Detection of miR-133a-5p Using a Molecular Beacon Probe for Investigating Postmortem Intervals
Abstract
1. Introduction
2. Results
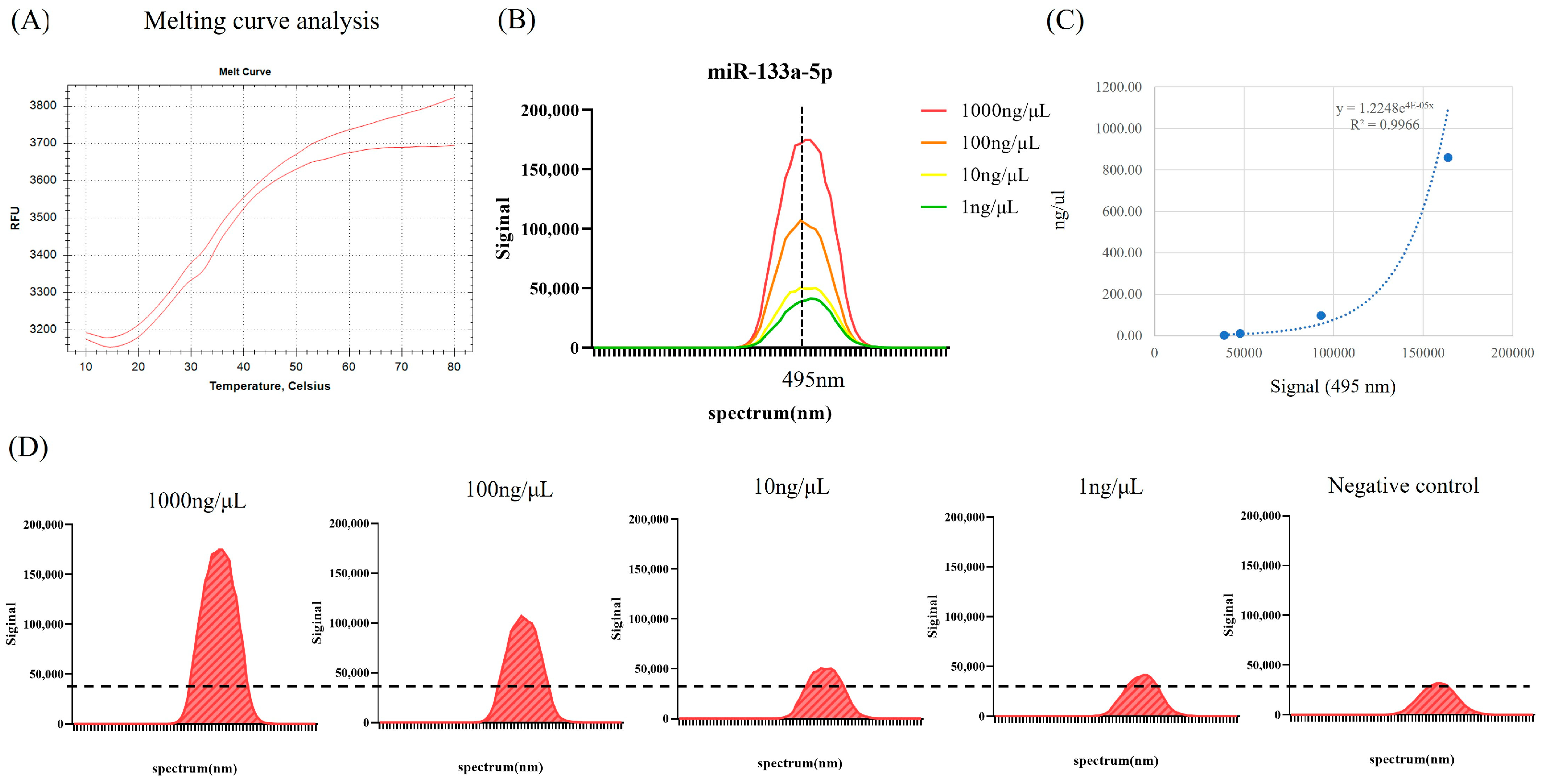
2.1. Molecular Beacon Probe Assay for mmu-miR-133a-5p
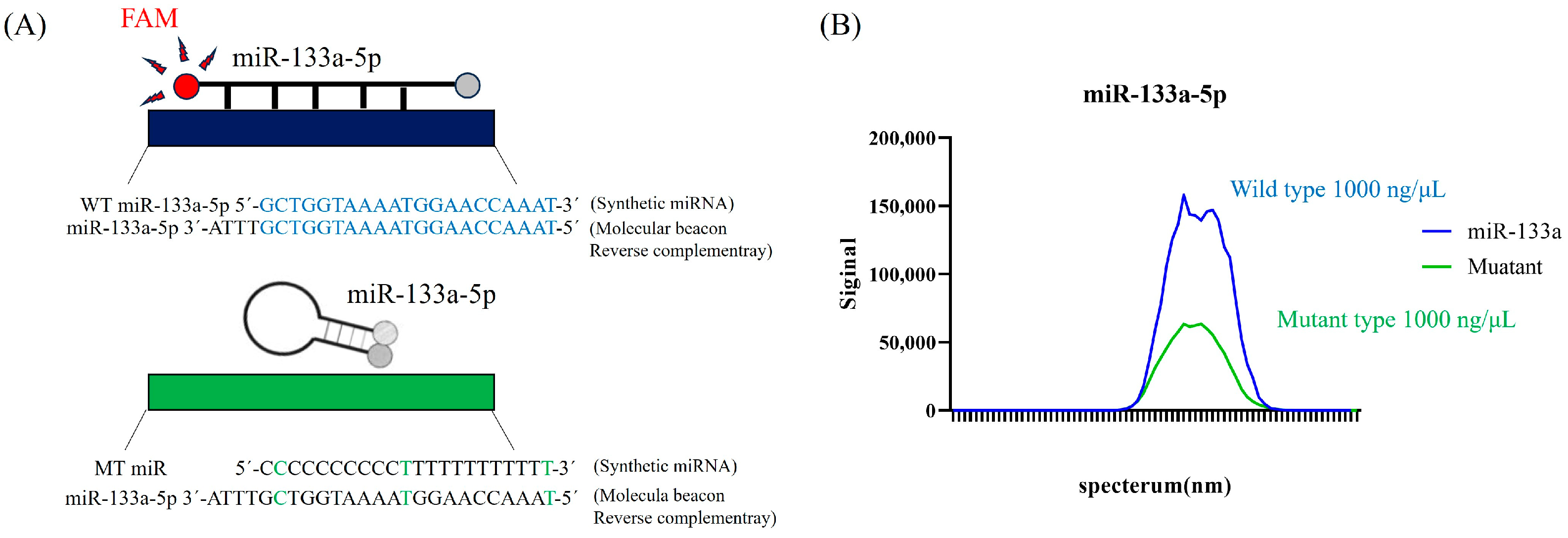
2.2. Specificity of the Molecular Beacon Probe for mmu-miR-133a-5p
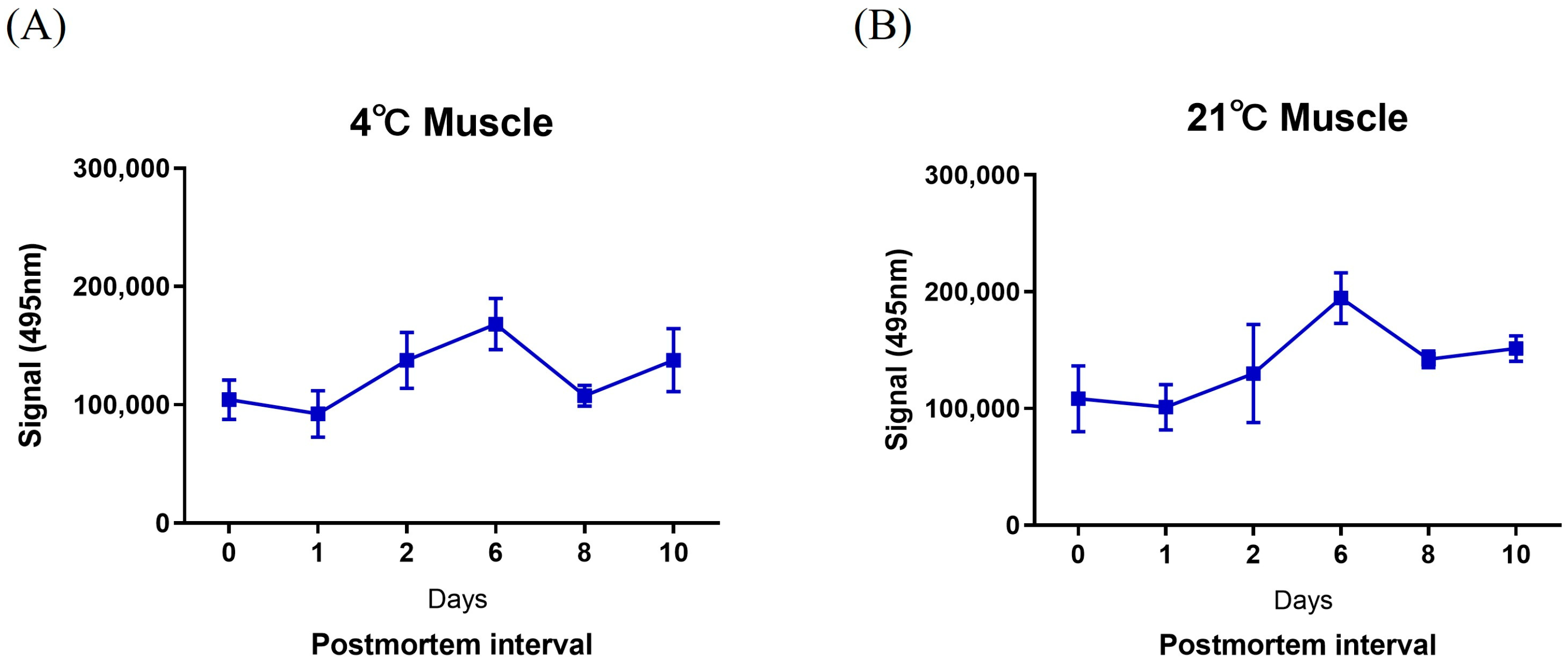
2.3. Detection of mmu-miR-133a-5p Using a FAM-Labeled MB Probe for PMI
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Extraction of Total RNA
4.3. Design of a Molecular Beacon for mmu-miR-133a-5p
4.4. Beacon Hybridization Assay
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henssge, C.; Althaus, L.; Bolt, J.; Freislederer, A.; Haffner, H.T.; Henssge, C.A.; Hoppe, B.; Schneider, V. Experiences with a compound method for estimating the time since death. II. Integration of non-temperature-based methods. Int. J. Leg. Med. 2000, 113, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Poloz, Y.O.; O’Day, D.H. Determining time of death: Temperature-dependent postmortem changes in calcineurin A, MARCKS, CaMKII, and protein phosphatase 2A in mouse. Int. J. Leg. Med. 2009, 123, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Maile, A.E.; Inoue, C.G.; Barksdale, L.E.; Carter, D.O. Toward a universal equation to estimate postmortem interval. Forensic Sci. Int. 2017, 272, 150–153. [Google Scholar] [CrossRef]
- Lv, Y.H.; Ma, K.J.; Zhang, H.; He, M.; Zhang, P.; Shen, Y.W.; Jiang, N.; Ma, D.; Chen, L. A time course study demonstrating mRNA, microRNA, 18S rRNA, and U6 snRNA changes to estimate PMI in deceased rat’s spleen. J. Forensic Sci. 2014, 59, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.; Kanchan, T.; Krishan, K. Methods of estimation of time since death. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Wang, H.; Mao, J.; Li, Y.; Luo, H.; Wu, J.; Liao, M.; Liang, W.; Zhang, L. 5 miRNA expression analyze in postmortem interval (PMI) within 48 h. