Abstract
Following the acute phase of SARS-CoV-2 infection, certain individuals experience persistent symptoms referred to as long COVID. This study analyzed the patients categorized into three distinct groups: (1) individuals presenting rheumatological symptoms associated with long COVID, (2) patients who have successfully recovered from COVID-19, and (3) donors who have never contracted COVID-19. A notable decline in the expression of miR-200c-3p, miR-766-3p, and miR-142-3p was identified among patients exhibiting rheumatological symptoms of long COVID. The highest concentration of miR-142-3p was found in healthy donors. One potential way to reduce miRNA concentrations is through antibody-mediated hydrolysis. Not only can antibodies possessing RNA-hydrolyzing activity recognize the miRNA substrate specifically, but they also catalyze its hydrolysis. The analysis of the catalytic activity of plasma antibodies revealed that antibodies from patients with long COVID demonstrated lower hydrolysis activity against five fluorescently labeled oligonucleotide sequences corresponding to the Flu-miR-146b-5p, Flu-miR-766-3p, Flu-miR-4742-3p, and Flu-miR-142-3p miRNAs and increased activity against the Flu-miR-378a-3p miRNA compared to other patient groups. The changes in miRNA concentrations and antibody-mediated hydrolysis of miRNAs are assumed to have a complex regulatory mechanism that is linked to gene pathways associated with the immune system. We demonstrate that all six miRNAs under analysis are associated with a large number of signaling pathways associated with immune response-associated pathways.
1. Introduction
MiRNAs are a class of noncoding RNAs that regulate the expression of numerous genes through degradation, translational repression, or activation of mRNAs [1] as they are involved in various biological processes such as regulation of immune response, immune cell differentiation, metabolism, apoptosis, cell cycle, and oncogenesis [2]. Numerous studies have demonstrated that miRNAs can be released into extracellular fluids [3]. Hence, extracellular miRNAs represent promising biomarkers for early diagnosis and prognosis of various diseases, as well as targets for therapy [4]. Moreover, miRNA-based drugs are expected to become the next generation of drugs for the treatment and prevention of various human diseases [5].
Various diseases, including viral infections, cause significant changes in the expression profile of miRNAs [6,7]. Currently, a large number of differentially expressed miRNAs associated with COVID-19 have been identified [8,9]. These studies identify miRNAs associated with severe COVID-19, and gene regulatory pathways to understand the underlying biological processes associated with COVID-19 [10]. For example, Nicoletti et al. [11] identified 18 plasma miRNAs that are differentially expressed in COVID-19 patients and controls, including miR-4433b-5p, miR-320b, and miR-16-2-3p. Expression of the miR-320 family (320a-3p, 320b, 320c, and 320d) is increased in patients with COVID-19 compared to healthy controls [12], and lower miR-320 expression is associated with death in patients [11,13]. Changes in the expression level of some miRNAs, such as miR-142-3p, miR-146b-5p, miR-148a-3p, miR-200c-3p, miR-370, miR-378a-3p, miR-483, miR-1246, and others, have been described during the acute period of SARS-CoV-2 infection [14,15,16]. Several miRNAs, such as miR-21, miR-143, miR-122, miR-133, miR-155, miR-208a, miR-499, miR-625, and others, are associated with disease severity [8,16,17]. Data on the expression of some miRNAs in COVID-19 are contradictory (e.g., miR-4433b-5p, miR-16-2-3p) [12,14,18,19], which is probably due to differences in the patient sample.
MiRNAs regulating the expression of viral genes are interesting. Several studies indicate that the interaction of host miRNAs with viral genes limits viral replication [20]. MiRNAs capable of targeting various regions of the SARS-CoV-2 genome, such as miR-197-5p and miR-18b-5p, have been identified [21,22].
MiRNA-mediated regulation has been shown not only for higher eukaryotes. To date, miRBase reports 30 viral miRNAs encoded by RNA viruses [23,24,25,26]. Several studies have demonstrated miRNAs originating from the SARS-CoV-2 genome [27,28,29].
Although most patients with COVID-19 fully recover, some experience lingering symptoms that may be related to previous SARS-CoV-2 infection (long COVID) [30]. The clinical manifestations associated with long-term COVID are incredibly heterogeneous and involve the respiratory system, gastrointestinal tract, joints, nervous system, endocrine system, etc. [31,32]. During the course of COVID-19, an important proportion of cases suffer from severe pneumonia and tend to have long-term sequelae [33]. Ongoing fibrosis during the recovery period results in decreased diffusion capacity of the lung [34]. There are a significant number of reports of patients with demyelinating pathologies such as Guillain–Barré syndrome, Miller–Fisher syndrome and others [35]. There is growing published data on a wide range of autoimmune diseases associated with SARS-CoV-2 [36,37]. To date, it has been confirmed that viral infections are associated with the development of various rheumatological diseases [38,39]. Medical experts have provided detailed descriptions and characterizations of certain cases, but the specific molecular mechanisms remain insufficiently researched.
Dysregulation of some miRNAs was suggested to lead to long COVID [40]. For example, miR-34a, contained in circulating extracellular vesicles, is associated with the risk of diabetes after COVID-19 infection [41]. Some miRNAs are involved in the pathogenesis of thromboembolic complications in COVID-19 [42]. For example, decreased expression of miR-103a, miR-145, and miR-885 and increased expression of miR-424 and miR-320 were found in a group of patients with a high frequency of thrombotic and ischemic events [43,44]. It has been shown that the concentration of exosomal miR-145 and miR-885 significantly correlates with D-dimer levels [45]. In some patients, SARS-CoV-2 causes an excessive immune response, resulting in a cytokine storm [46]. Some miRNAs are involved in the regulation of mediators associated with inflammation and appear to be important in some inflammatory diseases [47]. Significantly decreased miRNA-106a and miRNA-20a expression are associated with increased IL-10, TNF-α, INF-γ, and TLR-4 levels in COVID-19 patients [12,14,48]. This is not surprising since these miRNAs target proinflammatory cytokine genes (e.g., TNF, CCL2, CXCL9, IL10) and cytokine and chemokine receptors (IL1R1, IL2RA, IFNAR2) [49].
In light of the COVID-19 pandemic, extensive research has been dedicated to studying the immune response to the SARS-CoV-2 virus. Involved in the immune response to viral infection are catalytically active antibodies. These antibodies exhibit a dual function of antigen binding and catalyzing the hydrolysis of particular substrates, including proteins, nucleic acids, and polysaccharides [50,51]. To illustrate, antibodies displaying proteolytic activity were reported in several studies [52,53]. Additionally, there have been advancements in the creation of monoclonal antibodies that can directly hydrolyze the RNA of coronavirus and influenza virus [54,55,56,57]. Previous studies have described the capability of catalytically active antibodies to hydrolyze miRNAs in the context of autoimmune and neurological diseases [58,59,60,61].
Our study focused on evaluating the expression patterns of ten miRNAs in the plasma of COVID-19 patients, differentiating between those who had recovered without any complaints and those who continued to experience lingering rheumatological symptoms associated with long COVID. The focus of this study was to analyze the role of IgG in miRNA hydrolysis among COVID-19 patients. We proposed the idea that the presence of catalytically active antibodies with a specific affinity for particular miRNAs might lead to changes in the plasma concentration of these miRNAs through their hydrolysis. Also, among a large number of miRNA signaling pathways, pathways associated with the immune response have been identified.
2. Results
2.1. General Characteristics of the Patient Groups
Following the acute phase of SARS-CoV-2 infection, certain individuals were found to experience persistent symptoms referred to as long COVID [62]. Numerous respiratory, cardiovascular, autoimmune, neurological, and psychiatric diseases, along with their associated symptoms, have been documented in relation to long COVID [63,64,65,66,67,68]. Additionally, there are reports on the incidence of autoimmune diseases after SARS-CoV-2 infection [69,70,71]. The focus of this study was to identify and analyze a particular group of patients who presented complaints related to rheumatological conditions, known as the Lon group. We established a cohort of COVID-19 survivors who exhibited no post-recovery symptoms (Cov) and a control group of healthy individuals who had not contracted COVID-19 (Neg). The collection of blood plasma from COVID-19 patients occurred during the circulation of the Wuhan strain of SARS-CoV-2. Between summer 2020 and spring 2021, the epidemic in Russia was characterized by the spread of three lineages: B.1.1.7, B.1.1.317, and a sublineage of B.1.1 including B.1.1.397 [72]. The study did not include subjects vaccinated against SARS-CoV-2.
2.2. Analysis of miRNA Concentration in the Plasma of Patients
The concentrations of ten miRNAs (miR-l7f-5p, miR-146b-5p, miR-148a-3p, miR-200c-3p, miR-378a-3p, miR-9-5p, miR-766-3p, miR-3125, miR-4742-3p, and miR-142-3p) were studied in the blood plasma of patients in three groups. For this purpose, a reverse transcription method using appropriate stem-loop (SL) primers followed by real-time PCR was used. A calibration line was constructed using a standard with a known concentration to quantify the miRNA concentration. A standard was prepared using synthetic miRNA miR-148a-3p at concentrations of 10−1 ng/μL, 10−3 ng/μL, and 10−5 ng/μL. The quantification of cycle (Cq) values were calculated based on the amplification curves for different concentrations of synthetic miRNA. These values were used to create a calibration curve, and an equation was derived to accurately calculate the concentration of the researched miRNAs.
