miRNAs as Interconnectors between Obesity and Cancer
Abstract
:1. Introduction
2. Adipogenesis Linked to Cancer Development
3. Biogenesis of MicroRNAs
4. miRNAs Involved in Adipogenesis
5. miRNAs Involved in Carcinogenesis
6. Adipose Tissue Dysregulation in Obesity Can Potentiate Cancer Development through miRNA Interplay
7. Participation of Macrophages in AT and Cancer
8. Biogenesis of lncRNAs
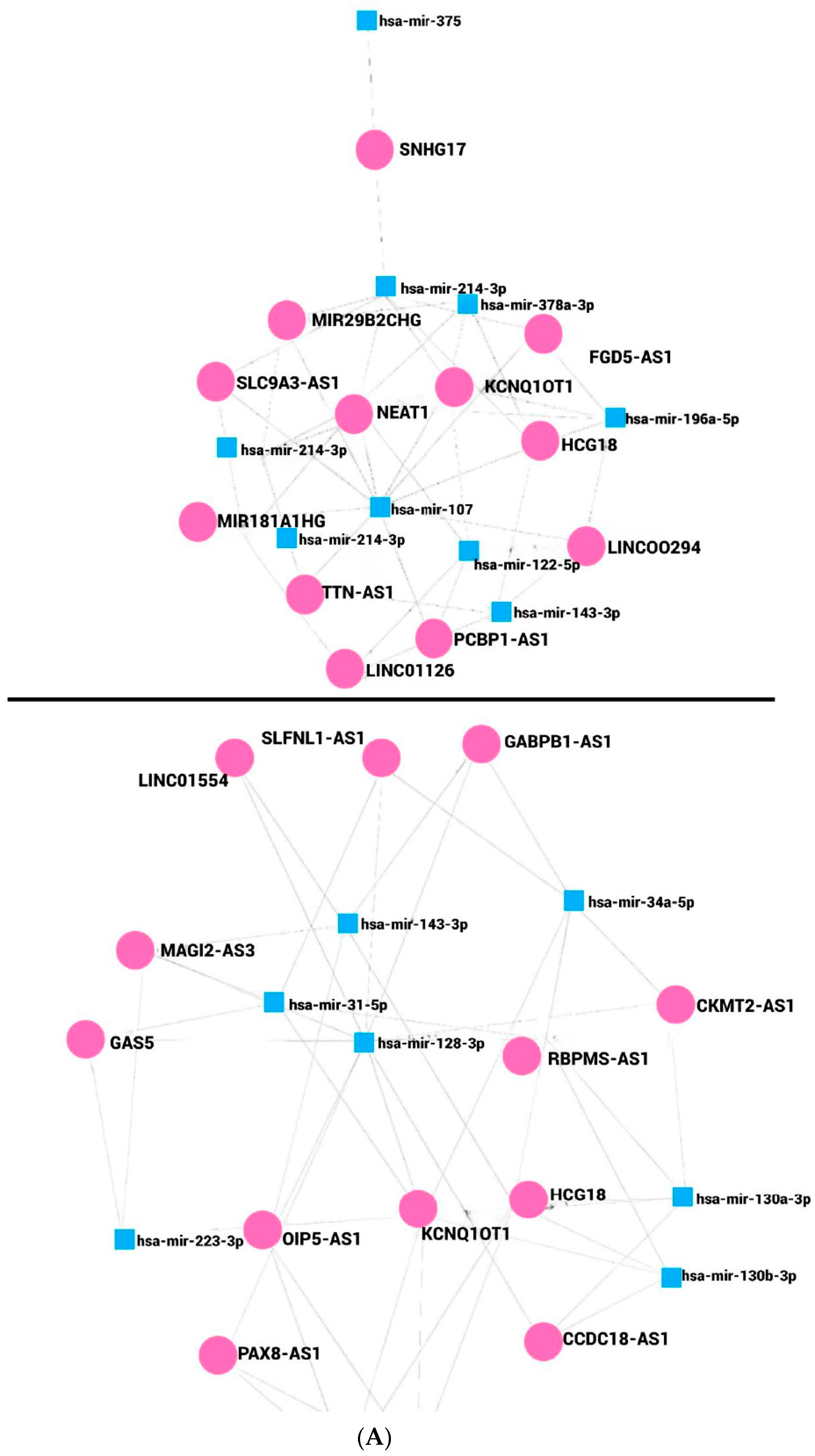
9. lncRNAs Regulate miRNA–mRNAs Axis in Obesity
10. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. World Health Organization: Obesity and Overweight Factsheet. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 19 October 2020).
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Bray, G.A.; Kim, K.K.; Wilding, J.P.H. Obesity: A chronic relapsing progressive disease process. A position statement of the World Obesity Federation. Obes. Rev. 2017, 18, 715–723. [Google Scholar] [CrossRef]
- NIH. National Cancer Institute: Obesity and Cancer. Available online: https://www.cancer.gov/about-cancer/causes-prevention/risk/obesity/obesity-fact-sheet (accessed on 21 October 2019).
