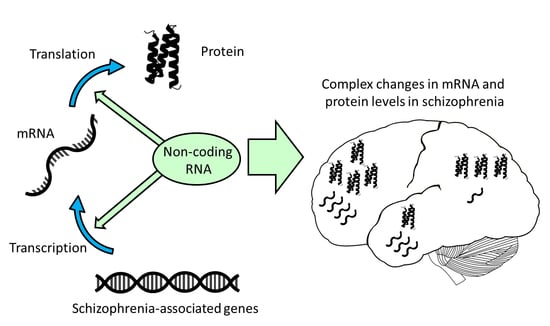
Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia
Abstract
:1. Introduction
2. MicroRNA in the Healthy Brain
3. MicroRNAs in Schizophrenia
4. miR-137 in Schizophrenia
5. Disruption of MicroRNA Processing Machinery
6. Short Non-Coding RNA in Schizophrenia
7. Long Non-Coding RNAs in Schizophrenia
8. Non-Coding RNAs as Therapeutic Targets
9. Non-Coding RNAs as Peripheral Biomarkers of Schizophrenia
10. Concluding Remarks
Acknowledgements
Author Contributions
Conflicts of Interest
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; American Psychiatric Press: Washington, DC, USA, 2013. [Google Scholar]
- Narayan, S.; Tang, B.; Head, S.R.; Gilmartin, T.J.; Sutcliffe, J.G.; Dean, B.; Thomas, E.A. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008, 1239, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Roy, M.; Sorokina, O.; Skene, N.; Simonnet, C.; Mazzo, F.; Zwart, R.; Sher, E.; Smith, C.; Armstrong, J.D.; Grant, S.G.N. Proteomic analysis of postsynaptic proteins in regions of the human neocortex. Nat. Neurosci. 2018, 21, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Ramaker, R.C.; Bowling, K.M.; Lasseigne, B.N.; Hagenauer, M.H.; Hardigan, A.A.; Davis, N.S.; Gertz, J.; Cartagena, P.M.; Walsh, D.M.; Vawter, M.P.; et al. Post-mortem molecular profiling of three psychiatric disorders. Genome Med. 2017, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Hertzberg, L.; Katsel, P.; Roussos, P.; Haroutunian, V.; Domany, E. Integration of gene expression and GWAS results supports involvement of calcium signaling in schizophrenia. Schizophr. Res. 2015, 164, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Shorter, K.R.; Miller, B.H. Epigenetic mechanisms in schizophrenia. Prog. Biophys. Mol. Biol. 2015, 118, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ibi, D.; Gonzalez-Maeso, J. Epigenetic signaling in schizophrenia. Cell. Signal. 2015, 27, 2131–2136. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. So much junk DNA in our genome. Brookhaven Symp. Biol. 1972, 23, 366–370. [Google Scholar] [PubMed]
- Fineberg, S.K.; Kosik, K.S.; Davidson, B.L. MicroRNAs potentiate neural development. Neuron 2009, 64, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Millan, M.J. An epigenetic framework for neurodevelopmental disorders: From pathogenesis to potential therapy. Neuropharmacology 2013, 68, 2–82. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, N.J.; Gardiner, E.; Carroll, A.P.; Tooney, P.A.; Cairns, M.J. Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol. Psychiatry 2010, 15, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.P.; Bruse, S.E.; David-Rus, R.; Buyske, S.; Brzustowicz, L.M. Altered microRNA expression profiles in postmortem brain samples from individuals with schizophrenia and bipolar disorder. Biol. Psychiatry 2011, 69, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, D.M.; Beveridge, N.J.; Tooney, P.A.; Cairns, M.J. Upregulation of Dicer and microRNA expression in the dorsolateral prefrontal cortex Brodmann area 46 in schizophrenia. Biol. Psychiatry 2011, 69, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Ragan, C.; Patel, K.; Edson, J.; Zhang, Z.H.; Gratten, J.; Mowry, B. Small non-coding RNA expression from anterior cingulate cortex in schizophrenia shows sex specific regulation. Schizophr. Res. 2017, 183, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.D.; Sun, X.Y.; Niu, W.; Kong, L.M.; He, M.J.; Li, W.S.; Zhong, A.F.; Lu, J.; Zhang, L.Y. Aberrant expression of long non-coding RNAs in schizophrenia patients. Med. Sci. Monit. 2016, 22, 3340–3351. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Xu, J.Y.; Pang, L.; Zhao, H.Y.; Li, F.; Deng, Y.L.; Liu, L.; Lan, Y.J.; Zhang, X.X.; Zhao, T.T.; et al. Systematically characterizing dysfunctional long intergenic non-coding RNAs in multiple brain regions of major psychosis. Oncotarget 2016, 7, 71087–71098. [Google Scholar] [CrossRef] [PubMed]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.P.; Liu, D.; Zhang, L.J.; Ingvarsson, S.; Chen, H.P. Quantitative analysis of miRNA expression in seven human foetal and adult organs. PLoS ONE 2011, 6, e28730. [Google Scholar] [CrossRef] [PubMed]