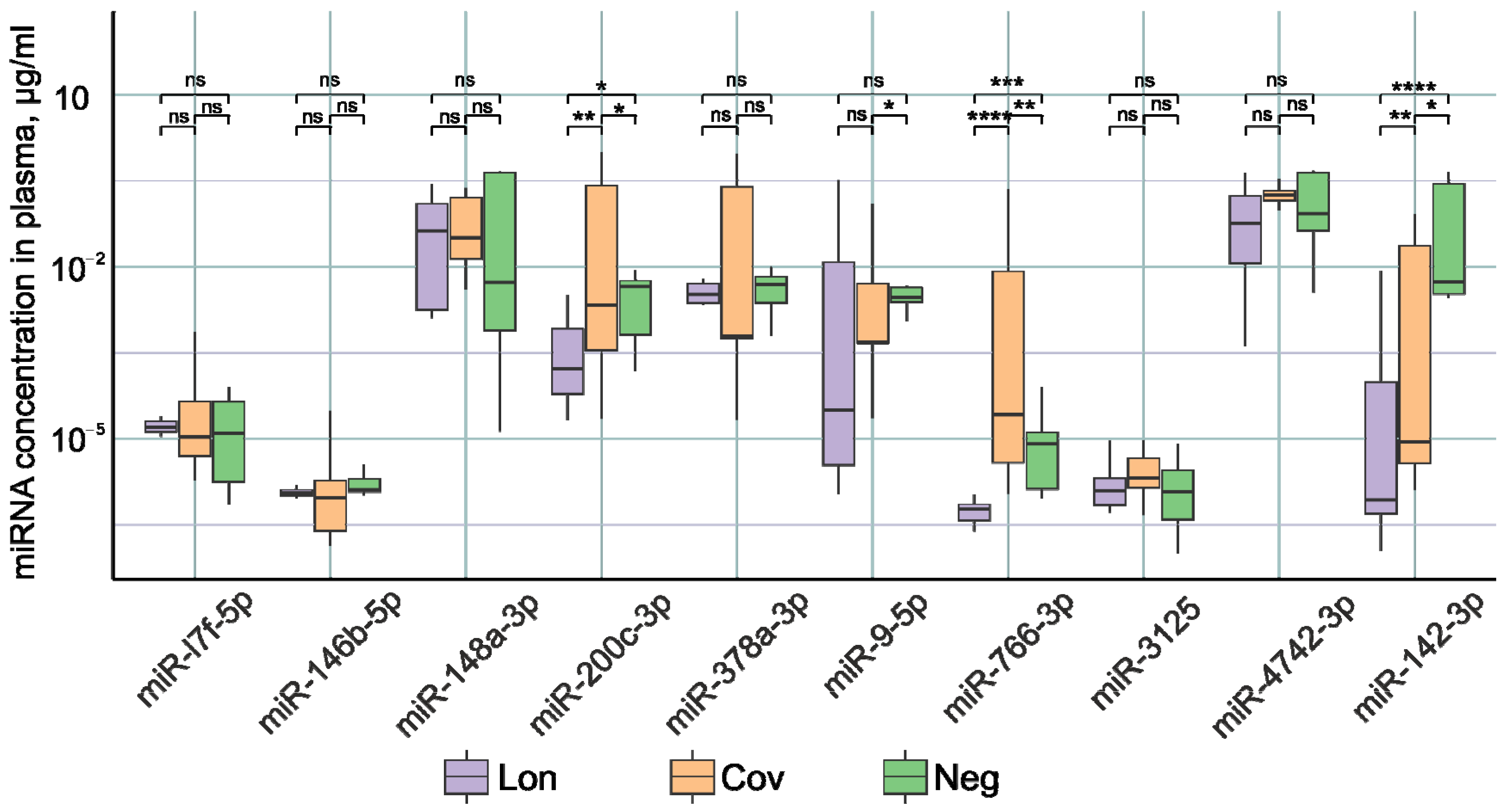
Figure 1 demonstrates statistically significant differences in the concentrations of miR-200c-3p, miR-766-3p, and miR-142-3p between patient groups. In the Lon group, there was a significant reduction in the expression of miR-200c-3p, miR-766-3p, and miR-142-3p compared to the other two groups. It was previously shown that in the acute phase of COVID-19 infection, patients experience the decreased expression of miR-142-3p [14,15,73]. Decreased miR-142-3p expression is associated with the inflammatory process [74,75,76]. Our results demonstrate that miR-142-3p levels remain lower after recovery compared to healthy patients. It is likely that normalization of miR-142-3p expression requires a longer time than 3–6 months after recovery.
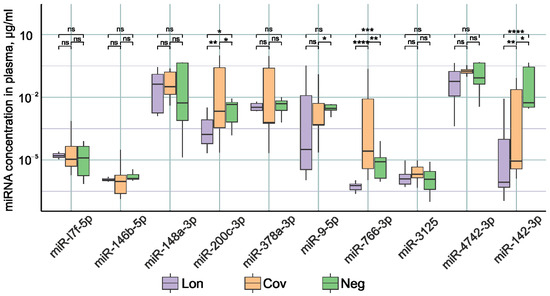
Figure 1.
Examination of miRNA levels in the plasma of individuals within the three study cohorts: Lon refers to patients who experienced persistent rheumatologic symptoms alongside COVID-19, Cov denotes patients who successfully recovered from COVID-19 without any lingering complaints, and Neg represents disease-free donors. *—p-value < 0.05; **—p-value < 0.01; ***—p-value < 0.001, ****—p-value < 0.0001, ns—not significant.
The highest concentration of miR-200c-3p and miR-766-3p was identified in the group of patients with no complaints after COVID-19. In the post-recovery period, we observed a reduced expression of these miRNAs. However, an increase in miR-200c-3p was reported in studies undertaken during the acute period of SARS-CoV-2 infection [77]. Moreover, the increase in the severity of the disease correlates with an increase in the expression of miR-200c-3p [78,79]. miR-200c-3p directly targets the 3′-untranslated region of ACE2 mRNA, resulting in decreased ACE2 expression [80]. miR-200c-3p expression is induced via the NF-κB pathway during infection. In particular, the NF-κB signaling pathway is hyperactivated during SARS-CoV-2 infection, leading to the overexpression of miR-200c-3p in COVID-19 patients with active disease. This leads to a decrease in ACE2 protein levels [81]. Decreased ACE2 expression leads to decreased susceptibility to SARS-CoV-2 entry into the host cell [82]. Thus, miR-200c-3p during COVID-19 infection by regulating ACE2 expression reduces further virus penetration into the cell, limits the spread of the virus, and has a positive effect on the recovery process. It is logical to assume that after recovery, normalization of ACE2 expression is necessary; therefore, miR-200c-3p expression decreases, which leads to an increase in ACE2 protein production until normal levels are reached. This is confirmed by our results of decreased miR-200c-3p expression in recovered patients from 3–6 months after COVID-19.
Data in the literature on the role of miR-766-3p in COVID-19 are scarce. Downregulation of this miRNA has been described in acute respiratory distress syndrome patients [19]. Our results demonstrate that miR-766-3p levels remain reduced 3–6 months after recovery.
2.3. Analysis of the Catalytic Activity of Antibodies in the Hydrolysis of miRNAs
The affinity chromatography method utilizing immobilized G-protein allows for the extraction of antibody preparations from different biological fluids, guaranteeing the absence of any protein or enzyme impurities [83]. In this study, IgG preparations were isolated from the blood plasma of the patients of the three study groups. To substantiate the specific catalytic function attributed to immunoglobulins, we employed the methodologies proposed by C. Paul [84] and further developed by the group of G. A. Nevinsky [50], in a similar way as described in [53].
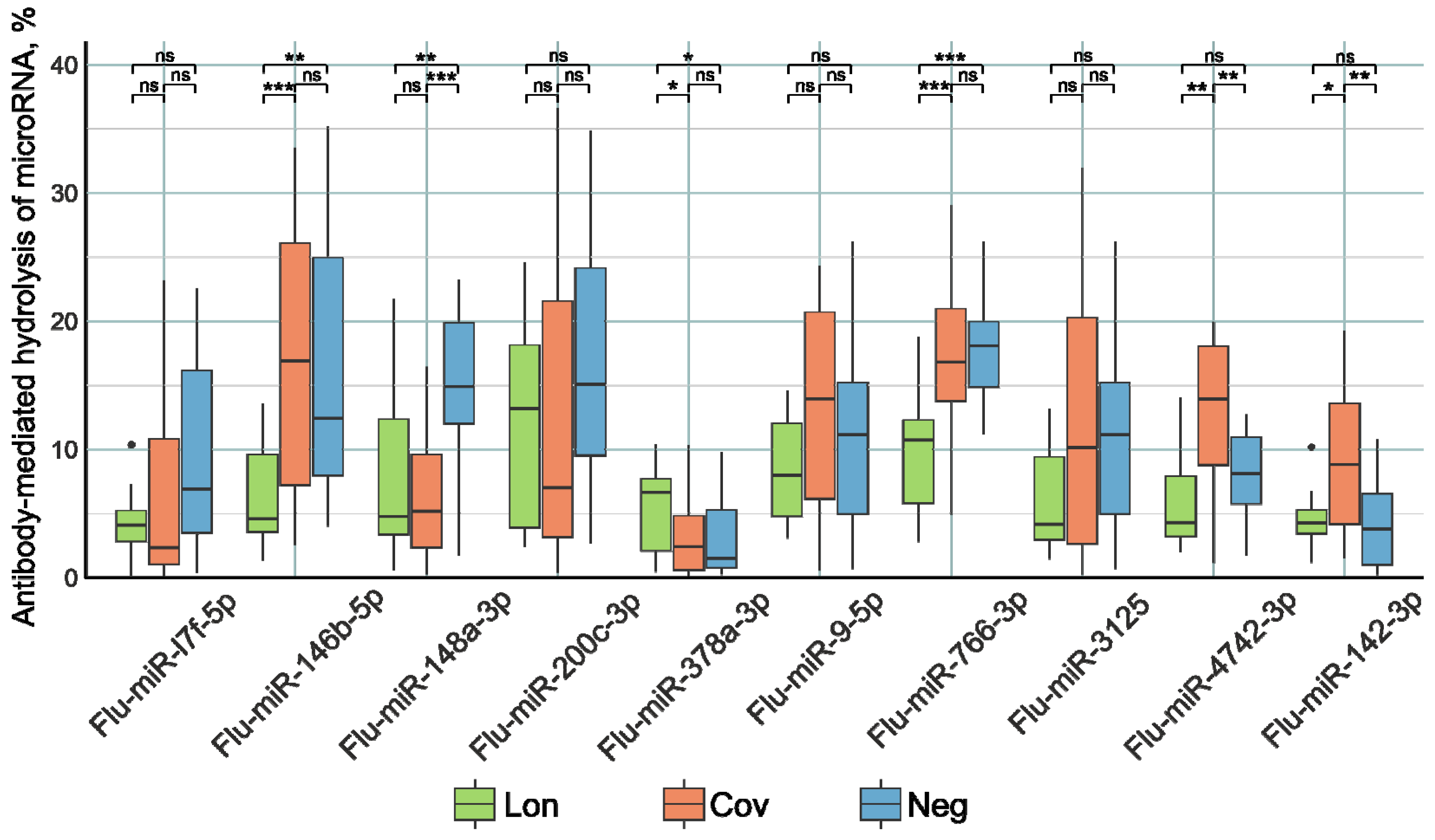
The examination of miRNA hydrolyzing activity involved the utilization of oligonucleotide sequences labeled with fluorescent tags, specifically designed to correspond to the tested miRNAs and distinguished by the prefix “Flu”. Oligonucleotide hydrolysis products were subjected to separation via a 20% polyacrylamide gel electrophoresis. Antibody activity in substrate hydrolysis was calculated by the decrease in the relative amount of the initial substrate in the reaction mixture compared to the control without IgG. The plasma antibodies of the patients were subjected to incubation with Flu-miRNAs. The nonparametric Kruskal–Wallis test (Figure 2) was utilized to evaluate the differentiation among the three patient groups.