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e190–e191. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- De Simone, S.; Giacani, E.; Bosco, M.A.; Vittorio, S.; Ferrara, M.; Bertozzi, G.; Cipolloni, L.; La Russa, R. The role of miRNAs as new molecular biomarkers for dating the age of wound production: A systematic review. Front. Med. 2022, 8, 803067. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Pasaribu, R.S.; Auerkari, E.I.; Suhartono, A.W.; Auerkari, P. A small RNA, microRNA as a potential biomolecular marker to estimate post mortem interval in forensic science: A systematic review. Int. J. Leg. Med. 2023, 137, 1313–1325. [Google Scholar] [CrossRef]
- Martínez-Rivera, V.; Cárdenas-Monroy, C.A.; Millan-Catalan, O.; González-Corona, J.; Huerta-Pacheco, N.S.; Martínez-Gutiérrez, A.; Villavicencio-Queijeiro, A.; Pedraza-Lara, C.; Hidalgo-Miranda, A.; Bravo-Gómez, M.E.; et al. Dysregulation of miR-381-3p and miR-23b-3p in skeletal muscle could be a possible estimator of early postmortem interval in rats. PeerJ 2021, 9, e11102. [Google Scholar] [CrossRef]
- Catuogno, S.; Esposito, C.L.; Quintavalle, C.; Condorelli, G.; de Franciscis, V.; Cerchia, L. Nucleic acids in human glioma treatment: Innovative approaches and recent results. J. Signal Transduct. 2012, 2012, 735135. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Ali, W.; Sandhu, S.; Mishra, S.; Singh, U.S.; Verma, A.K.; Singh, M.; Kaleem Ahmad, M.; Kumari, S. Postmortem interval estimation using miRNAs of road traffic accident cases: A forensic molecular approach. Sci. Justice 2023, 63, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Na, J.Y. Estimation of the postmortem interval using microRNA in the bones. J. Forensic Leg. Med. 2020, 75, 102049. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. MicroRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef]