- Beveridge, N.J.; Santarelli, D.M.; Wang, X.; Tooney, P.A.; Webster, M.J.; Weickert, C.S.; Cairns, M.J. Maturation of the human dorsolateral prefrontal cortex coincides with a dynamic shift in microRNA expression. Schizophr. Bull. 2014, 40, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Tau, G.Z.; Peterson, B.S. Normal development of brain circuits. Neuropsychopharmacology 2010, 35, 147–168. [Google Scholar] [CrossRef] [PubMed]
- Malmevik, J.; Petri, R.; Klussendorf, T.; Knauff, P.; Akerblom, M.; Johansson, J.; Soneji, S.; Jakobsson, J. Identification of the miRNA targetome in hippocampal neurons using RIP-Seq. Sci. Rep. 2015, 5, 12609. [Google Scholar] [CrossRef] [PubMed]
- Boudreau, R.L.; Jiang, P.; Gilmore, B.L.; Spengler, R.M.; Tirabassi, R.; Nelson, J.A.; Ross, C.A.; Xing, Y.; Davidson, B.L. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron 2014, 81, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A microRNA array reveals extensive regulation of microRNAs during brain development. RNA-Publ. RNA Soc. 2003, 9, 1274–1281. [Google Scholar] [CrossRef]
- Smith, B.; Treadwell, J.; Zhang, D.L.; Ly, D.; McKinnell, I.; Walker, P.R.; Sikorska, M. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS ONE 2010, 5, e11109. [Google Scholar] [CrossRef] [PubMed]
- Moszynska, A.; Gebert, M.; Collawn, J.F.; Bartoszewski, R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017, 7, 170019. [Google Scholar] [CrossRef] [PubMed]
- Hollins, S.L.; Goldie, B.J.; Carroll, A.P.; Mason, E.A.; Walker, F.R.; Eyles, D.W.; Cairns, M.J. Ontogeny of small RNA in the regulation of mammalian brain development. BMC Genom. 2014, 15, 777. [Google Scholar] [CrossRef] [PubMed]
- Zucchi, F.C.R.; Yao, Y.L.; Ward, I.D.; Ilnytskyy, Y.; Olson, D.M.; Benzies, K.; Kovalchuk, I.; Kovalchuk, O.; Metz, G.A.S. Maternal stress induces epigenetic signatures of psychiatric and neurological diseases in the offspring. PLoS ONE 2013, 8, e56967. [Google Scholar] [CrossRef] [PubMed]
- Scarr, E.; Udawela, M.; Dean, B. Changed frontal pole gene expression suggest altered interplay between neurotransmitter, developmental, and inflammatory pathways in schizophrenia. NPJ Schizophr. 2018, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Martins-de-Souza, D.; Gattaz, W.F.; Schmitt, A.; Novello, J.C.; Marangoni, S.; Turck, C.W.; Dias-Neto, E. Proteome analysis of schizophrenia patients Wernicke’s area reveals an energy metabolism dysregulation. BMC Psychiatry 2009, 9, 17. [Google Scholar] [CrossRef] [PubMed]
- Pennington, K.; Beasley, C.; Dicker, P.; Fagan, A.; English, J.; Pariante, C.; Wait, R.; Dunn, M.; Cotter, D. Prominent synaptic and metabolic abnormalities revealed by proteomic analysis of the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder. Mol. Psychiatry 2008, 13, 1102. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.; Dedova, I.; Cordwell, S.; Matsumoto, I. A proteome analysis of the anterior cingulate cortex gray matter in schizophrenia. Mol. Psychiatry 2006, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Dean, B.; Keriakous, D.; Thomas, E.; Scarr, E. Understanding the pathology of schizophrenia: The impact of high-throughput screening of the genome and proteome in postmortem CNS. Curr. Psychiatry Rev. 2005, 1, 1–9. [Google Scholar] [CrossRef]