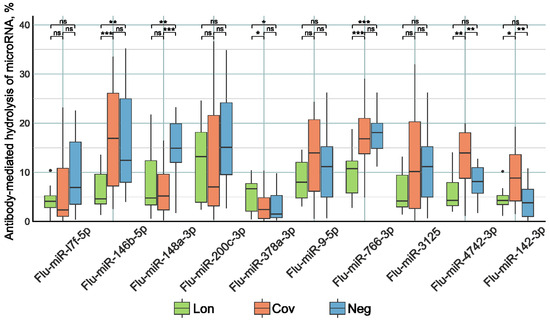
Figure 2.
The comparative activity of Flu-miRNA hydrolysis by plasma IgG preparations of patients in the three study groups. Lon refers to patients who experienced persistent rheumatologic symptoms alongside COVID-19, Cov denotes patients who successfully recovered from COVID-19 without any lingering complaints, and Neg represents disease-free donors. *—p-value < 0.05; **—p-value < 0.01; ***—p-value < 0.001, ns—not significant.
The IgG preparations of the Lon group patients demonstrated higher Flu-miR-378a-3p (p < 0.05) and lower Flu-miR-146b-5p, Flu-miR-148a-3p (p < 0.001), and Flu-miR-766-3p (p < 0.0001) hydrolysis activity compared to the control group of patients. Antibody preparations from Cov patients hydrolyzed Flu-miR-4742-3p and Flu-miR-142-3p (p < 0.001) with higher activity and hydrolyzed Flu-miR-148a-3p (p < 0.0001) with lower activity compared to the control group of patients.
3. Discussion
Although miRNAs have attracted attention as potential biomarkers, their practical use in diagnostics has faced obstacles. With miRNAs being short sequences that exhibit a significant degree of similarity [85], a high-specificity method is required for accurately quantifying each miRNA. Currently, the RT-qPCR method is considered the most reliable technique for detecting miRNA. The sensitivity of RT-qPCR is remarkable, allowing for the detection of miRNA molecules at the single nucleotide level, even when present in very low concentrations in the attomolar range [86,87]. Typically, the quantitative analysis of RNA by RT-qPCR involves a two-step procedure: (i) Reverse transcription (RT) employed to create complementary DNA from the initial RNA and (ii) subsequent amplification of the DNA by PCR. The amplification process is monitored in real time using a specific dye (e.g., SYBR Green I) or a specific fluorescent probe. RT-qPCR was initially developed to quantitatively analyze long RNA sequences. This involved using PCR primers that are typically 20 bases long, equivalent to the size of a full-length miRNA. The issue was effectively addressed through the implementation of stem-loop (SL) primers [87]. SL primers enable reliable analysis of miRNAs at low concentrations, enhance result accuracy by ensuring sequence specificity, and mitigate the influence of genomic DNA contaminants. The accuracy of the method is greatly enhanced by its resistance to contaminants [87,88]. In this study, we used this method to determine the concentrations of miRNAs in the plasma of patients, with the focus on convalescent individuals. Throughout the period of recovery, a complex mechanism is implemented to control inflammatory processes, with miRNA serving as a single component of the overall puzzle. We detected an alteration in the expression of three miRNAs out of 10 miRNAs, namely, miR-200c-3p, miR-766-3p, and miR-142-3p. It is worth mentioning that among the patients experiencing rheumatologic complications of long COVID, a notable decrease in the expression of these miRNAs was observed.
One potential method for reducing miRNA concentrations is antibody-mediated hydrolysis. In addition to their ability to recognize specific substrates, antibodies with catalytic functions can also hydrolyze the substrate [89]. Hence, the isolation of IgG from the blood plasma of patients was followed by an assessment of their catalytic activity in the degradation of fluorescently labeled oligonucleotides corresponding to ten miRNAs. The analysis of antibody catalytic activity revealed that patients in the Lon group exhibited reduced activity in hydrolyzing Flu-miR-146b-5p, Flu-miR-766-3p, Flu-miR-4742-3p, and Flu-miR-142-3p sequences, but increased activity against Flu-miR-378a-3p compared to other patient groups. Lower concentrations of miRNAs correspond to lower activity of antibodies in miRNA degradation. Thus, changes in miRNA concentrations are not associated with their hydrolysis by antibodies, but are regulated by more intricate mechanisms. There is reason to believe that a decrease in the concentration of miRNAs in blood plasma is associated with a decrease in the concentration of corresponding antibodies with miRNA-hydrolyzing activity.
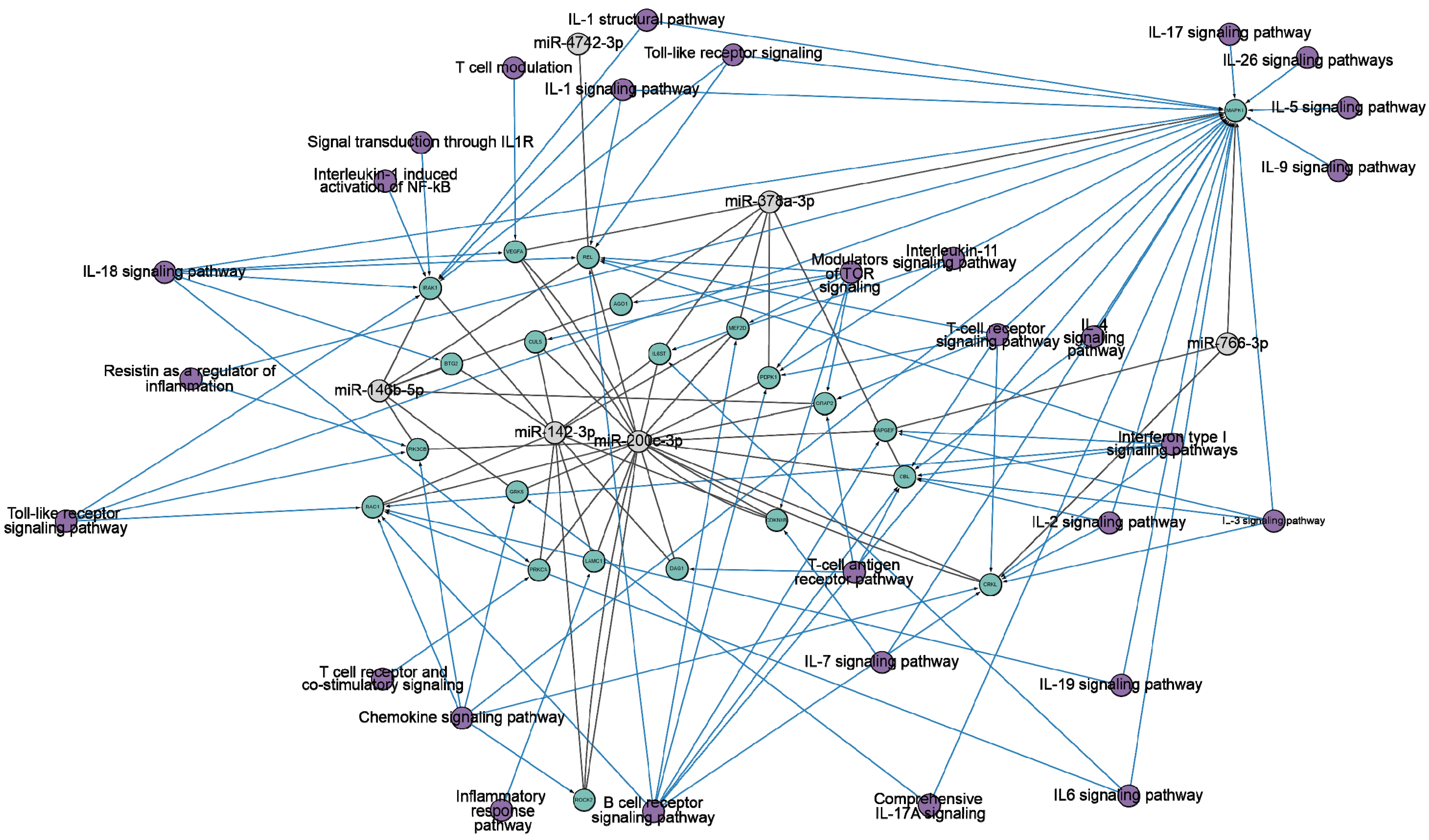
We hypothesized that changes in miRNA concentrations and antibody-mediated hydrolysis of miRNAs have a complex regulatory mechanism associated with gene pathways concerned with the immune system. Consequently, we conducted a more in-depth analysis of the target genes and pathways related to miR-200c-3p, miR-766-3p, miR-142-3p, miR-146b-5p, miR-4742-3p, and miR-378a-3p. The analysis framework is outlined in Figure S1 (Supplementary Materials). We searched for specific miRNA genes in the miRTarBase [90] and TargetScan [91] databases (accessed on 16 June 2024). Interactions between six specific miRNAs were observed with 1621 genes in the miRTarBase database and 1702 genes in the TargetScan database. Further analysis was conducted on a set of 338 common target genes, which were selected based on their association with six miRNAs. Then, gene-pathway associations were analyzed using the WikiPathways database [92,93] and pathways related to immune response regulation were selected (Figure 3).
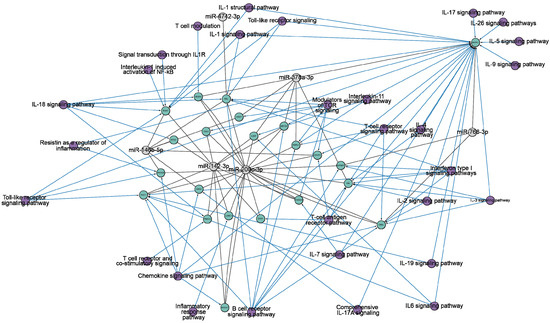
Figure 3.