- Odriozola, A.; Riancho, J.A.; de la Vega, R.; Agudo, G.; García-Blanco, A.; de Cos, E.; Fernández, F.; Sañudo, C.; Zarrabeitia, M.T. miRNA analysis in vitreous humor to determine the time of death: A proof-of-concept pilot study. Int. J. Leg. Med. 2013, 127, 573–578. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Caporali, S.; Calabrese, C.; Minieri, M.; Pieri, M.; Tarantino, U.; Marini, M.; D’Ottavio, S.; Angeletti, S.; Mauriello, A.; Cortese, C.; et al. The miR-133a, TPM4 and TAp63γ Role in Myocyte Differentiation Microfilament Remodelling and Colon Cancer Progression. Int. J. Mol. Sci. 2021, 22, 9818. [Google Scholar] [CrossRef]
- Carpi, S.; Polini, B.; Nieri, D.; Dubbini, N.; Celi, A.; Nieri, P.; Neri, T. Expression Analysis of Muscle-Specific miRNAs in Plasma-Derived Extracellular Vesicles from Patients with Chronic Obstructive Pulmonary Disease. Diagnostics 2020, 10, 502. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.H.; Ma, J.L.; Pan, H.; Zeng, Y.; Tao, L.; Zhang, H.; Li, W.C.; Ma, K.J.; Chen, L. Estimation of the human postmortem interval using an established rat mathematical model and multi-RNA markers. Forensic Sci. Med. Pathol. 2017, 13, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Bidar, N.; Amini, M.; Oroojalian, F.; Baradaran, B.; Hosseini, S.S.; Shahbazi, M.A.; Hashemzaei, M.; Mokhtarzadeh, A.; Hamblin, M.R.; de la Guardia, M. Molecular beacon strategies for sensing purpose. TrAC Trends Anal. Chem. 2021, 134, 116143. [Google Scholar] [CrossRef]
- Tyagi, S.; Kramer, F.R. Molecular beacons: Probes that fluoresce upon hybridization. Nat. Biotechnol. 1996, 14, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.; Rhee, W.J.; Tsourkas, A. Fluorescent probes for live-cell RNA detection. Annu. Rev. Biomed. Eng. 2009, 11, 25–47. [Google Scholar] [CrossRef]