- Perkins, D.O.; Jeffries, C.D.; Jarskog, L.F.; Thomson, J.M.; Woods, K.; Newman, M.A.; Parker, J.S.; Jin, J.P.; Hammond, S.M. MicroRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder. Genome Biol. 2007, 8, R27. [Google Scholar] [CrossRef] [PubMed]
- Merico, D.; Zarrei, M.; Costain, G.; Ogura, L.; Alipanahi, B.; Gazzellone, M.J.; Butcher, N.J.; Thiruvahindrapuram, B.; Nalpathamkalam, T.; Chow, E.W.C.; et al. Whole-genome sequencing suggests schizophrenia risk mechanisms in humans with 22q11.2 deletion syndrome. G3-Genes Genomes Genet. 2015, 5, 2453–2461. [Google Scholar] [CrossRef] [PubMed]
- Van, L.; Boot, E.; Bassett, A.S. Update on the 22q11.2 deletion syndrome and its relevance to schizophrenia. Curr. Opin. Psychiatry 2017, 30, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Castro, T.B.; Hernandez-Diaz, Y.; Juarez-Rojop, I.E.; Lopez-Narvaez, M.L.; Tovilla-Zarate, C.A.; Fresan, A. The role of a catechol-O-methyltransferase (COMT) Val158Met genetic polymorphism in schizophrenia: A systematic review and updated meta-analysis on 32,816 subjects. Neuromol. Med. 2016, 18, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Murphy, K.C.; Jones, L.A.; Owen, M.J. High rates of schizophrenia in adults with velo-cardio-facial syndrome. Arch. Gen. Psychiatry 1999, 56, 940–945. [Google Scholar] [CrossRef] [PubMed]
- Burmistrova, O.A.; Goltsov, A.Y.; Abramova, L.I.; Kaleda, V.G.; Orlova, V.A.; Rogaev, E.I. MicroRNA in schizophrenia: Genetic and expression analysis of miR-130b (22q 11). Biochemistry (Moscow) 2007, 72, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Yuan, Y.B.; Liu, S.; Wang, C.; Yang, F.D.; Lu, Z.; Wang, C.Y.; Deng, H.; Zhao, J.P.; Shen, Y.; et al. Detection of circulating miRNA levels in schizophrenia. Am. J. Psychiatry 2015, 172, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Forstner, A.J.; Basmanav, F.B.; Mattheisen, M.; Bohmer, A.C.; Hollegaard, M.V.; Janson, E.; Strengman, E.; Priebe, L.; Degenhardt, F.; Hoffmann, P.; et al. Investigation of the involvement of MIR185 and its target genes in the development of schizophrenia. J. Psychiatry Neurosci. 2014, 39, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.L.; Xu, B.; Bagchi, A.; Lai, W.S.; Liu, H.; Hsu, R.; Wan, X.; Pavlidis, P.; Mills, A.A.; Karayiorgou, M.; et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008, 40, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lang, N.; Chen, X.Z.; Tang, Q.L.; Liu, S.R.; Huang, J.A.; Zheng, Y.; Bi, F. MiR-185 targets RhoA and Cdc42 expression and inhibits the proliferation potential of human colorectal cells. Cancer Lett. 2011, 301, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.J.; Hashimoto, T.; Lewis, D.A. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry 2006, 11, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Ide, M.; Lewis, D.A. Altered cortical Cdc42 signaling pathways in schizophrenia: Implications for dendritic spine deficits. Biol. Psychiatry 2010, 68, 25–32. [Google Scholar] [CrossRef] [PubMed]
- De la Morena, M.T.; Eitson, J.L.; Dozmorov, I.M.; Belkaya, S.; Hoover, A.R.; Anguiano, E.; Pascual, M.V.; van Oers, N.S.C. Signature microRNA expression patterns identified in humans with 22q11.2 deletion/DiGeorge syndrome. Clin. Immunol. 2013, 147, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Sellier, C.; Hwang, V.J.; Dandekar, R.; Durbin-Johnson, B.; Charlet-Berguerand, N.; Ander, B.P.; Sharp, F.R.; Angkustsiri, K.; Simon, T.J.; Tassone, F. Decreased DGCR8 expression and miRNA dysregulation in individuals with 22q11.2 deletion syndrome. PLoS ONE 2014, 9, e103884. [Google Scholar] [CrossRef] [PubMed]