The network diagram of the interactions between target genes (highlighted in green) and six miRNAs (miR-200c-3p, miR-766-3p, miR-142-3p, miR-146b-5p, miR-4742-3p, miR-378a-3p) (highlighted in gray). The information is based on miRTarBase [90] and TargetScan [91] (accessed on 15 June 2024). The representation is limited to genes that regulate the immune response. Purple indicates the ontology of genes. The network is visualized using Cytoscape version 3.10.2.
Figure 3 clearly demonstrates the impact of all miRNAs on pathways related to the immune response. To illustrate, miR-200c-3p effectively targets 18 genes that are associated with a wide range of signaling pathways, including IL-1, IL-2, IL-3, IL-4, IL-6, IL-7, IL-18, IL-19, interferon type I, chemokine, T-cell, B-cell, and toll-like receptors signaling pathways. Consequently, the scientific literature has noted an increase in the expression of miR-200c-3p in viral infections, including influenza A [94]. miR-200c-3p is involved in the regulation of angiotensin-converting enzyme II (ACE2). ACE2 has many physiological functions in lung tissue, among which it physiologically hydrolyzes angiotensin II [95,96,97]. Overexpression of miR-200c-3p leads to increased angiotensin II concentrations [98]. It has proinflammatory and pro-oxidant effects, which causes vasoconstriction, increased inflammation, thrombosis, and increased collagen synthesis in lung fibroblasts [99,100]. This leads to acute lung injury, alveolar edema, and increased incidence of pulmonary fibrosis [101,102]. Pulmonary fibrosis is a well-known long-term complication of COVID-19, which may be caused by the same mechanism. It was shown that miR-200c-3p expression level increases with increasing disease severity in COVID-19 patients [78,79].
miR-142-3p is associated with 11 genes that influence the immune response through IL-1, IL-7, IL-11, IL-18, chemokine, T- and B-cell, toll-like receptor signaling pathways. miR-142-3p is an evolutionarily conserved vertebrate miRNA that exhibits expression in a variety of hematopoietic cells, including dendritic cells, monocytes, T cells, and B cells [103,104]. miR-142-3p plays an important role in the modulation of immune responses [105,106]. It was shown that miR-142-3p is part of a detrimental regulatory axis with proinflammatory cytokines IL-1β [107,108] and IL-6 [109] in COVID-19 patients, inducing a response associated with respiratory failure and death. miR-142 regulates the expression of occludin, which affects endothelial permeability [110,111,112], contributing to endothelial dysfunction [113,114,115]. This leads to thromboembolic events [116,117] in patients with severe COVID-19 [118,119,120].
miR-146b-5p, miR-378a-3p, and miR-766-3p are found to be associated with 7, 7, and 3 genes, respectively, and are known to be connected to diverse pathways involved in the immune response. Although miR-4742-3p is associated with just one gene, it shares common associations with IL-1, IL-18, IL-19, interferon type I, T- and B-cell, and toll-like receptor signaling pathways, just like the other miRNAs being considered. MiR-146b-5p and miR-378a-3p have been described as a tumor suppressor and anti-inflammatory effector [121,122]. Furthermore, the computational analysis determined the capacity of miR-4742-3p to bind to the RNA of SARS-CoV-2 and hinder the production of viral proteins [123].
In conclusion, our findings have revealed the changes in the expression of three specific miRNAs (miR-200c-3p, miR-766-3p, miR-142-3p) among COVID-19 survivors and individuals with long-COVID-related rheumatologic issues. These patients exhibited changes in the antibody-mediated hydrolysis activity of five specific miRNAs (Flu-miR-146b-5p, Flu-miR-378a-3p, Flu-miR-766-3p, Flu-miR-4742-3p, and Flu-miR-142-3p). Viral infection activates or represses the expression of cellular miRNAs, which in turn modulate the host response to infections [124]. MiRNAs are post-transcriptional regulators of gene expression. They target specific mRNA sequences and control protein production by binding to the untranslated region of the mRNA [125]. Many miRNAs are associated with the regulation of a large number of pathways. The bioinformatics methods allow us to identify the genes that miRNAs influence. In this work, we have shown that all miRNAs participate in a complex network of interactions. Numerous signaling pathways related to the immune system function have been associated with all these miRNAs. The study of the interplay between miRNAs and immune reactions in organisms holds significance in the field of modern biomedicine. It has the potential to greatly enhance our understanding of pathogenesis mechanisms, facilitate the development of diagnostic biomarkers, and inform treatment strategies for the consequences of not only COVID-19 but also other viral infections.
4. Materials and Methods
4.1. Donors and Patients
This study protocol underwent a thorough review and received approval from the local ethical committee at the ICBFM SB RAS, (the protocol of 15 August 2021). In accordance with the Helsinki Ethics Committee’s recommendations, patients and healthy donors were duly informed and gave consent for their blood donation for scientific purposes.
Venous blood was collected on an empty stomach using vacuum tubes that contained coagulation activators. The blood samples in the tubes underwent centrifugation at 3000× g for 10 min using a refrigerated Centrifuge 5810. The resulting plasma, which had been separated from red blood cells, was divided into 1 mL aliquots and stored at a temperature of −70 °C.
The patients involved in this study were categorized into 3 study groups:
Cov refers to patients who successfully recovered from COVID-19 without any complaints, with a total number of 24 donors. The blood plasma of the Cov group patients was collected in 2020–2021.
Lon refers to patients who experienced persistent rheumatologic symptoms alongside COVID-19. The Lon group was formed in 2021, with a total number of 16 donors. The patients were examined by a rheumatologist in dynamics, with most patients in this group complaining of joint and muscle pain (n = 16), fever to subfebrile digits (n = 15), fatigue and weakness (n = 13), shooting pains in the body (n = 7), increased anxiety related to their condition (n = 6), and numbness of extremities (n = 5).
Neg refers to conditionally healthy donors who did not have COVID-19, with a total number of 18 donors.
The characteristics of the study groups of patients are presented in Table S1. The composition of the groups was adjusted to ensure gender and age parity. ELISA was used to confirm the presence of persistent SARS-CoV-2 infection and the absence of infection in the control group, targeting the S- and N-proteins of the virus [53,126].
4.2. Isolation of miRNAs
MiRNA isolation from blood plasma samples was performed using the reagents from the “Total RNA and miRNA isolation kit” (LRU-100-50, Biolabmix, Russia, Novosibirsk) according to the manufacturer’s instructions. The isolated miRNA pool was stored in low sorption tubes at −20 °C. The RNA concentration was determined spectrophotometrically by measuring the absorbance at 260 nm.
4.3. Reverse Transcription Using SL-Primers
The reverse transcription reaction was performed using the OT kit M-MuLV-RH (Biolabmix, Novosibirsk, Russia) in combination with a specific SL-primer (Table 1). The reaction mixture comprised 2 μL of SL-primer (1 μM), 2 μL of the miRNA being tested, and 8 μL of DEPC pretreated water. The mixture was heated at 70 °C for 3 min to disrupt secondary structures and immediately cooled in ice. The subsequent addition involved a reaction mixture composed of 4 μL of 5 × OT buffer, 1 μL of M-MuLV-RH revertase (concentration: 10 units/μL), and 3 μL of DEPK-pretreated water.

Table 1.
SL-primers used in the study.
Reverse transcription was performed in 45 cycles according to the following protocol:
1. 30 min at 16 °C;
2. (30 s at 30 °C, 30 s at 42 °C, 1 s at 50 °C) × 45;
3. 5 min at 85 °C.
4.4. Real-Time PCR
The cDNA generated by reverse transcription was analyzed using the BioMaster HS-qPCR PCR kit (Biolabmix, Novosibirsk, Russia). The reaction mixture (20 µL) contained 10 µL of BioMaster HS-qPCR (2×), 0.4 µL of forward primer (10 µM), 0.4 µL of universal reverse primer SL_rev (Table 2), 0.2 µL of universal probe with FAM dye (10 µM), 2 µL of cDNA (SL-OT product), and 7 µL of sterile water.

Table 2.
Primers used in the study.
The amplification procedure included the following steps: 5 min at 95 °C, 45 cycles (20 s at 95 °C and 1 min at 55 °C). The fluorescent signal detection was performed through the FAM at the end of each cycle.
MiRNAs were quantified in the blood plasma using a synthetic miR-148a-3p with known concentration as normalization. A calibration curve was generated using synthetic miRNA at concentrations of 10–1 ng/µL, 10–3 ng/µL, and 10–5 ng/µL. Subsequently, OT-PCR and real-time PCR reactions were conducted. The construction of calibration curves involved the utilization of Microsoft Excel 2016 software and the analysis of four repeated measurements. The equation for the calibration curve was derived using CFX Maestro software version 2.3 (BioRad, Singapore, Tower): y = −2.384 ln(x) − 1.983, with y representing the Cq value obtained post-amplification, and x denoting the concentration.
4.5. Identification of IgG Activity in the miRNA Hydrolysis Reaction
IgGs were isolated from the blood plasma of patients using affinity chromatography on Protein-G-Sepharose similar to [127]. The efficiency of miRNA cleavage in 20% PAAG was used to determine the relative catalytic activity of IgG, as described in [58,59]. The reaction mixture (10 µL) contained 20 mM tris–HCl, pH 7.5; IgG preparation (0.1 mg/mL), and fluorescently labeled synthetic miRNA (Table 3), with a concentration from 0.09 to 0.39 f.u./mL (depending on the miRNA). The reaction mixture was incubated for 1 h at 37 °C. A reaction mixture without IgG was used as a control. Following the incubation, the reaction mixture was supplemented with 10 μL of denaturing buffer containing 8 M urea and 0.025% xylenecyanol. The marker (Leader) was procured via a process of limited alkaline hydrolysis, employing a bicarbonate buffer with a concentration of 0.05 M and miRNA concentrations ranging from 0.08 to 0.35 f.u./mL. The mixture was incubated for 15 min at 90 °C. After that, 10 µL of denaturing buffer was added. The hydrolysis products were analyzed using electrophoresis under denaturing conditions (20% acrylamide, 8 M urea, 1× TBE pH 8.3) at 800 V, 40 mA, for 2 h. The hydrolysis products were visualized using the iBright™ CL1500 Imaging System (Invitrogen™, Waltham, MA, USA), and the analysis was performed using the GelAnalyzer 23.1 software.