- Baker, M.B.; Bao, G.; Searles, C.D. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012, 40, e13. [Google Scholar] [CrossRef]
- Vet, J.A.; Marras, S.A. Design and optimization of molecular beacon real-time polymerase chain reaction assays. Methods Mol. Biol. 2005, 288, 273–290. [Google Scholar] [CrossRef]
- Santangelo, P.; Nitin, N.; Bao, G. Nanostructured probes for RNA detection in living cells. Ann. Biomed. Eng. 2006, 34, 39–50. [Google Scholar] [CrossRef]
- Huang, K.; Martí, A.A. Recent trends in molecular beacon design and applications. Anal. Bioanal. Chem. 2012, 402, 3091–3102. [Google Scholar] [CrossRef]
- Tan, W.; Wang, K.; Drake, T.J. Molecular beacons. Curr. Opin. Chem. Biol. 2004, 8, 547–553. [Google Scholar] [CrossRef]
- Wang, K.; Tang, Z.; Yang, C.J.; Kim, Y.; Fang, X.; Li, W.; Wu, Y.; Medley, C.D.; Cao, Z.; Li, J.; et al. Molecular engineering of DNA: Molecular beacons. Angew. Chem. Int. Ed. Engl. 2009, 48, 856–870. [Google Scholar] [CrossRef] [PubMed]
- Maiese, A.; Scatena, A.; Costantino, A.; Di Paolo, M.; La Russa, R.; Turillazzi, E.; Frati, P.; Fineschi, V. MicroRNAs as useful tools to estimate time since death. A systematic review of current literature. Diagnostics 2021, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Montanari, E.; Giorgetti, R.; Busardò, F.P.; Giorgetti, A.; Tagliabracci, A.; Alessandrini, F. Suitability of miRNA assessment in postmortem interval estimation. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1774–1787. [Google Scholar] [CrossRef]
- Tu, C.; Du, T.; Ye, X.; Shao, C.; Xie, J.; Shen, Y. Using miRNAs and circRNAs to estimate PMI in advanced stage. Leg. Med. 2019, 38, 51–57. [Google Scholar] [CrossRef]
- Vass, A.A. The elusive universal postmortem interval formula. Forensic Sci. Int. 2011, 204, 34–40. [Google Scholar] [CrossRef]
- Brooks, J.W. Postmortem changes in animal carcasses and estimation of the postmortem interval. Vet. Pathol. 2016, 53, 929–940. [Google Scholar] [CrossRef]