- Banigan, M.G.; Kao, P.F.; Kozubek, J.A.; Winslow, A.R.; Medina, J.; Costa, J.; Schmitt, A.; Schneider, A.; Cabral, H.; Cagsal-Getkin, O.; et al. Differential expression of exosomal microRNAs in prefrontal cortices of schizophrenia and bipolar disorder patients. PLoS ONE 2013, 8, e48814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beveridge, N.J.; Tooney, P.A.; Carroll, A.P.; Gardiner, E.; Bowden, N.; Scott, R.J.; Tran, N.; Dedova, I.; Cairns, M.J. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum. Mol. Genet. 2008, 17, 1156–1168. [Google Scholar] [CrossRef] [PubMed]
- Wojtalik, J.A.; Smith, M.J.; Keshavan, M.S.; Eack, S.M. A systematic and meta-analytic review of neural correlates of functional outcome in schizophrenia. Schizophr. Bull. 2017, 43, 1329–1347. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.W.; Kim, Y.H.; Jeong, G.W. Whole brain volume changes and its correlation with clinical symptom severity in patients with schizophrenia: A DARTEL-based VBM study. PLoS ONE 2017, 12, e0177251. [Google Scholar] [CrossRef] [PubMed]
- Dirnberger, G.; Fuller, R.; Frith, C.; Jahanshahi, M. Neural correlates of executive dysfunction in schizophrenia: Failure to modulate brain activity with task demands. Neuroreport 2014, 25, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; Zeier, Z.; Xi, L.; Lanz, T.A.; Deng, S.B.; Strathmann, J.; Willoughby, D.; Kenny, P.J.; Elsworth, J.D.; Lawrence, M.S.; et al. MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl. Acad. Sci. USA 2012, 109, 3125–3130. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.H.; Reimers, M.; Mahera, B.; Williamson, V.; McMichael, O.I.; McClay, J.L.; van den Oord, E.; Riley, B.P.; Kendler, K.S.; Vladimirov, V.I. MicroRNA expression profiling in the prefrontal cortex of individuals affected with schizophrenia and bipolar disorders. Schizophr. Res. 2010, 124, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Strazisar, M.; Cammaerts, S.; van der Ven, K.; Forero, D.A.; Lenaerts, A.S.; Nordin, A.; Almeida-Souza, L.; Genovese, G.; Timmerman, V.; Liekens, A.; et al. MIR137 variants identified in psychiatric patients affect synaptogenesis and neuronal transmission gene sets. Mol. Psychiatry 2015, 20, 472–481. [Google Scholar] [CrossRef] [PubMed]
- Guan, F.L.; Zhang, B.; Yan, T.L.; Li, L.; Liu, F.; Li, T.; Feng, Z.F.; Zhang, B.; Liu, X.S.; Li, S.B. MIR137 gene and target gene CACNA1C of miR-137 contribute to schizophrenia susceptibility in Han Chinese. Schizophr. Res. 2014, 152, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Kuswanto, C.N.; Sum, M.Y.; Qiu, A.Q.; Sitoh, Y.Y.; Liu, J.J.; Sim, K. The impact of genome wide supported microRNA-137 (MIR137) risk variants on frontal and striatal white matter integrity, neurocognitive functioning, and negative symptoms in schizophrenia. Am. J. Med. Genet. B 2015, 168, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.D.; Yin, J.W.; Fu, J.W.; Luo, X.D.; Zhou, H.H.; Tao, H.; Cui, L.L.; Li, Y.; Lin, Z.X.; Zhao, B.; et al. Association of a miRNA-137 polymorphism with schizophrenia in a Southern Chinese Han Population. BioMed Res. Int. 2014, 2014, 751267. [Google Scholar] [CrossRef] [PubMed]
- Egawa, J.; Nunokawa, A.; Shibuya, M.; Watanabe, Y.; Kaneko, N.; Igeta, H.; Someya, T. Resequencing and association analysis of MIR137 with schizophrenia in a Japanese Population. Psychiatry Clin. Neurosci. 2013, 67, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.J.; Yu, Y.; Zhu, G.C.; Sun, Z.H.; Xu, J.; Cao, J.H.; Ge, J.X. Association between single nucleotide polymorphisms in MiR219-1 and MiR137 and susceptibility to schizophrenia in a Chinese Population. FEBS Open Bio 2015, 5, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.M.; Cheng, Z.H.; Zhang, F.Q.; Zhou, Z.H.; Yu, S.; Jin, C.H. Lack of association between microRNA-137 SNP rs1625579 and schizophrenia in a replication study of Han Chinese. Mol. Genet. Genom. 