- Cowey, S.; Hardy, R.W. The Metabolic Syndrome: A High-Risk State for Cancer? Am. J. Pathol. 2006, 169, 1505–1522. [Google Scholar] [CrossRef]
- Calle, E.E.; Kaaks, R. Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 2004, 4, 579–591. [Google Scholar] [CrossRef]
- Rohde, K.; Keller, M.; la Cour Poulsen, L.; Blüher, M.; Kovacs, P.; Böttcher, Y. Genetics and epigenetics in obesity. Metab. Clin. Exp. 2019, 92, 37–50. [Google Scholar] [CrossRef]
- Maes, H.H.M.; Neale, M.C.; Eaves, L.J. Genetic and Environmental Factors in Relative Body Weight and Human Adiposity. Behav. Genet. 1997, 27, 325–351. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Lorente-Cebrián, S.; González-Muniesa, P.; Milagro, F.I.; Martínez, J.A. MicroRNAs and other non-coding RNAs in adipose tissue and obesity: Emerging roles as biomarkers and therapeutic targets. Clin. Sci. 2019, 133, 23–40. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Lo, P.-K.; Wolfson, B.; Zhou, X.; Duru, N.; Gernapudi, R.; Zhou, Q. Noncoding RNAs in breast cancer. Brief. Funct. Genom. 2016, 15, 200–221. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Stapleton, K.; Das, S.; Reddy, M.A.; Leung, A.; Amaram, V.; Lanting, L.; Chen, Z.; Zhang, L.; Palanivel, R.; Deiuliis, J.A.; et al. Novel Long Noncoding RNA, Macrophage Inflammation-Suppressing Transcript (MIST), Regulates Macrophage Activation During Obesity. Arter. Thromb. Vasc. Biol. 2020, 40, 914–928. [Google Scholar] [CrossRef]
- Pradas-Juni, M.; Hansmeier, N.R.; Link, J.C.; Schmidt, E.; Larsen, B.D.; Klemm, P.; Meola, N.; Topel, H.; Loureiro, R.; Dhaouadi, I.; et al. A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat. Commun. 2020, 11, 644. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.; Dhaouadi, I.; Gaziano, I.; Oliverio, M.; Klemm, P.; Awazawa, M.; Mitterer, G.; Fernandez-Rebollo, E.; Pradas-Juni, M.; Wagner, W.; et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat. Commun. 2018, 9, 3622. [Google Scholar] [CrossRef]
- Huang, X.; Fu, C.; Liu, W.; Liang, Y.; Li, P.; Liu, Z.; Sheng, Q.; Liu, P. Chemerin-induced angiogenesis and adipogenesis in 3 T3-L1 preadipocytes is mediated by lncRNA Meg3 through regulating Dickkopf-3 by sponging miR-217. Toxicol. Appl. Pharmacol. 2019, 385, 114815. [Google Scholar] [CrossRef]
- Zhu, E.; Zhang, J.; Li, Y.; Yuan, H.; Zhou, J.; Wang, B. Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator-activated receptor γ. FASEB J. 2019, 33, 2396–2408. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Taheri, M. The expression profile and role of non-coding RNAs in obesity. Eur. J. Pharmacol. 2021, 892, 173809. [Google Scholar] [CrossRef]
- Hu, X.-t.; Xing, W.; Zhao, R.-s.; Tan, Y.; Wu, X.-f.; Ao, L.-q.; Li, Z.; Yao, M.-w.; Yuan, M.; Guo, W.; et al. HDAC2 inhibits EMT-mediated cancer metastasis by downregulating the long noncoding RNA H19 in colorectal cancer. J. Exp. Clin. Cancer Res. 2020, 39, 270. [Google Scholar] [CrossRef] [PubMed]
- Ni, W.; Yao, S.; Zhou, Y.; Liu, Y.; Huang, P.; Zhou, A.; Liu, J.; Che, L.; Li, J. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol. Cancer 2019, 18, 143. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Flier, J.S. Adipose Tissue as an Endocrine Organ. J. Clin. Endocrinol. Metab. 2004, 89, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Arch. Med. Sci. 2013, 9, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Eckel, J. Chapter 2—Adipose Tissue: A Major Secretory Organ. In The Cellular Secretome and Organ Crosstalk; Eckel, J., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 9–63. [Google Scholar]
- Lopez, J.A.; Granados-Lopez, A.J. Future directions of extracellular vesicle-associated miRNAs in metastasis. Ann. Transl. Med. 2017, 5, 115. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, N.; Lupien, L.; Kuemmerle, N.B.; Kinlaw, W.B.; Swinnen, J.V.; Smans, K. Lipogenesis and lipolysis: The pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 2013, 52, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.Q.; Lane, M.D. Adipogenesis: From stem cell to adipocyte. Annu. Rev. Biochem. 2012, 81, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Sabol, R.A.; Bowles, A.C.; Côté, A.; Wise, R.; Pashos, N.; Bunnell, B.A. Therapeutic Potential of Adipose Stem Cells. Adv. Exp. Med. Biol. 2021, 1341, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, Y.; Sun, Y.; Wang, B.; Xiong, Y.; Lin, W.; Wei, Q.; Wang, H.; He, W.; Wang, B.; et al. Tissue source determines the differentiation potentials of mesenchymal stem cells: A comparative study of human mesenchymal stem cells from bone marrow and adipose tissue. Stem Cell Res. Ther. 2017, 8, 275. [Google Scholar] [CrossRef]
- Badimon, L.; Oñate, B.; Vilahur, G. Adipose-derived Mesenchymal Stem Cells and Their Reparative Potential in Ischemic Heart Disease. Rev. Esp. Cardiol. 2015, 68, 599–611. [Google Scholar] [CrossRef]
- Katagiri, T.; Watabe, T. Bone Morphogenetic Proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021899. [Google Scholar] [CrossRef]
- Tang, Q.Q.; Otto, T.C.; Lane, M.D. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. USA 2004, 101, 9607–9611. [Google Scholar] [CrossRef] [PubMed]