Table 3.
5′-Flu-labeled oligoribonucleotides used as substrates in this study.
4.6. Statistical Analysis
The calculation of the significance of differences between patient groups was performed utilizing the Mann–Whitney U-test from the Python library SciPy v 1.11.4 [128] and the Kruskal–Wallis criterion from the standard R package v 4.3.2. The correlation coefficients between groups were computed using the nonparametric Spearman rank correlation method available in the Python library SciPy. The p-value was used to determine statistical significance, with a minimum threshold set at 0.05.
4.7. Target Predictions of the miRNAs Analyzed
The miRTarBase [90] and TargetScan [91] databases were utilized to search for target miRNA genes. This was performed through the CyTargetLinker 4.0.0+ application [129] (accessed on 15 June 2024). Subsequently, we utilized the WikiPathways database [92,93] to perform pathway analysis on the identified genes. Genes associated with the control of the immune response were specifically selected. The interaction network was visualized using Cytoscape 3.10.2.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ncrna10050048/s1, Table S1. Characteristics of the patients. Figure S1: Interaction networks between target genes and six miRNAs.
Author Contributions
Conceptualization, A.M.T.; methodology, A.O.N.; validation, A.M.T.; investigation, A.O.N. and A.M.T.; resources, A.O.N. and A.M.T.; writing—original draft preparation, A.O.N.; writing—review and editing, A.M.T. and G.A.N.; visualization, A.O.N. and A.M.T.; project administration, A.M.T.; funding acquisition G.A.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was maintained by the Russian Science Foundation (Project 22-15-00103 to Georgy Nevinsky—all the research except for statistical analysis) and the Russian State-funded budget project of ICBFM SB RAS 0245-2021-0009 (121031300041-4) to Georgy Nevinsky—statistical analysis).
Institutional Review Board Statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Local Ethics Committee of the Institute of Chemical Biology and Fundamental Medicine (Protocol Number 21-4 from 15 August 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent was obtained from all patients before their inclusion in the study.
Data Availability Statement
Empirical data that do not relate to the personal data of donors can be provided by Anna Timofeeva upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kincaid, R.P.; Sullivan, C.S. Virus-Encoded MicroRNAs: An Overview and a Look to the Future. PLoS Pathog. 2012, 8, e1003018. [Google Scholar] [CrossRef] [PubMed]
- Intartaglia, D.; Giamundo, G.; Conte, I. The Impact of MiRNAs in Health and Disease of Retinal Pigment Epithelium. Front. Cell Dev. Biol. 2021, 8, 589985. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Galvão-Lima, L.J.; Morais, A.H.F.; Valentim, R.A.M.; Barreto, E.J.S.S. MiRNAs as Biomarkers for Early Cancer Detection and Their Application in the Development of New Diagnostic Tools. Biomed. Eng. Online 2021, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Dash, C.; Pandhare, J. Therapeutic Significance of MicroRNA-Mediated Regulation of PARP-1 in SARS-CoV-2 Infection. Non-Coding RNA 2021, 7, 60. [Google Scholar] [CrossRef]
- Belmonte, T.; Perez-Pons, M.; Benítez, I.D.; Molinero, M.; García-Hidalgo, M.C.; Rodríguez-Muñoz, C.; Gort-Paniello, C.; Moncusí-Moix, A.; Madè, A.; Devaux, Y.; et al. Addressing the Unsolved Challenges in MicroRNA-Based Biomarker Development: Suitable Endogenous Reference MicroRNAs for SARS-CoV-2 Infection Severity. Int. J. Biol. Macromol. 2024, 269, 131926. [Google Scholar] [CrossRef]
- Wicik, Z.; Eyileten, C.; Nowak, A.; Keshwani, D.; Simões, S.N.; Martins, D.C.; Klos, K.; Wlodarczyk, W.; Assinger, A.; Soldacki, D.; et al. Alteration of Circulating ACE2-Network Related MicroRNAs in Patients with COVID-19. Sci. Rep. 2024, 14, 13573. [Google Scholar] [CrossRef]
- Garcia-Giralt, N.; Du, J.; Marin-Corral, J.; Bódalo-Torruella, M.; Blasco-Hernando, F.; Muñoz-Bermúdez, R.; Clarós, M.; Nonell, L.; Perera-Bel, J.; Fernandez-González, M.; et al. Circulating MicroRNA Profiling Is Altered in the Acute Respiratory Distress Syndrome Related to SARS-CoV-2 Infection. Sci. Rep. 2022, 12, 6929. [Google Scholar] [CrossRef] [PubMed]
- Togami, Y.; Matsumoto, H.; Yoshimura, J.; Matsubara, T.; Ebihara, T.; Matsuura, H.; Mitsuyama, Y.; Kojima, T.; Ishikawa, M.; Sugihara, F.; et al. Significance of Interferon Signaling Based on MRNA-MicroRNA Integration and Plasma Protein Analyses in Critically Ill COVID-19 Patients. Mol. Ther.-Nucleic Acids 2022, 29, 343–353. [Google Scholar] [CrossRef]
- Gao, L.; Kyubwa, E.M.; Starbird, M.A.; Diaz de Leon, J.; Nguyen, M.; Rogers, C.J.; Menon, N. Circulating MiRNA Profiles in COVID-19 Patients and Meta-Analysis: Implications for Disease Progression and Prognosis. Sci. Rep. 2023, 13, 21656. [Google Scholar] [CrossRef]
- de Souza Nicoletti, A.; Berlofa Visacri, M.; Regina da Silva Correa da Ronda, C.; Tiemi Siguemoto, J.; Motta Neri, C.; Nogueira de Souza, R.; de Souza Ventura, D.; Eguti, A.; Ferreira de Souza Silva, L.; Wesley Perroud Junior, M.; et al. Increased Expression of MiR-320b in Blood Plasma of Patients in Response to SARS-CoV-2 Infection. Sci. Rep. 2024, 14, 13702. [Google Scholar] [CrossRef] [PubMed]
- Giannella, A.; Riccetti, S.; Sinigaglia, A.; Piubelli, C.; Razzaboni, E.; Di Battista, P.; Agostini, M.; Dal Molin, E.; Manganelli, R.; Gobbi, F.; et al. Circulating MicroRNA Signatures Associated with Disease Severity and Outcome in COVID-19 Patients. Front. Immunol. 2022, 13, 968991. [Google Scholar] [CrossRef] [PubMed]
- Duecker, R.P.; Adam, E.H.; Wirtz, S.; Gronau, L.; Khodamoradi, Y.; Eberhardt, F.J.; Donath, H.; Gutmann, D.; Vehreschild, M.J.G.T.; Zacharowski, K.; et al. The MiR-320 Family Is Strongly Downregulated in Patients with COVID-19 Induced Severe Respiratory Failure. Int. J. Mol. Sci. 2021, 22, 10351. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hu, X.; Li, L.; Li, J. Differential MicroRNA Expression in the Peripheral Blood from Human Patients with COVID-19. J. Clin. Lab. Anal. 2020, 34, e23590. [Google Scholar] [CrossRef] [PubMed]
- Meidert, A.S.; Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Billaud, J.-N.; Klein, M.; Lindemann, A.; Aue, E.; Schelling, G.; et al. Extracellular Vesicle Associated MiRNAs Regulate Signaling Pathways Involved in COVID-19 Pneumonia and the Progression to Severe Acute Respiratory Corona Virus-2 Syndrome. Front. Immunol. 2021, 12, 784028. [Google Scholar] [CrossRef]
- Zeng, Q.; Qi, X.; Ma, J.; Hu, F.; Wang, X.; Qin, H.; Li, M.; Huang, S.; Yang, Y.; Li, Y.; et al. Distinct MiRNAs Associated with Various Clinical Presentations of SARS-CoV-2 Infection. iScience 2022, 25, 104309. [Google Scholar] [CrossRef]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating Cardiovascular microRNAs in Critically Ill COVID-19 Patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef]
- Nicoletti, A.d.S.; Visacri, M.B.; da Ronda, C.R.d.S.C.; Vasconcelos, P.E.d.N.S.; Quintanilha, J.C.F.; de Souza, R.N.; Ventura, D.d.S.; Eguti, A.; Silva, L.F.d.S.; Perroud Junior, M.W.; et al. Differentially Expressed Plasmatic MicroRNAs in Brazilian Patients with Coronavirus Disease 2019 (COVID-19): Preliminary Results. Mol. Biol. Rep. 2022, 49, 6931–6943. [Google Scholar] [CrossRef]
- Najafipour, R.; Mohammadi, D.; Estaki, Z.; Zarabadi, K.; Jalilvand, M.; Moghbelinejad, S. Screening for Differentially Expressed MicroRNAs in BALF and Blood Samples of Infected COVID-19 ARDS Patients by Small RNA Deep Sequencing. J. Clin. Lab. Anal. 2022, 36, e24672. [Google Scholar] [CrossRef]
- tenOever, B.R. RNA Viruses and the Host MicroRNA Machinery. Nat. Rev. Microbiol. 2013, 11, 169–180. [Google Scholar] [CrossRef]
- Rad, S.M.A.H.; Wannigama, D.L.; Hirankarn, N.; McLellan, A.D. The Impact of Non-Synonymous Mutations on MiRNA Binding Sites within the SARS-CoV-2 NSP3 and NSP4 Genes. Sci. Rep. 2023, 13, 16945. [Google Scholar] [CrossRef] [PubMed]