2015, 290, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.L.; Liu, G.; Xiao, D.; Zhang, B.H.; Guo, C.C.; Ye, X.G.; Liu, Y.; Zhang, N.; Wang, M.; Han, Y.J.; et al. Association between miR-137 polymorphism and risk of schizophrenia: A meta-analysis. Genet. Mol. Res. 2016, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Bian, Y.; Liu, N.; Tang, Y.X.; Pan, C.; Hu, Y.; Tang, Z.P. The SNP rs1625579 in miR-137 gene and risk of schizophrenia in Chinese Population: A meta-analysis. Compr. Psychiatry 2016, 67, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Guella, I.; Sequeira, A.; Rollins, B.; Morgan, L.; Torri, F.; van Erp, T.G.M.; Myers, R.M.; Barchas, J.D.; Schatzberg, A.F.; Watson, S.J.; et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J. Psychiatry Res. 2013, 47, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Vita, A.; De Peri, L.; Deste, G.; Sacchetti, E. Progressive loss of cortical gray matter in schizophrenia: A meta-analysis and meta-regression of longitudinal MRI studies. Transl. Psychiatry 2012, 2, e190. [Google Scholar] [CrossRef]
- Vitolo, E.; Tatu, M.K.; Pignolo, C.; Cauda, F.; Costa, T.; Ando, A.; Zennaro, A. White matter and schizophrenia: A meta-analysis of voxel-based morphometry and diffusion tensor imaging studies. Psychiatry Res. Neuroimaging 2017, 270, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Rose, E.J.; Morris, D.W.; Fahey, C.; Cannon, D.; McDonald, C.; Scanlon, C.; Kelly, S.; Gill, M.; Corvin, A.; Donohoe, G. The miR-137 schizophrenia susceptibility variant rs1625579 does not predict variability in brain volume in a sample of schizophrenic patients and healthy individuals. Am. J. Med. Genet. B 2014, 165, 467–471. [Google Scholar] [CrossRef]
- Kelly, S.; Morris, D.W.; Mothersill, O.; Rose, E.J.; Fahey, C.; O’Brien, C.; O’Hanlon, E.; Gill, M.; Corvin, A.P.; Donohoe, G. Genome-wide schizophrenia variant at MIR137 does not impact white matter microstructure in healthy participants. Neurosci. Lett. 2014, 574, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.S.; Kelly, S.; Wright, C.; Gupta, C.N.; Arias-Vasquez, A.; Perrone-Bizzozero, N.; Ehrlich, S.; Wang, L.; Bustillo, J.R.; Morris, D.; et al. MIR137HG risk variant rs1625579 genotype is related to corpus callosum volume in schizophrenia. Neurosci. Lett. 2015, 602, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.; Gupta, C.N.; Chen, J.; Patel, V.; Calhoun, V.D.; Ehrlich, S.; Wang, L.; Bustillo, J.R.; Perrone-Bizzozero, N.I.; Turner, J.A. Polymorphisms in MIR137HG and microRNA-137-regulated genes influence gray matter structure in schizophrenia. Transl. Psychiatry 2016, 6, e724. [Google Scholar] [CrossRef]
- Siegert, S.; Seo, J.; Kwon, E.J.; Rudenko, A.; Cho, S.; Wang, W.Y.; Flood, Z.; Martorell, A.J.; Ericsson, M.; Mungenast, A.E.; et al. The schizophrenia risk gene product miR-137 alters presynaptic plasticity. Nat. Neurosci. 2015, 18, 1008–1016. [Google Scholar] [CrossRef] [PubMed]
- Van Erp, T.G.M.; Guella, I.; Vawter, M.P.; Turner, J.; Brown, G.G.; McCarthy, G.; Greve, D.N.; Glover, G.H.; Calhoun, V.D.; Lim, K.O.; et al. Schizophrenia miR-137 locus risk genotype is associated with dorsolateral prefrontal cortex hyperactivation. Biol. Psychiatry 2014, 75, 398–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhang, X.L.; Hou, B.; Li, J.; Qiu, C.X.; Qin, W.; Yu, C.S.; Jiang, T.Z. The impact of MIR137 on dorsolateral prefrontal-hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology 2014, 39, 2153–2160. [Google Scholar] [CrossRef] [PubMed]