- Moldes, M.; Zuo, Y.; Morrison, R.F.; Silva, D.; Park, B.H.; Liu, J.; Farmer, S.R. Peroxisome-proliferator-activated receptor gamma suppresses Wnt/beta-catenin signalling during adipogenesis. Biochem. J. 2003, 376, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Ambele, M.A.; Dessels, C.; Durandt, C.; Pepper, M.S. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016, 16, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef]
- Bowers, R.R.; Lane, M.D. A role for bone morphogenetic protein-4 in adipocyte development. Cell Cycle 2007, 6, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Gregoire, F.M.; Smas, C.M.; Sul, H.S. Understanding adipocyte differentiation. Physiol. Rev. 1998, 78, 783–809. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Lazar, M.A. New developments in adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef]
- Cordova-Rivas, S.; Fraire-Soto, I.; Mercado-Casas Torres, A.; Servin-Gonzalez, L.S.; Granados-Lopez, A.J.; Lopez-Hernandez, Y.; Reyes-Estrada, C.A.; Gutierrez-Hernandez, R.; Castaneda-Delgado, J.E.; Ramirez-Hernandez, L.; et al. 5p and 3p Strands of miR-34 Family Members Have Differential Effects in Cell Proliferation, Migration, and Invasion in Cervical Cancer Cells. Int. J. Mol. Sci. 2019, 20, 545. [Google Scholar] [CrossRef]
- Wang, L.; Ouyang, M.; Xing, S.; Zhao, S.; Liu, S.; Sun, L.; Yu, H. Mesenchymal Stem Cells and their Derived Exosomes Promote Malignant Phenotype of Polyploid Non-Small-Cell Lung Cancer Cells through AMPK Signaling Pathway. Anal. Cell. Pathol. 2022, 2022, 8708202. [Google Scholar] [CrossRef]
- Velasco, R.M.; Garcia, A.G.; Sanchez, P.J.; Sellart, I.M.; Sanchez-Arevalo Lobo, V.J. Tumour microenvironment and heterotypic interactions in pancreatic cancer. J. Physiol. Biochem. 2023, 79, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno, M.; Mercader, J.M.; Moreno-Navarrete, J.M.; Fuentes-Batllevell, N.; Sabater, M.; Ricart, W.; Fernández-Real, J.M. Inflammation triggers specific microRNA profiles in human adipocytes and macrophages and in their supernatants. Clin. Epigenetics 2015, 7, 49. [Google Scholar] [CrossRef]
- Almalki, S.G.; Agrawal, D.K. Key transcription factors in the differentiation of mesenchymal stem cells. Differentiation 2016, 92, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Hamam, D.; Ali, D.; Kassem, M.; Aldahmash, A.; Alajez, N.M. microRNAs as regulators of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2015, 24, 417–425. [Google Scholar] [CrossRef]
- Lin, Z.; He, H.; Wang, M.; Liang, J. MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 2019, 52, e12688. [Google Scholar] [CrossRef]
- Chen, C.; Deng, Y.; Hu, X.; Ren, H.; Zhu, J.; Fu, S.; Xie, J.; Peng, Y. miR-128-3p regulates 3T3-L1 adipogenesis and lipolysis by targeting Pparg and Sertad2. J. Physiol. Biochem. 2018, 74, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Lin, H.; Wang, L.; Wang, B.; Hao, X.; Shi, Y. miR-130a regulates macrophage polarization and is associated with non-small cell lung cancer. Oncol. Rep. 2015, 34, 3088–3096. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Luo, X. Lipid droplet and its implication in cancer progression. Am. J. Cancer Res. 2020, 10, 4112–4122. [Google Scholar] [PubMed]
- Yi, X.; Liu, J.; Wu, P.; Gong, Y.; Xu, X.; Li, W. The key microRNA on lipid droplet formation during adipogenesis from human mesenchymal stem cells. J. Cell. Physiol. 2020, 235, 328–338. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, Y.; Li, W.; Hu, A.J.; Luo, C.; Zhou, W.; Hu, J.K.; Daniele, S.G.; Wang, J.; Sheng, J.; et al. CDK6 inhibits white to beige fat transition by suppressing RUNX1. Nat. Commun. 2018, 9, 1023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Sun, X.; Zhang, X.; Yang, H.; Jiang, Z.; Luo, Q.; Liu, Y.; Wang, G. miR-107 is involved in the regulation of NEDD9-mediated invasion and metastasis in breast cancer. BMC Cancer 2022, 22, 533. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Lemecha, M.; Morino, K.; Imamura, T.; Iwasaki, H.; Ohashi, N.; Ida, S.; Sato, D.; Sekine, O.; Ugi, S.; Maegawa, H. MiR-494-3p regulates mitochondrial biogenesis and thermogenesis through PGC1-α signalling in beige adipocytes. Sci. Rep. 2018, 8, 15096. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Shi, C.M.; Chen, L.; Pang, L.X.; Xu, G.F.; Gu, N.; Zhu, L.J.; Guo, X.R.; Ni, Y.H.; Ji, C.B. The biological effects of hsa-miR-1908 in human adipocytes. Mol. Biol. Rep. 2015, 42, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs Induced During Adipogenesis that Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes 2009, 58, 1050. [Google Scholar] [CrossRef] [PubMed]
- Viallard, C.; Larrivée, B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis 2017, 20, 409–426. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 388354. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Vasudevan, S. Posttranscriptional upregulation by microRNAs. RNA 2012, 3, 311–330. [Google Scholar] [CrossRef]
- Brennecke, J.; Hipfner, D.R.; Stark, A.; Russell, R.B.; Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 2003, 113, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Im, H.-I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends Neurosci 2012, 35, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Huang, Y.; Shen, X.-J.; Zou, Q.; Wang, S.; Tang, S.; Zhang, G. Biological functions of MicroRNAs: A review. J. Physiol. Biochem. 2010, 67, 129–139. [Google Scholar] [CrossRef]
