29, 399–434. [Google Scholar] [CrossRef]
- Taebi, A.; Janibek, N.; Goldman, R.; Pillai, R.; Vu, C.T.; Roncali, E. On the Impact of Injection Distance to Bifurcations on Yttrium-90 Distribution in Liver Cancer Radioembolization. J. Vasc. Interv. Radiol. 2022, 33, 668–677.e1. [Google Scholar] [CrossRef]

| Application | DL Model | CFD Assumptions (Solver) | DL Input | DL Output | Reference |
|---|---|---|---|---|---|
| Coronary artery lesion | DNN | 1D, US, AF, EW, N, IC | Q & P at each centerline node + Q & P of the upstream and downstream nodes | Fractional flow reserve along the vessel centerline | [66] |
| Coronary blood flow | DNN regressor | 1D, SS, RW, N, IC (custom-built solver) | Geometry, SS P drop | P drop | [67] |
| Coronary bifurcations | 2D CNN | 3D, US, n-N (ANSYS) | Geometric features (e.g., vessel radii, bifurcation angles, etc.), shear stress for SS flow in a constant-radius straight tube | Time-averaged WSS | [71] |
| Aortic aneurysm | DNN | 3D, SS, RW, N, IC (STAR-CCM+) | 3D geometry, CFD results | v & P distributions, v magnitude | [61] |
| Coronary stenosis (coronary bypass surgery) | PointNet (based on [72]) | 3D, SS, N, IC (ANSYS) | 3D geometry, CFD results | v & P distributions | [73] |
| Aortic coarctation | NN | 3D, US, RW, N, IC (HARVEY [74]) | 3D geometry, CFD results | P, WSS | [30] |
| Aortic coarctation | DNN (LSTM RNN + DenseNet) | SS, IC (STAR-CCM+) | Vessel centerline, BC | P, time-averaged WSS, secondary flow degree, kinetic energy (averaged at the centerline) | [75] |
| Thrombus formation (in left arterial appendage) | DNN | 3D, US, RW, N, IC (ANSYS) | 3D geometry, CFD results | Endothelial cell activation potential | [76] |
| Hepatic artery (liver cancer radioembolization) | CNN | 3D, RW, N, IC (SimVascular [29]) | CFD results, outlet BC | Outlet flow rate | [77,78] |
| Intracranial aneurysm (right internal carotid artery) | PINN | 3D, US, IC | Concentration of the passive scalar, 3D geometry | v & P distributions, WSS | [46] |
| Near-wall blood flow (in aneurysm and stenosis models) | PINN | 1D-3D, SS, N, IC (FEniCS [79]) | Geometry, CFD results | v & P distributions | [33] |
| Cerebral vasospasm | PINN | 1D, US, EW, N, IC (SimVascular) | 3D angiography, 4D flow MRI, or ultrasound measurements | v & P distributions, vessel cross-sectional area | [68] |
| Cerebral aneurysm (before and after flow-diverting stent) | DNN | 3D, SS, N, IC, stent modeled by porous media (ANSYS) | 3D geometry, CFD results | v & P distributions | [70] |
| Aorta/carotid bifurcation | PINN | 1D, US, EW, N, IC | Reduced-order pulsatile flow results, 4D flow MRI at some cross sections | P wave propagation | [34] |
| Cerebral aneurysm | CNN | 3D, US, RW, N, IC (CONVERGE) | 3D geometry, CFD results, 4D flow MRI | Enhanced 4D flow MRI | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taebi, A. Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances. Fluids 2022, 7, 197. https://doi.org/10.3390/fluids7060197
Taebi A. Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances. Fluids. 2022; 7(6):197. https://doi.org/10.3390/fluids7060197
Chicago/Turabian StyleTaebi, Amirtahà. 2022. "Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances" Fluids 7, no. 6: 197. https://doi.org/10.3390/fluids7060197
APA StyleTaebi, A. (2022). Deep Learning for Computational Hemodynamics: A Brief Review of Recent Advances. Fluids, 7(6), 197. https://doi.org/10.3390/fluids7060197