- Mothersill, O.; Morris, D.W.; Kelly, S.; Rose, E.J.; Fahey, C.; O’Brien, C.; Lyne, R.; Reilly, R.; Gill, M.; Corvin, A.P.; et al. Effects of MIR137 on fronto-amygdala functional connectivity. Neuroimage 2014, 90, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Green, M.J.; Cairns, M.J.; Wu, J.; Dragovic, M.; Jablensky, A.; Tooney, P.A.; Scott, R.J.; Carr, V.J. Genome-wide supported variant MIR137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol. Psychiatry 2013, 18, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Cummings, E.; Donohoe, G.; Hargreaves, A.; Moore, S.; Fahey, C.; Dinan, T.G.; McDonald, C.; O’Callaghan, E.; O’Neill, F.A.; Waddington, J.L.; et al. Mood congruent psychotic symptoms and specific cognitive deficits in carriers of the novel schizophrenia risk variant at MIR-137. Neurosci. Lett. 2013, 532, 33–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenelon, K.; Mukai, J.; Xu, B.; Hsu, P.K.; Drew, L.J.; Karayiorgou, M.; Fischbach, G.D.; MacDermott, A.B.; Gogos, J.A. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion, results in altered short-term plasticity in the prefrontal cortex. Proc. Natl. Acad. Sci. USA 2011, 108, 4447–4452. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, J.; Lu, X.J.; Song, X.B.; Ye, Y.X.; Zhou, J.; Ying, B.W.; Wang, L.L. Evaluation of six SNPs of microRNA machinery genes and risk of schizophrenia. J. Mol. Neurosci. 2013, 49, 594–599. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.Q.; Xu, Y.; Shugart, Y.Y.; Yue, W.H.; Qi, G.Y.; Yuan, G.Z.; Cheng, Z.H.; Yao, J.J.; Wang, J.D.; Wang, G.Q.; et al. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr. Bull. 2015, 41, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.Q.; Chen, Y.G.; Liu, C.X.; Lu, T.L.; Yan, H.; Ruan, Y.Y.; Yue, W.H.; Wang, L.F.; Zhang, D. Systematic association analysis of microRNA machinery genes with schizophrenia informs further study. Neurosci. Lett. 2012, 520, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Dean, B.; Gibbons, A.; Gogos, A.; Udawela, M.; Thomas, E.; Scarr, E. Studies on prostaglandin-endoperoxide synthase 1: Lower levels in schizophrenia and after treatment with antipsychotic drugs in conjunction with aspirin. Int. J. Neuropsychopharmacol. 2017, pyx092. [Google Scholar] [CrossRef]
- Smalheiser, N.R.; Lugli, G.; Zhang, H.; Rizavi, H.; Cook, E.H.; Dwivedi, Y. Expression of microRNAs and other small RNAs in prefrontal cortex in schizophrenia, bipolar disorder and depressed subjects. PLoS ONE 2014, 9, e86469. [Google Scholar] [CrossRef] [PubMed]
- Krude, T.; Christov, C.P.; Hyrien, O.; Marheineke, K. Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J. Cell Sci. 2009, 122, 2836–2845. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.; Ihling, C.; Sinz, A.; Krohn, K.; Huttelmaier, S. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev. 2015, 29, 1998–2003. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.A.; Laufer, B.I.; Melka, M.G.; Diehl, E.J.; O’Reilly, R.L.; Singh, S.M. DNA methylation differences in monozygotic twin pairs discordant for schizophrenia identifies psychosis related genes and networks. BMC Med. Genom. 2015, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Kishore, S.; Khanna, A.; Zhang, Z.Y.; Hui, J.Y.; Balwierz, P.J.; Stefan, M.; Beach, C.; Nicholls, R.D.; Zavolan, M.; Stamm, S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum. Mol. Genet. 2010, 19, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Bazeley, P.S.; Shepelev, V.; Talebizadeh, Z.; Butler, M.G.; Fedorova, L.; Filatov, V.; Fedorov, A. SnoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene 2008, 408, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Q.; Wang, X.; Beveridge, N.J.; Tooney, P.A.; Scott, R.J.; Carr, V.J.; Cairns, M.J. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS ONE 2012, 7, e36351. [Google Scholar] [CrossRef] [PubMed]