- Bushati, N.; Cohen, S.M. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Arner, P.; Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Lowe, S.W. The microcosmos of cancer. Nature 2012, 482, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Landrier, J.-F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef]
- Reilly, J.J.; El-Hamdouchi, A.; Diouf, A.; Monyeki, A.; Somda, S.A. Determining the worldwide prevalence of obesity. Lancet 2018, 391, 1773–1774. [Google Scholar] [CrossRef] [PubMed]
- Tareen, S.H.K.; Kutmon, M.; de Kok, T.M.; Mariman, E.C.M.; van Baak, M.A.; Evelo, C.T.; Adriaens, M.E.; Arts, I.C.W. Stratifying cellular metabolism during weight loss: An interplay of metabolism, metabolic flexibility and inflammation. Sci. Rep. 2020, 10, 1651. [Google Scholar] [CrossRef]
- Ma, X.; Sun, J.; Zhu, S.; Du, Z.; Li, D.; Li, W.; Li, Z.; Tian, Y.; Kang, X.; Sun, G. MiRNAs and mRNAs Analysis during Abdominal Preadipocyte Differentiation in Chickens. Animals 2020, 10, 468. [Google Scholar] [CrossRef]
- Cruz, K.J.C.; de Oliveira, A.R.S.; Morais, J.B.S.; Severo, J.S.; Marreiro Ph, D.D. Role of microRNAs on adipogenesis, chronic low-grade inflammation, and insulin resistance in obesity. Nutrition 2017, 35, 28–35. [Google Scholar] [CrossRef]
- Saini, S.K.; Singh, A.; Saini, M.; Gonzalez-Freire, M.; Leeuwenburgh, C.; Anton, S.D. Time-Restricted Eating Regimen Differentially Affects Circulatory miRNA Expression in Older Overweight Adults. Nutrients 2022, 14, 1843. [Google Scholar] [CrossRef]
- Xi, F.X.; Wei, C.S.; Xu, Y.T.; Ma, L.; He, Y.L.; Shi, X.E.; Yang, G.S.; Yu, T.Y. MicroRNA-214-3p Targeting Ctnnb1 Promotes 3T3-L1 Preadipocyte Differentiation by Interfering with the Wnt/β-Catenin Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 1816. [Google Scholar] [CrossRef]
- Ahonen, M.A.; Haridas, P.A.N.; Mysore, R.; Wabitsch, M.; Fischer-Posovszky, P.; Olkkonen, V.M. miR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol. Cell Endocrinol. 2019, 479, 110–116. [Google Scholar] [CrossRef]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef]
- Chen, L.; Hou, J.; Ye, L.; Chen, Y.; Cui, J.; Tian, W.; Li, C.; Liu, L. MicroRNA-143 Regulates Adipogenesis by Modulating the MAP2K5–ERK5 Signaling. Sci. Rep. 2014, 4, 3819. [Google Scholar] [CrossRef]
- Ling, H.Y.; Wen, G.B.; Feng, S.D.; Tuo, Q.H.; Ou, H.S.; Yao, C.H.; Zhu, B.Y.; Gao, Z.P.; Zhang, L.; Liao, D.F. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin. Exp. Pharmacol. Physiol. 2011, 38, 239–246. [Google Scholar] [CrossRef]
- Hilton, C.; Neville, M.J.; Wittemans, L.B.L.; Todorcevic, M.; Pinnick, K.E.; Pulit, S.L.; Luan, J.; Kulyté, A.; Dahlman, I.; Wareham, N.J.; et al. MicroRNA-196a links human body fat distribution to adipose tissue extracellular matrix composition. EBioMedicine 2019, 44, 467–475. [Google Scholar] [CrossRef]
- Afonso, M.S.; Verma, N.; Van Solingen, C.; Cyr, Y.; Sharma, M.; Perie, L.; Corr, E.M.; Schlegel, M.; Shanley, L.C.; Peled, D.; et al. MicroRNA-33 Inhibits Adaptive Thermogenesis and Adipose Tissue Beiging. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 1360–1373. [Google Scholar] [CrossRef]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.; Wang, Y.X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 4725. [Google Scholar] [CrossRef]
- Trohatou, O.; Zagoura, D.; Orfanos, N.K.; Pappa, K.I.; Marinos, E.; Anagnou, N.P.; Roubelakis, M.G. miR-26a Mediates Adipogenesis of Amniotic Fluid Mesenchymal Stem/Stromal Cells via PTEN, Cyclin E1, and CDK6. Stem. Cells. Dev. 2017, 26, 482–494. [Google Scholar] [CrossRef]
- Chen, Y.; Li, K.; Zhang, X.; Chen, J.; Li, M.; Liu, L. The novel long noncoding RNA lncRNA-Adi regulates adipogenesis. Stem. Cells Transl. Med. 2020, 9, 1053–1067. [Google Scholar] [CrossRef]
- Feng, L.; Xie, Y.; Zhang, H.; Wu, Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med. Oncol. 2012, 29, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Guan, X.; Shi, A.; Zou, Y.; Sun, M.; Zhan, Y.; Dong, Y.; Fan, Z. EZH2-Mediated microRNA-375 Upregulation Promotes Progression of Breast Cancer via the Inhibition of FOXO1 and the p53 Signaling Pathway. Front. Genet. 2021, 12, 633756. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Yang, Q.; Han, B.; Li, Y.; Yao, K.; Yang, X.; Chen, Z.; Yang, S.; Zhou, J.; Li, M.; et al. microRNA-375 inhibits colorectal cancer cells proliferation by downregulating JAK2/STAT3 and MAP3K8/ERK signaling pathways. Oncotarget 2017, 8, 16633–16641. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Sharma, A.R.; Patra, B.C.; Bhattacharya, M.; Sharma, G.; Lee, S.S. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget 2016, 7, 42683–42697. [Google Scholar] [CrossRef]
- Hu, B.; Tang, W.G.; Fan, J.; Xu, Y.; Sun, H.X. Differentially expressed miRNAs in hepatocellular carcinoma cells under hypoxic conditions are associated with transcription and phosphorylation. Oncol. Lett. 2018, 15, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Yuan, Y.; Shanmugam, M.K.; Anbalagan, D.; Tan, T.Z.; Sethi, G.; Kumar, A.P.; Lim, L.H.K. MicroRNA-196a promotes renal cancer cell migration and invasion by targeting BRAM1 to regulate SMAD and MAPK signaling pathways. Int. J. Biol. Sci. 2021, 17, 4254–4270. [Google Scholar] [CrossRef] [PubMed]