- Hosseini Rad SM, A.; McLellan, A.D. Implications of SARS-CoV-2 Mutations for Genomic RNA Structure and Host MicroRNA Targeting. Int. J. Mol. Sci. 2020, 21, 4807. [Google Scholar] [CrossRef] [PubMed]
- Nanbo, A.; Furuyama, W.; Lin, Z. RNA Virus-Encoded MiRNAs: Current Insights and Future Challenges. Front. Microbiol. 2021, 12, 679210. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. MiRBase: Integrating MicroRNA Annotation and Deep-Sequencing Data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Grundhoff, A.; Sullivan, C.S. Virus-Encoded MicroRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef]
- Cullen, B.R. Five Questions about Viruses and MicroRNAs. PLoS Pathog. 2010, 6, e1000787. [Google Scholar] [CrossRef] [PubMed]
- Greco, F.; Lorefice, E.; Carissimi, C.; Laudadio, I.; Ciccosanti, F.; Di Rienzo, M.; Colavita, F.; Meschi, S.; Maggi, F.; Fimia, G.M.; et al. A MicroRNA Arising from the Negative Strand of SARS-CoV-2 Genome Targets FOS to Reduce AP-1 Activity. Non-Coding RNA 2023, 9, 33. [Google Scholar] [CrossRef]
- Morales, L.; Oliveros, J.C.; Fernandez-Delgado, R.; TenOever, B.R.; Enjuanes, L.; Sola, I. SARS-CoV-Encoded Small RNAs Contribute to Infection-Associated Lung Pathology. Cell Host Microbe 2017, 21, 344–355. [Google Scholar] [CrossRef]
- Meng, F.; Siu, G.K.-H.; Mok, B.W.-Y.; Sun, J.; Fung, K.S.C.; Lam, J.Y.-W.; Wong, N.K.; Gedefaw, L.; Luo, S.; Lee, T.M.H.; et al. Viral MicroRNAs Encoded by Nucleocapsid Gene of SARS-CoV-2 Are Detected during Infection, and Targeting Metabolic Pathways in Host Cells. Cells 2021, 10, 1762. [Google Scholar] [CrossRef]
- Gracia-Ramos, A.E.; Martin-Nares, E.; Hernández-Molina, G. New Onset of Autoimmune Diseases Following COVID-19 Diagnosis. Cells 2021, 10, 3592. [Google Scholar] [CrossRef]
- Talotta, R.; Robertson, E. Autoimmunity as the Comet Tail of COVID-19 Pandemic. World J. Clin. Cases 2020, 8, 3621–3644. [Google Scholar] [CrossRef]
- Gollapudi, S.; Chimurkar, V. Comprehensive Insights Into the Multi-Faceted Manifestations of COVID-19: A Narrative Review. Cureus 2024, 16, e63493. [Google Scholar] [CrossRef] [PubMed]
- Sugino, K.; Ono, H.; Haraguchi, S.; Igarashi, S.; Hebisawa, A.; Tsuboi, E. Post-coronavirus Disease 2019 Organizing Pneumonia Confirmed Pathologically by Video-assisted Thoracoscopic Surgery. Respirol. Case Rep. 2021, 9, e0871. [Google Scholar] [CrossRef] [PubMed]
- Bazdyrev, E.; Rusina, P.; Panova, M.; Novikov, F.; Grishagin, I.; Nebolsin, V. Lung Fibrosis after COVID-19: Treatment Prospects. Pharmaceuticals 2021, 14, 807. [Google Scholar] [CrossRef]
- Abu-Rumeileh, S.; Abdelhak, A.; Foschi, M.; Tumani, H.; Otto, M. Guillain–Barré Syndrome Spectrum Associated with COVID-19: An up-to-Date Systematic Review of 73 Cases. J. Neurol. 2021, 268, 1133–1170. [Google Scholar] [CrossRef]
- Kozłowski, P.; Leszczyńska, A.; Ciepiela, O. Long COVID Definition, Symptoms, Risk Factors, Epidemiology and Autoimmunity: A Narrative Review. Am. J. Med. Open 2024, 11, 100068. [Google Scholar] [CrossRef]
- Roghani, S.A.; Dastbaz, M.; Lotfi, R.; Shamsi, A.; Abdan, Z.; Rostampour, R.; Soleymani, B.; Zamanian, M.H.; Soufivand, P.; Pournazari, M.; et al. The Development of Anticyclic Citrullinated Peptide (Anti-CCP) Antibody Following Severe COVID-19. Immun. Inflamm. Dis. 2024, 12, e1276. [Google Scholar] [CrossRef] [PubMed]
- Ciaffi, J.; Vanni, E.; Mancarella, L.; Brusi, V.; Lisi, L.; Pignatti, F.; Naldi, S.; Assirelli, E.; Neri, S.; Reta, M.; et al. Post-Acute COVID-19 Joint Pain and New Onset of Rheumatic Musculoskeletal Diseases: A Systematic Review. Diagnostics 2023, 13, 1850. [Google Scholar] [CrossRef]
- Migliorini, F.; Bell, A.; Vaishya, R.; Eschweiler, J.; Hildebrand, F.; Maffulli, N. Reactive Arthritis Following COVID-19 Current Evidence, Diagnosis, and Management Strategies. J. Orthop. Surg. Res. 2023, 18, 205. [Google Scholar] [CrossRef]
- Reyes-Long, S.; Cortés-Altamirano, J.L.; Bandala, C.; Avendaño-Ortiz, K.; Bonilla-Jaime, H.; Bueno-Nava, A.; Ávila-Luna, A.; Sánchez-Aparicio, P.; Clavijo-Cornejo, D.; Dotor-LLerena, A.L.; et al. Role of the MicroRNAs in the Pathogenic Mechanism of Painful Symptoms in Long COVID: Systematic Review. Int. J. Mol. Sci. 2023, 24, 3574. [Google Scholar] [CrossRef]
- Mone, P.; Jankauskas, S.S.; Manzi, M.V.; Gambardella, J.; Coppola, A.; Kansakar, U.; Izzo, R.; Fiorentino, G.; Lombardi, A.; Varzideh, F.; et al. Endothelial Extracellular Vesicles Enriched in MicroRNA-34a Predict New-Onset Diabetes in Coronavirus Disease 2019 (COVID-19) Patients: Novel Insights for Long COVID Metabolic Sequelae. J. Pharmacol. Exp. Ther. 2024, 389, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Izzo, C.; Visco, V.; Gambardella, J.; Ferruzzi, G.J.; Rispoli, A.; Rusciano, M.R.; Toni, A.L.; Virtuoso, N.; Carrizzo, A.; Di Pietro, P.; et al. Cardiovascular Implications of MicroRNAs in Coronavirus Disease 2019. J. Pharmacol. Exp. Ther. 2023, 384, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, J.; Santulli, G. What Is Linking COVID-19 and Endothelial Dysfunction? Updates on Nanomedicine and Bioengineering from the 2020 AHA Scientific Sessions. Eur. Hear. J.-Cardiovasc. Pharmacother. 2021, 7, e2–e3. [Google Scholar] [CrossRef] [PubMed]
- Fayyad-Kazan, M.; Makki, R.; Skafi, N.; El Homsi, M.; Hamade, A.; El Majzoub, R.; Hamade, E.; Fayyad-Kazan, H.; Badran, B. Circulating MiRNAs: Potential Diagnostic Role for Coronavirus Disease 2019 (COVID-19). Infect. Genet. Evol. 2021, 94, 105020. [Google Scholar] [CrossRef]
- Gambardella, J.; Kansakar, U.; Sardu, C.; Messina, V.; Jankauskas, S.S.; Marfella, R.; Maggi, P.; Wang, X.; Mone, P.; Paolisso, G.; et al. Exosomal MiR-145 and MiR-885 Regulate Thrombosis in COVID-19. J. Pharmacol. Exp. Ther. 2023, 384, 109–115. [Google Scholar] [CrossRef]
- Ye, Q.; Wang, B.; Mao, J. The Pathogenesis and Treatment of the `Cytokine Storm’ in COVID-19. J. Infect. 2020, 80, 607–613. [Google Scholar] [CrossRef]
- Chandan, K.; Gupta, M.; Sarwat, M. Role of Host and Pathogen-Derived MicroRNAs in Immune Regulation During Infectious and Inflammatory Diseases. Front. Immunol. 2020, 10, 3081. [Google Scholar] [CrossRef]
- Mohamed, H.A.; Abdelkafy, A.E.; Khairy, R.M.M.; Abdelraheim, S.R.; Kamel, B.A.; Marey, H. MicroRNAs and Cytokines as Potential Predictive Biomarkers for COVID-19 Disease Progression. Sci. Rep. 2023, 13, 3531. [Google Scholar] [CrossRef]
- Liu, Z.; Yu, H.; Guo, Q. MicroRNA-20a Promotes Inflammation via the Nuclear Factor-κB Signaling Pathway in Pediatric Pneumonia. Mol. Med. Rep. 2017, 17, 612–617. [Google Scholar] [CrossRef]
- Nevinsky, G.A.; Buneva, V.N. Natural Catalytic Antibodies in Norm, Autoimmune, Viral, and Bacterial Diseases. Sci. World J. 2010, 10, 1203–1233. [Google Scholar] [CrossRef]
- Tolmacheva, A.S.; Onvumere, M.K.; Sedykh, S.E.; Timofeeva, A.M.; Nevinsky, G.A. Catalase Activity of IgGs of Patients Infected with SARS-CoV-2. Int. J. Mol. Sci. 2023, 24, 10081. [Google Scholar] [CrossRef] [PubMed]
- McConnell, S.A.; Sachithanandham, J.; Mudrak, N.J.; Zhu, X.; Farhang, P.A.; Cordero, R.J.B.; Wear, M.P.; Shapiro, J.R.; Park, H.-S.; Klein, S.L.; et al. Spike-Protein Proteolytic Antibodies in COVID-19 Convalescent Plasma Contribute to SARS-CoV-2 Neutralization. Cell Chem. Biol. 2023, 30, 726–738.e4. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Shayakhmetova, L.S.; Nikitin, A.O.; Sedykh, T.A.; Matveev, A.L.; Shanshin, D.V.; Volosnikova, E.A.; Merkuleva, I.A.; Shcherbakov, D.N.; Tikunova, N.V.; et al. Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Hydrolyze Recombinant RBD and Nucleocapsid (N) Proteins. Biomedicines 2024, 12, 1007. [Google Scholar] [CrossRef]
- Lee, G.; Budhathoki, S.; Lee, G.-Y.; Oh, K.; Ham, Y.; Kim, Y.-J.; Lim, Y.; Hoang, P.; Lee, Y.; Lim, S.-W.; et al. Broad-Spectrum Antiviral Activity of 3D8, a Nucleic Acid-Hydrolyzing Single-Chain Variable Fragment (ScFv), Targeting SARS-CoV-2 and Multiple Coronaviruses In Vitro. Viruses 2021, 13, 650. [Google Scholar] [CrossRef] [PubMed]