- Brockbals, L.; Garrett-Rickman, S.; Fu, S.; Ueland, M.; McNevin, D.; Padula, M.P. Estimating the time of human decomposition based on skeletal muscle biopsy samples utilizing an untargeted LC-MS/MS-based proteomics approach. Anal. Bioanal. Chem. 2023, 415, 5487–5498. [Google Scholar] [CrossRef]
- Wenzlow, N.; Mills, D.; Byrd, J.; Warren, M.; Long, M.T. Review of the current and potential use of biological and molecular methods for the estimation of the postmortem interval in animals and humans. J. Vet. Diagn. Investig. 2023, 35, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.T. Single-labeled oligonucleotides showing fluorescence changes upon hybridization with target nucleic acids. Molecules 2018, 23, 124. [Google Scholar] [CrossRef]
- Nie, Y.; Sato, Y.; Wang, C.; Yue, F.; Kuang, S.; Gavin, T.P. Impaired exercise tolerance, mitochondrial biogenesis, and muscle fiber maintenance in miR-133a-deficient mice. FASEB J. 2016, 30, 3745–3758. [Google Scholar] [CrossRef]
- Alexandre, D.; Fernandes, A.R.; Baptista, P.V.; Cruz, C. Evaluation of miR-155 silencing using a molecular beacon in human lung adenocarcinoma cell line. Talanta 2024, 274, 126052. [Google Scholar] [CrossRef] [PubMed]
- Napoletano, S.; Battista, E.; Martone, N.; Netti, P.A.; Causa, F. Direct, precise, enzyme-free detection of miR-103-3p in real samples by microgels with highly specific molecular beacons. Talanta 2023, 259, 124468. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhou, J.; Zheng, Y.; Gamson, A.S.; Roembke, B.T.; Nakayama, S.; Sintim, H.O. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 2014, 10, 970–1003. [Google Scholar] [CrossRef]
- Roderburg, C.; Luedde, M.; Vargas Cardenas, D.; Vucur, M.; Mollnow, T.; Zimmermann, H.W.; Koch, A.; Hellerbrand, C.; Weiskirchen, R.; Frey, N.; et al. miR-133a mediates TGF-β-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J. Hepatol. 2023, 58, 736–742. [Google Scholar] [CrossRef]