- Kang, L.; Zhang, Z.H.; Zhao, Y. SCAMP3 is regulated by miR-128-3p and promotes the metastasis of hepatocellular carcinoma cells through EGFR-MAPK p38 signaling pathway. Am. J. Transl. Res. 2020, 12, 7870–7884. [Google Scholar]
- Zhu, L.; Wang, X.; Wang, T.; Zhu, W.; Zhou, X. miR-494-3p promotes the progression of endometrial cancer by regulating the PTEN/PI3K/AKT pathway. Mol. Med. Rep. 2019, 19, 581–588. [Google Scholar] [CrossRef]
- Wu, C.; Yang, J.; Li, R.; Lin, X.; Wu, J.; Wu, J. LncRNA WT1-AS/miR-494-3p Regulates Cell Proliferation, Apoptosis, Migration and Invasion via PTEN/PI3K/AKT Signaling Pathway in Non-Small Cell Lung Cancer. Onco. Targets Ther. 2021, 14, 891–904. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Q.; Xia, Y.; Xiong, X.; Weng, S.; Ni, H.; Ye, Y.; Chen, L.; Lin, J.; Chen, Y.; et al. Evaluation of MiR-1908-3p as a novel serum biomarker for breast cancer and analysis its oncogenic function and target genes. BMC Cancer 2020, 20, 644. [Google Scholar] [CrossRef]
- Xia, X.; Li, Y.; Wang, W.; Tang, F.; Tan, J.; Sun, L.; Li, Q.; Sun, L.; Tang, B.; He, S. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol. Cancer 2015, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Lian, D.; Wang, Z.Z.; Liu, N.S. MicroRNA-1908 is a biomarker for poor prognosis in human osteosarcoma. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 1258–1262. [Google Scholar] [PubMed]
- Zhuang, W.; Liu, J.; Li, W. hsa-miR-33-5p as a Therapeutic Target Promotes Apoptosis of Breast Cancer Cells via Selenoprotein T. Front. Med. 2021, 8, 651473. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, X.; Shan, B.; Han, J.; Wang, F.; Fan, X.; Lv, Y.; Chang, L.; Liu, W. Downregulation of microRNA-33a promotes cyclin-dependent kinase 6, cyclin D1 and PIM1 expression and gastric cancer cell proliferation. Mol. Med. Rep. 2015, 12, 6491–6500. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Milde-Langosch, K.; Steinbach, B.; Muller, V.; Pantel, K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer. Res. Treat. 2012, 134, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Chen, J.; Cheng, X.; Huang, J.; Xiang, T.; Li, Q.; Long, H.; Zhu, B. Targeting the Wnt-Regulatory Protein CTNNBIP1 by microRNA-214 Enhances the Stemness and Self-Renewal of Cancer Stem-Like Cells in Lung Adenocarcinomas. Stem. Cells 2015, 33, 3423–3436. [Google Scholar] [CrossRef] [PubMed]
- Granados-Lopez, A.J.; Ruiz-Carrillo, J.L.; Servin-Gonzalez, L.S.; Martinez-Rodriguez, J.L.; Reyes-Estrada, C.A.; Gutierrez-Hernandez, R.; Lopez, J.A. Use of Mature miRNA Strand Selection in miRNAs Families in Cervical Cancer Development. Int. J. Mol. Sci. 2017, 18, 407. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Song, J.; Ding, B.; Cui, Y.; Liang, J.; Han, S. miR-146a promotes cervical cancer cell viability via targeting IRAK1 and TRAF6. Oncol. Rep. 2018, 39, 3015–3024. [Google Scholar] [CrossRef]
- Xiang, W.; Wu, X.; Huang, C.; Wang, M.; Zhao, X.; Luo, G.; Li, Y.; Jiang, G.; Xiao, X.; Zeng, F. PTTG1 regulated by miR-146a-3p promotes bladder cancer migration, invasion, metastasis and growth. Oncotarget 2017, 8, 664–678. [Google Scholar] [CrossRef]
- Zhong, R.; Chen, Z.; Mo, T.; Li, Z.; Zhang, P. Potential Role of circPVT1 as a proliferative factor and treatment target in esophageal carcinoma. Cancer Cell. Int. 2019, 19, 267. [Google Scholar] [CrossRef]
- Yang, X.; Lou, Y.; Wang, M.; Liu, C.; Liu, Y.; Huang, W. miR-675 promotes colorectal cancer cell growth dependent on tumor suppressor DMTF1. Mol. Med. Rep. 2019, 19, 1481–1490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shao, W. MicroRNA-143-3p Inhibits Wilms’ Tumor Cell Growth By Targeting the Ras/Raf/MEK/ERK Pathway. Altern. Ther. Health Med. 2023, 29, 140–147. [Google Scholar] [PubMed]
- Xie, F.; Li, C.; Zhang, X.; Peng, W.; Wen, T. MiR-143-3p suppresses tumorigenesis in pancreatic ductal adenocarcinoma by targeting KRAS. Biomed. Pharmacother. 2019, 119, 109424. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, Z.M.; Jiang, C.F.; Liu, X.; Chen, Q.D.; Qian, X.; Li, D.M.; Ge, X.; Wang, X.F.; Liu, L.Z.; et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget 2014, 5, 5416–5427. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Li, Y.; Wang, F.; Gu, J.; Isaji, T.; Li, J.; Qi, R.; Zhu, X.; Zhao, Y. Transforming growth factor (TGF) beta1 acted through miR-130b to increase integrin alpha5 to promote migration of colorectal cancer cells. Tumour. Biol. 2016, 37, 10763–10773. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, L.; Shang, P.; Song, X. LncRNA MEG8 promotes tumor progression of non-small cell lung cancer via regulating miR-107/CDK6 axis. Anticancer Drugs 2020, 31, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hou, L.; Liang, R.; Chen, X.; Zhang, R.; Chen, W.; Zhu, J. CircDLST promotes the tumorigenesis and metastasis of gastric cancer by sponging miR-502-5p and activating the NRAS/MEK1/ERK1/2 signaling. Mol. Cancer 2019, 18, 80. [Google Scholar] [CrossRef]
- Pan, Y.; Hui, X.; Hoo, R.L.C.; Ye, D.; Chan, C.Y.C.; Feng, T.; Wang, Y.; Lam, K.S.L.; Xu, A. Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Invest. 2019, 129, 834–849. [Google Scholar] [CrossRef]