- Luong, Q.X.T.; Hoang, P.T.; Lee, Y.; Ayun, R.Q.; Na, K.; Park, S.; Lin, C.; Ho, P.T.; Lee, T.-K.; Lee, S. An RNA-Hydrolyzing Recombinant Minibody Prevents Both Influenza A Virus and Coronavirus in Co-Infection Models. Sci. Rep. 2024, 14, 8472. [Google Scholar] [CrossRef] [PubMed]
- Hoang, P.T.; Luong, Q.X.T.; Ayun, R.Q.; Lee, Y.; Oh, K.-J.; Kim, T.; Lee, T.-K.; Lee, S. A Synergistic Therapy against Influenza Virus A/H1N1/PR8 by a HA1 Specific Neutralizing Single-Domain VL and an RNA Hydrolyzing ScFv. Front. Microbiol. 2024, 15, 1355599. [Google Scholar] [CrossRef]
- Lee, Y.; Hoang, P.; Kim, D.; Ayun, R.; Luong, Q.; Na, K.; Kim, T.; Oh, Y.; Kim, W.-K.; Lee, S. A Therapeutically Active Minibody Exhibits an Antiviral Activity in Oseltamivir-Resistant Influenza-Infected Mice via Direct Hydrolysis of Viral RNAs. Viruses 2022, 14, 1105. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Kabirova, E.M.; Sizikov, A.E.; Buneva, V.N.; Nevinsky, G.A. IgGs-Abzymes from the Sera of Patients with Systemic Lupus Erythematosus Hydrolyzed MiRNAs. J. Inflamm. Res. 2020, 13, 681–699. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Kabirova, E.M.; Buneva, V.N.; Nevinsky, G.A. IgGs-Abzymes from the Sera of Patients with Multiple Sclerosis Recognize and Hydrolyze MiRNAs. Int. J. Mol. Sci. 2021, 22, 2812. [Google Scholar] [CrossRef]
- Nevinsky, G.A.; Urusov, A.E.; Aulova, K.S.; Ermakov, E.A. Experimental Autoimmune Encephalomyelitis of Mice: IgGs from the Sera of Mice Hydrolyze MiRNAs. Int. J. Mol. Sci. 2023, 24, 4433. [Google Scholar] [CrossRef]
- Ermakov, E.A.; Ivanova, S.A.; Buneva, V.N.; Nevinsky, G.A. Hydrolysis by Catalytic IgGs of MicroRNA Specific for Patients with Schizophrenia. IUBMB Life 2018, 70, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Tesch, F.; Ehm, F.; Vivirito, A.; Wende, D.; Batram, M.; Loser, F.; Menzer, S.; Jacob, J.; Roessler, M.; Seifert, M.; et al. Incident Autoimmune Diseases in Association with SARS-CoV-2 Infection: A Matched Cohort Study. Clin. Rheumatol. 2023, 42, 2905–2914. [Google Scholar] [CrossRef] [PubMed]
- Al-Aly, Z.; Xie, Y.; Bowe, B. High-Dimensional Characterization of Post-Acute Sequelae of COVID-19. Nature 2021, 594, 259–264. [Google Scholar] [CrossRef]
- Taquet, M.; Geddes, J.R.; Husain, M.; Luciano, S.; Harrison, P.J. 6-Month Neurological and Psychiatric Outcomes in 236 379 Survivors of COVID-19: A Retrospective Cohort Study Using Electronic Health Records. Lancet Psychiatry 2021, 8, 416–427. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, E.; Bowe, B.; Al-Aly, Z. Long-Term Cardiovascular Outcomes of COVID-19. Nat. Med. 2022, 28, 583–590. [Google Scholar] [CrossRef]
- Xie, Y.; Xu, E.; Al-Aly, Z. Risks of Mental Health Outcomes in People with COVID-19: Cohort Study. BMJ 2022, 376, e068993. [Google Scholar] [CrossRef]
- Xu, E.; Xie, Y.; Al-Aly, Z. Long-Term Neurologic Outcomes of COVID-19. Nat. Med. 2022, 28, 2406–2415. [Google Scholar] [CrossRef]
- Roessler, M.; Tesch, F.; Batram, M.; Jacob, J.; Loser, F.; Weidinger, O.; Wende, D.; Vivirito, A.; Toepfner, N.; Ehm, F.; et al. Post-COVID-19-Associated Morbidity in Children, Adolescents, and Adults: A Matched Cohort Study Including More than 157,000 Individuals with COVID-19 in Germany. PLOS Med. 2022, 19, e1004122. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.Y.; Mao, T.; Klein, J.; Dai, Y.; Huck, J.D.; Jaycox, J.R.; Liu, F.; Zhou, T.; Israelow, B.; Wong, P.; et al. Diverse Functional Autoantibodies in Patients with COVID-19. Nature 2021, 595, 283–288. [Google Scholar] [CrossRef]
- Yazdanpanah, N.; Rezaei, N. Autoimmune Complications of COVID-19. J. Med. Virol. 2022, 94, 54–62. [Google Scholar] [CrossRef]
- Chang, R.; Yen-Ting Chen, T.; Wang, S.-I.; Hung, Y.-M.; Chen, H.-Y.; Wei, C.-C.J. Risk of Autoimmune Diseases in Patients with COVID-19: A Retrospective Cohort Study. eClinicalMedicine 2023, 56, 101783. [Google Scholar] [CrossRef] [PubMed]
- Klink, G.V.; Safina, K.R.; Garushyants, S.K.; Moldovan, M.; Nabieva, E.; Komissarov, A.B.; Lioznov, D.; Bazykin, G.A. Spread of Endemic SARS-CoV-2 Lineages in Russia before April 2021. PLoS ONE 2022, 17, e0270717. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The Noncoding and Coding Transcriptional Landscape of the Peripheral Immune Response in Patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. MicroRNA Regulation of Inflammatory Responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J. Turning 21: Induction of MiR-21 as a Key Switch in the Inflammatory Response. Front. Immunol. 2015, 6, 19. [Google Scholar] [CrossRef]
- Cheng, H.S.; Sivachandran, N.; Lau, A.; Boudreau, E.; Zhao, J.L.; Baltimore, D.; Delgado-Olguin, P.; Cybulsky, M.I.; Fish, J.E. MicroRNA-146 Represses Endothelial Activation by Inhibiting Pro-inflammatory Pathways. EMBO Mol. Med. 2013, 5, 1017–1034. [Google Scholar] [CrossRef]
- Pimenta, R.; Viana, N.I.; Dos Santos, G.A.; Candido, P.; Guimarães, V.R.; Romão, P.; Silva, I.A.; de Camargo, J.A.; Hatanaka, D.M.; Queiroz, P.G.S.; et al. MiR-200c-3p Expression May Be Associated with Worsening of the Clinical Course of Patients with COVID-19. Mol. Biol. Res. Commun. 2021, 10, 141–147. [Google Scholar] [CrossRef]
- Shaker, O.; El Amir, M.; Elfatah, Y.A.; Elwi, H.M. Expression Patterns of LncRNA MALAT-1 in SARS-CoV-2 Infection and Its Potential Effect on Disease Severity via MiR-200c-3p and SIRT1. Biochem. Biophys. Rep. 2023, 36, 101562. [Google Scholar] [CrossRef]
- Roustai Geraylow, K.; Hemmati, R.; Kadkhoda, S.; Ghafouri-Fard, S. MiRNA Expression in COVID-19. Gene Rep. 2022, 28, 101641. [Google Scholar] [CrossRef]
- Nersisyan, S.; Shkurnikov, M.; Turchinovich, A.; Knyazev, E.; Tonevitsky, A. Integrative Analysis of MiRNA and MRNA Sequencing Data Reveals Potential Regulatory Mechanisms of ACE2 and TMPRSS2. PLoS ONE 2020, 15, e0235987. [Google Scholar] [CrossRef]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and Viruses Induce a Novel Truncated ACE2 Isoform and Not the Full-Length SARS-CoV-2 Receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Sensi, S.; Goebel, A. Human Auto-IgG Purification from High Volume Serum Sample by Protein G Affinity Purification. Bio-Protocol 2022, 12, e4562. [Google Scholar] [CrossRef]
- Paul, S.; Li, L.; Kalaga, R.; Wilkins-Stevens, P.; Stevens, F.J.; Solomon, A. Natural Catalytic Antibodies: Peptide-Hydrolyzing Activities of Bence Jones Proteins and VL Fragment. J. Biol. Chem. 1995, 270, 15257–15261. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The Let-7 Family of MicroRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef]
- Shi, R.; Chiang, V.L. Facile Means for Quantifying Microrna Expression by Real-Time PCR. Biotechniques 2005, 39, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Chen, C. Real-Time Quantification of MicroRNAs by Stem-Loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Mei, Q.; Li, X.; Meng, Y.; Wu, Z.; Guo, M.; Zhao, Y.; Fu, X.; Han, W. A Facile and Specific Assay for Quantifying MicroRNA by an Optimized RT-QPCR Approach. PLoS ONE 2012, 7, e46890. [Google Scholar] [CrossRef] [PubMed]
- Ermakov, E.A.; Nevinsky, G.A.; Buneva, V.N. Immunoglobulins with Non-Canonical Functions in Inflammatory and Autoimmune Disease States. Int. J. Mol. Sci. 2020, 21, 5392. [Google Scholar] [CrossRef]
- Papadopoulos, G.L.; Reczko, M.; Simossis, V.A.; Sethupathy, P.; Hatzigeorgiou, A.G. The Database of Experimentally Supported Targets: A Functional Update of TarBase. Nucleic Acids Res. 2009, 37, D155–D158. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates That Thousands of Human Genes Are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Balcı, H.; Hanspers, K.; Coort, S.L.; Martens, M.; Slenter, D.N.; Ehrhart, F.; Digles, D.; Waagmeester, A.; Wassink, I.; et al. WikiPathways 2024: Next Generation Pathway Database. Nucleic Acids Res. 2024, 52, D679–D689. [Google Scholar] [CrossRef] [PubMed]
- Pico, A.R.; Kelder, T.; van Iersel, M.P.; Hanspers, K.; Conklin, B.R.; Evelo, C. WikiPathways: Pathway Editing for the People. PLoS Biol. 2008, 6, e184. [Google Scholar] [CrossRef]