- Sun, L.; Guo, Z.; Sun, J.; Li, J.; Dong, Z.; Zhang, Y.; Chen, J.; Kan, Q.; Yu, Z. MiR-133a acts as an anti-oncogene in Hepatocellular carcinoma by inhibiting FOSL2 through TGF-β/Smad3 signaling pathway. Biomed. Pharmacother. 2018, 107, 168–176. [Google Scholar] [CrossRef]

| Probe | Sequence | Dye | Quencher | Probe Length | Stem Length | Tm (°C) |
|---|---|---|---|---|---|---|
| MB for mmu-miR-133a-5p | 5′-ATTTGGTTCCATTTTACCAGCAAAT-3′ | FAM | BHQ1 | 25 | 5 | 54 |
| mmu-miR-133a-5p (target) | 5′-GCTGGTAAAATGGAACCAAAT-3′ | |||||
| Mutant miR-133a-5p | 5′-CCCCCCCCCCTTTTTTTTTTTT-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.H.; Jeong, M.; Park, K.; Lee, D.G.; Lee, E.J.; Lee, H.; Kim, A.Y.; Ahn, J.W.; Woo, H.J.; Kim, S.; et al. Detection of miR-133a-5p Using a Molecular Beacon Probe for Investigating Postmortem Intervals. Non-Coding RNA 2024, 10, 58. https://doi.org/10.3390/ncrna10060058
Lee EH, Jeong M, Park K, Lee DG, Lee EJ, Lee H, Kim AY, Ahn JW, Woo HJ, Kim S, et al. Detection of miR-133a-5p Using a Molecular Beacon Probe for Investigating Postmortem Intervals. Non-Coding RNA. 2024; 10(6):58. https://doi.org/10.3390/ncrna10060058
Chicago/Turabian StyleLee, Eun Hye, Mingyoung Jeong, Kwangmin Park, Dong Geon Lee, Eun Ju Lee, Haneul Lee, Ah Yeoung Kim, Jae Won Ahn, Hyun Jun Woo, Sunghyun Kim, and et al. 2024. "Detection of miR-133a-5p Using a Molecular Beacon Probe for Investigating Postmortem Intervals" Non-Coding RNA 10, no. 6: 58. https://doi.org/10.3390/ncrna10060058
APA StyleLee, E. H., Jeong, M., Park, K., Lee, D. G., Lee, E. J., Lee, H., Kim, A. Y., Ahn, J. W., Woo, H. J., Kim, S., Lim, J., & Kim, J. (2024). Detection of miR-133a-5p Using a Molecular Beacon Probe for Investigating Postmortem Intervals. Non-Coding RNA, 10(6), 58. https://doi.org/10.3390/ncrna10060058