- Wang, Y.C.; Li, Y.; Wang, X.Y.; Zhang, D.; Zhang, H.; Wu, Q.; He, Y.Q.; Wang, J.Y.; Zhang, L.; Xia, H.; et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013, 56, 2275–2285. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, L.; Sun, W.; Guan, W.; Lin, Q.; Ren, W.; Zhang, J.; Xu, G. Cytidine monophosphate kinase is inhibited by the TGF-beta signalling pathway through the upregulation of miR-130b-3p in human epithelial ovarian cancer. Cell Signal 2017, 35, 197–207. [Google Scholar] [CrossRef]
- Tian, F.; Wang, P.; Lin, D.; Dai, J.; Liu, Q.; Guan, Y.; Zhan, Y.; Yang, Y.; Wang, W.; Wang, J.; et al. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021, 112, 3744–3755. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Sun, L.; Xu, F.; Liu, L.; Hu, F.; Song, D.; Hou, Z.; Wu, W.; Luo, X.; Wang, J.; et al. M2 Macrophage-Derived Exosomes Promote Cell Migration and Invasion in Colon Cancer. Cancer Res. 2019, 79, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Mouton, A.J.; Li, X.; Hall, M.E.; Hall, J.E. Obesity, Hypertension, and Cardiac Dysfunction: Novel Roles of Immunometabolism in Macrophage Activation and Inflammation. Circ. Res. 2020, 126, 789–806. [Google Scholar] [CrossRef]
- Engin, A.B. Adipocyte-Macrophage Cross-Talk in Obesity. Adv. Exp. Med. Biol. 2017, 960, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Montastier, E.; Beuzelin, D.; Martins, F.; Mir, L.; Marqués, M.A.; Thalamas, C.; Iacovoni, J.; Langin, D.; Viguerie, N. Niacin induces miR-502-3p expression which impairs insulin sensitivity in human adipocytes. Int. J. Obes. 2019, 43, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Evers, B.M.; Zhou, Z.; Celano, P.; Li, J. The neurotensin gene is a downstream target for Ras activation. J. Clin. Investig. 1995, 95, 2822–2830. [Google Scholar] [CrossRef] [PubMed]
- Rock, S.; Li, X.; Song, J.; Townsend, C.M., Jr.; Weiss, H.L.; Rychahou, P.; Gao, T.; Li, J.; Evers, B.M. Kinase suppressor of Ras 1 and Exo70 promote fatty acid-stimulated neurotensin secretion through ERK1/2 signaling. PLoS ONE 2019, 14, e0211134. [Google Scholar] [CrossRef] [PubMed]
- Yehuda-Shnaidman, E.; Schwartz, B. Mechanisms linking obesity, inflammation and altered metabolism to colon carcinogenesis. Obes. Rev. 2012, 13, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Boden, G. Obesity and free fatty acids. Endocrinol. Metab. Clin. N. Am. 2008, 37, 635–646. [Google Scholar] [CrossRef]
- Blucher, C.; Stadler, S.C. Obesity and Breast Cancer: Current Insights on the Role of Fatty Acids and Lipid Metabolism in Promoting Breast Cancer Growth and Progression. Front. Endocrinol. 2017, 8, 293. [Google Scholar] [CrossRef]
- Bartsch, H.; Nair, J.; Owen, R.W. Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: Emerging evidence for their role as risk modifiers. Carcinogenesis 1999, 20, 2209–2218. [Google Scholar] [CrossRef]
- Azrad, M.; Turgeon, C.; Demark-Wahnefried, W. Current evidence linking polyunsaturated Fatty acids with cancer risk and progression. Front. Oncol. 2013, 3, 224. [Google Scholar] [CrossRef]
- Yucel Polat, A.; Ayva, E.S.; Gurdal, H.; Ozdemir, B.H.; Gur Dedeoglu, B. MiR-25 and KLF4 relationship has early prognostic significance in the development of cervical cancer. Pathol. Res. Pract. 2021, 222, 153435. [Google Scholar] [CrossRef]
- Nagata, T.; Shimada, Y.; Sekine, S.; Moriyama, M.; Hashimoto, I.; Matsui, K.; Okumura, T.; Hori, T.; Imura, J.; Tsukada, K. KLF4 and NANOG are prognostic biomarkers for triple-negative breast cancer. Breast Cancer 2017, 24, 326–335. [Google Scholar] [CrossRef]
- Arendt, L.M.; McCready, J.; Keller, P.J.; Baker, D.D.; Naber, S.P.; Seewaldt, V.; Kuperwasser, C. Obesity promotes breast cancer by CCL2-mediated macrophage recruitment and angiogenesis. Cancer Res. 2013, 73, 6080–6093. [Google Scholar] [CrossRef]
- Zhao, H.; Shang, Q.; Pan, Z.; Bai, Y.; Li, Z.; Zhang, H.; Zhang, Q.; Guo, C.; Zhang, L.; Wang, Q. Exosomes From Adipose-Derived Stem Cells Attenuate Adipose Inflammation and Obesity Through Polarizing M2 Macrophages and Beiging in White Adipose Tissue. Diabetes 2018, 67, 235–247. [Google Scholar] [CrossRef]
- Genin, M.; Clement, F.; Fattaccioli, A.; Raes, M.; Michiels, C. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer 2015, 15, 577. [Google Scholar] [CrossRef]
- Frank, A.C.; Raue, R.; Fuhrmann, D.C.; Sirait-Fischer, E.; Reuse, C.; Weigert, A.; Lutjohann, D.; Hiller, K.; Syed, S.N.; Brune, B. Lactate dehydrogenase B regulates macrophage metabolism in the tumor microenvironment. Theranostics 2021, 11, 7570–7588. [Google Scholar] [CrossRef]
- Ribeiro, J.; Marinho-Dias, J.; Monteiro, P.; Loureiro, J.; Baldaque, I.; Medeiros, R.; Sousa, H. miR-34a and miR-125b Expression in HPV Infection and Cervical Cancer Development. Biomed. Res. Int. 2015, 2015, 304584. [Google Scholar] [CrossRef] [PubMed]
- Kahraman, M.; Röske, A.; Laufer, T.; Fehlmann, T.; Backes, C.; Kern, F.; Kohlhaas, J.; Schrörs, H.; Saiz, A.; Zabler, C.; et al. MicroRNA in diagnosis and therapy monitoring of early-stage triple-negative breast cancer. Sci. Rep. 2018, 8, 11584. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Su, Y.; Wu, H.; Dai, Y.; Zhao, M.; Lu, Q. Characters, functions and clinical perspectives of long non-coding RNAs. Mol. Genet. Genom. 2016, 291, 1013–1033. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Avino, M.; Wellinger, R.J.; Laurent, B. Distinct regulatory functions and biological roles of lncRNA splice variants. Mol. Ther. Nucleic Acids 2023, 32, 127–143. [Google Scholar] [CrossRef] [PubMed]