- Buggele, W.A.; Johnson, K.E.; Horvath, C.M. Influenza A Virus Infection of Human Respiratory Cells Induces Primary MicroRNA Expression. J. Biol. Chem. 2012, 287, 31027–31040. [Google Scholar] [CrossRef] [PubMed]
- Lelis, D.d.F.; Freitas, D.F.d.; Machado, A.S.; Crespo, T.S.; Santos, S.H.S. Angiotensin-(1-7), Adipokines and Inflammation. Metabolism 2019, 95, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Yu, C.-H.; Li, W.; Li, T.; Luo, W.; Huang, S.; Wu, P.-S.; Cai, S.-X.; Li, X. Angiotensin-Converting Enzyme 2/Angiotensin-(1-7)/Mas Axis Protects against Lung Fibrosis by Inhibiting the MAPK/NF-ΚB Pathway. Am. J. Respir. Cell Mol. Biol. 2014, 50, 723–736. [Google Scholar] [CrossRef]
- Dalan, R.; Bornstein, S.R.; El-Armouche, A.; Rodionov, R.N.; Markov, A.; Wielockx, B.; Beuschlein, F.; Boehm, B.O. The ACE-2 in COVID-19: Foe or Friend? Horm. Metab. Res. 2020, 52, 257–263. [Google Scholar] [CrossRef]
- Uhal, B.D.; Dang, M.; Dang, V.; Llatos, R.; Cano, E.; Abdul-Hafez, A.; Markey, J.; Piasecki, C.C.; Molina-Molina, M. Cell Cycle Dependence of ACE-2 Explains Downregulation in Idiopathic Pulmonary Fibrosis. Eur. Respir. J. 2013, 42, 198–210. [Google Scholar] [CrossRef]
- Verdecchia, P.; Cavallini, C.; Spanevello, A.; Angeli, F. The Pivotal Link between ACE2 Deficiency and SARS-CoV-2 Infection. Eur. J. Intern. Med. 2020, 76, 14–20. [Google Scholar] [CrossRef]
- Nemeth, Z.; Kiss, E.; Takacs, I. The Role of Epigenetic Regulator SIRT1 in Balancing the Homeostasis and Preventing the Formation of Specific “Soil” of Metabolic Disorders and Related Cancers. Front. Biosci. 2022, 27, 253. [Google Scholar] [CrossRef]
- Rossi, G.A.; Sacco, O.; Capizzi, A.; Mastromarino, P. Can Resveratrol-Inhaled Formulations Be Considered Potential Adjunct Treatments for COVID-19? Front. Immunol. 2021, 12, 670955. [Google Scholar] [CrossRef] [PubMed]
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting Enzyme 2 (ACE2), SARS-CoV-2 and the Pathophysiology of Coronavirus Disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248. [Google Scholar] [CrossRef]
- Sun, Y.; Varambally, S.; Maher, C.A.; Cao, Q.; Chockley, P.; Toubai, T.; Malter, C.; Nieves, E.; Tawara, I.; Wang, Y.; et al. Targeting of MicroRNA-142-3p in Dendritic Cells Regulates Endotoxin-Induced Mortality. Blood 2011, 117, 6172–6183. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Neilson, J.R.; Kumar, P.; Manocha, M.; Shankar, P.; Sharp, P.A.; Manjunath, N. MiRNA Profiling of Naïve, Effector and Memory CD8 T Cells. PLoS ONE 2007, 2, e1020. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA Profiling of Multiple Sclerosis Lesions Identifies Modulators of the Regulatory Protein CD47. Brain 2009, 132, 3342–3352. [Google Scholar] [CrossRef]
- Ma, X.; Zhou, J.; Zhong, Y.; Jiang, L.; Mu, P.; Li, Y.; Singh, N.; Nagarkatti, M.; Nagarkatti, P. Expression, Regulation and Function of MicroRNAs in Multiple Sclerosis. Int. J. Med. Sci. 2014, 11, 810–818. [Google Scholar] [CrossRef]
- Mandolesi, G.; De Vito, F.; Musella, A.; Gentile, A.; Bullitta, S.; Fresegna, D.; Sepman, H.; Di Sanza, C.; Haji, N.; Mori, F.; et al. MiR-142-3p Is a Key Regulator of IL-1β-Dependent Synaptopathy in Neuroinflammation. J. Neurosci. 2017, 37, 546–561. [Google Scholar] [CrossRef] [PubMed]
- De Vito, F.; Musella, A.; Fresegna, D.; Rizzo, F.R.; Gentile, A.; Stampanoni Bassi, M.; Gilio, L.; Buttari, F.; Procaccini, C.; Colamatteo, A.; et al. MiR-142-3p Regulates Synaptopathy-driven Disease Progression in Multiple Sclerosis. Neuropathol. Appl. Neurobiol. 2022, 48, e12765. [Google Scholar] [CrossRef]
- Chen, L.Y.C.; Hoiland, R.L.; Stukas, S.; Wellington, C.L.; Sekhon, M.S. Confronting the Controversy: Interleukin-6 and the COVID-19 Cytokine Storm Syndrome. Eur. Respir. J. 2020, 56, 2003006. [Google Scholar] [CrossRef]
- Chen, Z.; Li, G. Immune Response and Blood–Brain Barrier Dysfunction during Viral Neuroinvasion. Innate Immun. 2021, 27, 109–117. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L.; Liang, X.; Liu, X.; Zhao, Y.; Ma, C.; Gao, L. Tim-4 in Health and Disease: Friend or Foe? Front. Immunol. 2020, 11, 537. [Google Scholar] [CrossRef]
- Santiago, C.; Ballesteros, A.; Martínez-Muñoz, L.; Mellado, M.; Kaplan, G.G.; Freeman, G.J.; Casasnovas, J.M. Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site Where Phosphatidylserine Binds. Immunity 2007, 27, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Perico, L.; Benigni, A.; Casiraghi, F.; Ng, L.F.P.; Renia, L.; Remuzzi, G. Immunity, Endothelial Injury and Complement-Induced Coagulopathy in COVID-19. Nat. Rev. Nephrol. 2021, 17, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.; Raju, S.; Wu, R.; Ching, C.; Veitch, S.; Rathnakumar, K.; Boudreau, E.; Howe, K.L.; Fish, J.E. Overcoming Barriers. Arterioscler. Thromb. Vasc. Biol. 2020, 40, 1818–1829. [Google Scholar] [CrossRef]
- Kang, S.; Tanaka, T.; Inoue, H.; Ono, C.; Hashimoto, S.; Kioi, Y.; Matsumoto, H.; Matsuura, H.; Matsubara, T.; Shimizu, K.; et al. IL-6 Trans-Signaling Induces Plasminogen Activator Inhibitor-1 from Vascular Endothelial Cells in Cytokine Release Syndrome. Proc. Natl. Acad. Sci. USA 2020, 117, 22351–22356. [Google Scholar] [CrossRef]
- Malas, M.B.; Naazie, I.N.; Elsayed, N.; Mathlouthi, A.; Marmor, R.; Clary, B. Thromboembolism Risk of COVID-19 Is High and Associated with a Higher Risk of Mortality: A Systematic Review and Meta-Analysis. eClinicalMedicine 2020, 29–30, 100639. [Google Scholar] [CrossRef]
- Piazza, G.; Morrow, D.A. Diagnosis, Management, and Pathophysiology of Arterial and Venous Thrombosis in COVID-19. JAMA 2020, 324, 2548. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Quintili, A.L.; Karamchandani, K.; Bose, S. Thromboembolic Disease in COVID-19 Patients: A Brief Narrative Review. J. Intensive Care 2020, 8, 70. [Google Scholar] [CrossRef]
- Bilaloglu, S.; Aphinyanaphongs, Y.; Jones, S.; Iturrate, E.; Hochman, J.; Berger, J.S. Thrombosis in Hospitalized Patients with COVID-19 in a New York City Health System. JAMA 2020, 324, 799. [Google Scholar] [CrossRef]
- Zhang, L.; Feng, X.; Zhang, D.; Jiang, C.; Mei, H.; Wang, J.; Zhang, C.; Li, H.; Xia, X.; Kong, S.; et al. Deep Vein Thrombosis in Hospitalized Patients with COVID-19 in Wuhan, China. Circulation 2020, 142, 114–128. [Google Scholar] [CrossRef]
- Gjorgjieva, M.; Sobolewski, C.; Dolicka, D.; Correia de Sousa, M.; Foti, M. MiRNAs and NAFLD: From Pathophysiology to Therapy. Gut 2019, 68, 2065–2079. [Google Scholar] [CrossRef]
- Mosca, N.; Pezzullo, M.; De Leo, I.; Truda, A.; Marchese, G.; Russo, A.; Potenza, N. A Novel CeRNET Relying on the LncRNA JPX, MiR-378a-3p, and Its MRNA Targets in Lung Cancer. Cancers 2024, 16, 1526. [Google Scholar] [CrossRef] [PubMed]
- Moatar, A.I.; Chis, A.R.; Marian, C.; Sirbu, I.-O. Gene Network Analysis of the Transcriptome Impact of SARS-CoV-2 Interacting MicroRNAs in COVID-19 Disease. Int. J. Mol. Sci. 2022, 23, 9239. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, D.; Nayeem, M.; Vanderven, H.A.; Sarker, S. Role of MiRNA in Highly Pathogenic H5 Avian Influenza Virus Infection: An Emphasis on Cellular and Chicken Models. Viruses 2024, 16, 1102. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Sedykh, S.E.; Sedykh, T.A.; Nevinsky, G.A. Natural Antibodies Produced in Vaccinated Patients and COVID-19 Convalescents Recognize and Hydrolyze Oligopeptides Corresponding to the S-Protein of SARS-CoV-2. Vaccines 2023, 11, 1494. [Google Scholar] [CrossRef]
- Timofeeva, A.M.; Sedykh, S.E.; Ermakov, E.A.; Matveev, A.L.; Odegova, E.I.; Sedykh, T.A.; Shcherbakov, D.N.; Merkuleva, I.A.; Volosnikova, E.A.; Nesmeyanova, V.S.; et al. Natural IgG against S-Protein and RBD of SARS-CoV-2 Do Not Bind and Hydrolyze DNA and Are Not Autoimmune. Int. J. Mol. Sci. 2022, 23, 13681. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Kutmon, M.; Ehrhart, F.; Willighagen, E.L.; Evelo, C.T.; Coort, S.L. CyTargetLinker App Update: A Flexible Solution for Network Extension in Cytoscape. F1000Research 2018, 7, 743. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).