- Tano, K.; Akimitsu, N. Long non-coding RNAs in cancer progression. Front. Genet. 2012, 3, 33822. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, J.K. The Emerging Role of Long Noncoding RNAs in Human Disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.T.; Yang, Y.M.; Sun, M.M.; He, Y.; Liao, L.; Chen, K.S.; Li, B. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun. 2022, 42, 117–140. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Garcia, J.T.; Hung, T.; Flynn, R.A.; Shen, Y.; Qu, K.; Payumo, A.Y.; Peres-da-Silva, A.; Broz, D.K.; Baum, R.; et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat. Genet. 2016, 48, 1370–1376. [Google Scholar] [CrossRef]
- Chiu, H.S.; Somvanshi, S.; Patel, E.; Chen, T.W.; Singh, V.P.; Zorman, B.; Patil, S.L.; Pan, Y.; Chatterjee, S.S.; Sood, A.K.; et al. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018, 23, 297–312.e212. [Google Scholar] [CrossRef]
- Yang, X.; Wang, C.C.; Lee, W.Y.W.; Trovik, J.; Chung, T.K.H.; Kwong, J. Long non-coding RNA HAND2-AS1 inhibits invasion and metastasis in endometrioid endometrial carcinoma through inactivating neuromedin U. Cancer Lett. 2018, 413, 23–34. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J. HAND2-AS1 inhibits invasion and metastasis of cervical cancer cells via microRNA-330-5p-mediated LDOC1. Cancer Cell Int. 2019, 19, 353. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Xu, Z.J.; Jiang, S.X.; Tang, D.S.; Yan, C.S.; Deng, Y.Y.; Zhao, F.Y. lncRNA FGD5-AS1 promotes breast cancer progression by regulating the hsa-miR-195-5p/NUAK2 axis. Mol. Med. Rep. 2021, 23, 460. [Google Scholar] [CrossRef] [PubMed]
- Park, M.K.; Zhang, L.; Min, K.W.; Cho, J.H.; Yeh, C.C.; Moon, H.; Hormaechea-Agulla, D.; Mun, H.; Ko, S.; Lee, J.W.; et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021, 33, 2380–2397.e9. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Xu, Y.; Wang, X.; Ren, M. KCNQ1OT1 affects cell proliferation, invasion, and migration through a miR-34a/Notch3 axis in breast cancer. Environ. Sci. Pollut. Res. Int. 2022, 29, 28480–28494. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, Q.; Xiao, W.; Zhang, B.; Jin, Y.; Lu, H. LncRNA TTN-AS1 Regulates miR-524-5p and RRM2 to Promote Breast Cancer Progression. Onco. Targets Ther. 2020, 13, 4799–4811. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qiao, K.; Zhu, K.; Li, X.; Zhao, C.; Li, J.; Feng, D.; Fang, Y.; Wang, P.; Qian, C.; et al. Long Noncoding RNA HCG18 Promotes Malignant Phenotypes of Breast Cancer Cells via the HCG18/miR-103a-3p/UBE2O/mTORC1/HIF-1α-Positive Feedback Loop. Front. Cell Dev. Biol. 2021, 9, 675082. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Liu, Y.; Wei, L.; Han, M. LncRNA OIP5-AS1 Promotes Breast Cancer Progression by Regulating miR-216a-5p/GLO1. J. Surg. Res. 2021, 257, 501–510. [Google Scholar] [CrossRef]
- Rishehri, M.; Etemadi, T.; Pisheh, L.; Koufigar, G.; Azadeh, M. Quantitative Expression of SFN, lncRNA CCDC18-AS1, and lncRNA LINC01343 in Human Breast Cancer as the Regulator Biomarkers in a Novel ceRNA Network: Based on Bioinformatics and Experimental Analyses. Genet. Res. 2022, 2022, 6787791. [Google Scholar] [CrossRef]
- Shafaroudi, A.M.; Sharifi-Zarchi, A.; Rahmani, S.; Nafissi, N.; Mowla, S.J.; Lauria, A.; Oliviero, S.; Matin, M.M. Expression and Function of C1orf132 Long-Noncoding RNA in Breast Cancer Cell Lines and Tissues. Int. J. Mol. Sci. 2021, 22, 6768. [Google Scholar] [CrossRef] [PubMed]
- Filippova, E.A.; Fridman, M.V.; Burdennyy, A.M.; Loginov, V.I.; Pronina, I.V.; Lukina, S.S.; Dmitriev, A.A.; Braga, E.A. Long Noncoding RNA GAS5 in Breast Cancer: Epigenetic Mechanisms and Biological Functions. Int. J. Mol. Sci. 2021, 22, 6810. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yang, B.; Zhang, J.; Chu, X. Long Noncoding RNA LINC01554 Inhibits the Progression of NSCLC Progression by Functioning as a ceRNA for miR-1267 and Regulating ING3/Akt/mTOR Pathway. Biomed. Res. Int. 2022, 2022, 7162623. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Lai, C.; Zhao, W.; Song, Y. GABPB1-AS1 acts as a tumor suppressor and inhibits non-small cell lung cancer progression by targeting miRNA-566/F-box protein 47. Oncol. Res. 2021, 29, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Huang, M.; Chen, M.; Chen, X. lncRNA SLC9A3-AS1 Promotes Oncogenesis of NSCLC via Sponging microRNA-760 and May Serve as a Prognosis Predictor of NSCLC Patients. Cancer Manag. Res. 2022, 14, 1087–1098. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Huang, X.; Chen, X.; Zhang, L.; Zhou, C.; Li, S.; Song, T.; Zhuang, L. Construction and validation of an angiogenesis-related lncRNA prognostic model in lung adenocarcinoma. Front. Genet. 2023, 14, 1083593. [Google Scholar] [CrossRef]
- Qi, P.; Yexie, Z.; Xue, C.; Huang, G.; Zhao, Z.; Zhang, X. LINC00294/miR-620/MKRN2 axis provides biomarkers and negatively regulates malignant progression in colorectal carcinoma. Hum. Exp. Toxicol. 2023, 42, 9603271231167577. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Zheng, S. The role of XBP-1-mediated unfolded protein response in colorectal cancer progression-a regulatory mechanism associated with lncRNA-miRNA-mRNA network. Cancer Cell Int. 2021, 21, 488. [Google Scholar] [CrossRef]
- Ramachandran, D.; Wang, Y.; Schürmann, P.; Hülse, F.; Mao, Q.; Jentschke, M.; Böhmer, G.; Strauß, H.G.; Hirchenhain, C.; Schmidmayr, M.; et al. Association of genomic variants at PAX8 and PBX2 with cervical cancer risk. Int. J. Cancer 2021, 149, 893–900. [Google Scholar] [CrossRef]
- Li, L.; Peng, Q.; Gong, M.; Ling, L.; Xu, Y.; Liu, Q. Using lncRNA Sequencing to Reveal a Putative lncRNA-mRNA Correlation Network and the Potential Role of PCBP1-AS1 in the Pathogenesis of Cervical Cancer. Front. Oncol. 2021, 11, 634732. [Google Scholar] [CrossRef]
- Tye, C.E.; Ghule, P.N.; Gordon, J.A.R.; Kabala, F.S.; Page, N.A.; Falcone, M.M.; Tracy, K.M.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; et al. LncMIR181A1HG is a novel chromatin-bound epigenetic suppressor of early stage osteogenic lineage commitment. Sci. Rep. 2022, 12, 7770. [Google Scholar] [CrossRef] [PubMed]
- Kazeminasab, F.; Marandi, S.M.; Baharlooie, M.; Safaeinejad, Z.; Nasr-Esfahani, M.H.; Ghaedi, K. Aerobic exercise modulates noncoding RNA network upstream of FNDC5 in the Gastrocnemius muscle of high-fat-diet-induced obese mice. J. Physiol. Biochem. 2021, 77, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Leija-Martínez, J.J.; Guzmán-Martín, C.A.; González-Ramírez, J.; Giacoman-Martínez, A.; Del-Río-Navarro, B.E.; Romero-Nava, R.; Villafaña, S.; Flores-Saenz, J.L.; Sánchez-Muñoz, F.; Huang, F. Whole Blood Expression Levels of Long Noncoding RNAs: HOTAIRM1, GAS5, MZF1-AS1, and OIP5-AS1 as Biomarkers in Adolescents with Obesity-Related Asthma. Int. J. Mol. Sci. 2023, 24, 6481. [Google Scholar] [CrossRef] [PubMed]



| miRNA | Effect | Targets | Model Study | Reference |
|---|---|---|---|---|
| miR-214-3p | Proadipogenic | ↓ Ctnnb1, Wnt/β-Catenin | 3T3-L1 cells | [84] |
| miR-130a | Antiadipogenic | ↓ Smurf2, PPARγ | Mouse BMSC | [48] |
| miR-146a-3p, miR-4495, miR-4663, miR-6069, miR-675-3p | Proadipogenic | ↑ ACSL1, APOB, METTL7A, PLIN1, PLIN4. A | Human MSCs | [52] |
| miR-107 | Antiadipogenic | ↓ CDK6, Notch3 | Human SGBS | [85] |
| miR-143 | Proadipogenic | ↓ ERK5, MAP2K5 | Human White preadipocytes | [86,87] |
| miR-375 | Proadipogenic | ↑ C/EBPa, PPARγ2, aP2 | 3T3-L1 cells | [88] |
| miR-196a | Proadipogenic | ↑ COL25A1 | ASAT biopsies | [89] |
| miR-128-3p | Antiadipogenic | ↓ Pparg, Sertad2 | 3T3-L1 cells | [49] |
| miR-494-3p | Antiadipogenic | ↓ PGC1-α | 3T3-L1 cells | [57] |
| miR-1908 | Antiadipogenic | ↓ PPARc, C/EBPa | hMADS cells | [58] |
| miR-33 | Antiadipogenic | ↓ Zfp516, Dio2, and Ppargc1a | 10T1/2 cells | [90] |
| miR-378 | Proadipogenic | ↓ Pde1b | C3H10T1/2 cells | [91] |
| miRNA | Dysregulation | Mediator Affected | Cancer Related | Reference |
|---|---|---|---|---|
| miR-214-3p | Overexpressed | ↓ CTNNBIP1 | Breast cancer | [108] |
| ↑ Nanog, EpCAM | Cancer stem-like cells | [109] | ||
| ↑ Oct-4, Nanog, Sox-2, CD133, and EpCAM | Lung adenocarcinoma | [109] | ||
| miR130a | Underexpressed | ↓ PPARγ | Non-small cell lung cancer | [50] |
| miR-146a-3p | Overexpressed | ↓ IRAK1 and TRAF6 | Cervical Cancer | [110,111] |
| Underexpressed | ↑ PTTG1 | Bladder cancer | [112] | |
| miR-4663 | Overexpressed | ↓ Paxs ↑ PPARs | Esophageal carcinoma | [113] |
| miR 675 | Overexpressed | ↓ DMTF1 | Colorectal cancer | [114] |
| mirR-107 | Overexpressed | ↓ NEDD9 | Breast cancer | [55] |
| miR-143-3p | Underexpressed | ↓ Ras/Raf/MEK/ERK | Wilms’ tumor | [115] |
| Underexpressed | ↓ KRAS | Pancreatic ductal adenocarcinoma | [116] | |
| Underexpressed | ↓ N-RAS | Glioma | [117] | |
| miR-375 | Overexpressed | ↓ FOXO1 | Breast cancer | [95] |
| Underexpressed | ↓ JAK2/STAT3 and MAP3K8/ERK | Colorectal cancer | [96] | |
| miR-196a | Overexpressed | ↓ Bram1 | Renal cancer | [99] |
| miR-128-3p | Underexpressed | ↓ Pparg and Sertad2 ↑ EGFR-MAPK p38 | Hepatocellular carcinoma | [49,100] |
| miR-494-3p | Overexpressed | ↓ PGC1-α ↑ PTEN/PI3K/AKT | Cancer cancer non-small cell lung and endometrial | [57,101] |
| miR-1908 | Overexpressed | ↓ PPARγ and C/EBPα | Glioblastoma, osteosarcoma and breast cancer tissues | [103,104,105] |
| miR-33 | Underexpressed | ↓ CCND1 and CDK6 | Breast cancer | [106] |
| miR-130b | Underexpressed | ↑ TGF-β1 and integrin α5 | Epithelial ovarian cancer cells | [118] |
| miR-107 | Underexpressed | ↑ CDK6 | Lung cancer | [119] |
| miR-502-5p | Overexpressed | ↓ NRAS/MEK1/ERK1/2 | Gastric cancer | [120] |
| miR-34a | Overexpressed | ↓ Klf4 | In cervical cancer | [121] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Sánchez, G.D.; Granados-López, A.J.; López-Hernández, Y.; Robles, M.J.G.; López, J.A. miRNAs as Interconnectors between Obesity and Cancer. Non-Coding RNA 2024, 10, 24. https://doi.org/10.3390/ncrna10020024
González-Sánchez GD, Granados-López AJ, López-Hernández Y, Robles MJG, López JA. miRNAs as Interconnectors between Obesity and Cancer. Non-Coding RNA. 2024; 10(2):24. https://doi.org/10.3390/ncrna10020024
Chicago/Turabian StyleGonzález-Sánchez, Grecia Denisse, Angelica Judith Granados-López, Yamilé López-Hernández, Mayra Judith García Robles, and Jesús Adrián López. 2024. "miRNAs as Interconnectors between Obesity and Cancer" Non-Coding RNA 10, no. 2: 24. https://doi.org/10.3390/ncrna10020024