- Cohen, O.S.; McCoy, S.Y.; Middleton, F.A.; Bialosuknia, S.; Zhang-James, Y.; Liu, L.; Tsuang, M.T.; Faraone, S.V.; Glatt, S.J. Transcriptomic analysis of post-mortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophr. Res. 2012, 142, 188–199. [Google Scholar] [CrossRef] [PubMed]
- Castensson, A.; Emilsson, L.; Sundberg, R.; Jazin, E. Decrease of serotonin receptor 2C in schizophrenia brains identified by high-resolution mRNA expression analysis. Biol. Psychiatry 2003, 54, 1212–1221. [Google Scholar] [CrossRef]
- Lee, M.A.; Jayathilake, K.; Sim, M.Y.; Meltzer, H.Y. Decreased serotonin2C receptor responses in male patients with schizophrenia. Psychiatry Res. 2015, 226, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, K.; Kato, T. RNA editing of serotonin 2C receptor in human postmortem brains of major mental disorders. Neurosci. Lett. 2003, 346, 169–172. [Google Scholar] [CrossRef]
- Dracheva, S.; Elhakem, S.L.; Marcus, S.M.; Siever, L.J.; McGurk, S.R.; Haroutunian, V. RNA editing and alternative splicing of human serotonin 2C receptor in schizophrenia. J. Neurochem. 2003, 87, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, P.A. The Role of Regulatory Long Non-Coding RNAs in Adaptive Behaviour. Ph.D. Thesis, The University of Queensland, Brisbane, Australia, November 2015. [Google Scholar]
- Rogelj, B.; Hartmann, C.E.A.; Yeo, C.H.; Hunt, S.P.; Giese, K.P. Contextual fear conditioning regulates the expression of brain-specific small nucleolar RNAs in hippocampus. Eur. J. Neurosci. 2003, 18, 3089–3096. [Google Scholar] [CrossRef] [PubMed]
- Burenina, O.Y.; Oretskaya, T.S.; Kubareva, E.A. Non-coding RNAs as transcriptional regulators in eukaryotes. Acta Nat. 2017, 9, 13–25. [Google Scholar]
- Long, Y.C.; Wang, X.Y.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional gene regulation by long noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Sawyer, I.A.; Dundr, M. Chromatin loops and causality loops: The influence of RNA upon spatial nuclear architecture. Chromosoma 2017, 126, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Wang, L.Z.; Ding, Y.; Lu, X.Y.; Zhang, G.S.; Yang, J.X.; Zheng, H.W.; Wang, H.; Jiang, Y.S.; Xu, L.D. LncRNA structural characteristics in epigenetic regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Shah, A.; Shan, G. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Cui, Y.H.; Li, X.R.; Wang, B.H.; Na, L.; Shi, J.Y.; Wang, L.; Qiu, L.X.; Zhang, K.R.; Liu, G.F.; et al. A co-expression network analysis reveals lncRNA abnormalities in peripheral blood in early-onset schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2015, 63, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.L.; Niu, W.; Kong, L.M.; He, M.J.; Jiang, K.H.; Chen, S.D.; Zhong, A.F.; Li, W.S.; Lu, J.; Zhang, L.Y. Can lncRNAs be indicators for the diagnosis of early onset or acute schizophrenia and distinguish major depressive disorder and generalized anxiety disorder?—A cross validation analysis. Am. J. Med. Genet. B 2017, 174, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Barry, G.; Briggs, J.A.; Vanichkina, D.P.; Poth, E.M.; Beveridge, N.J.; Ratnu, V.S.; Nayler, S.P.; Nones, K.; Hu, J.; Bredy, T.W.; et al. The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol. Psychiatry 2014, 19, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.Q.; Hui, H.L.; Ye, N.; Shen, Y.; Xu, Q. Genetic variants in long non-coding RNA miat contribute to risk of paranoid schizophrenia in a Chinese Han Population. Schizophr. Res. 2015, 166, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Sone, M.; Hayashi, T.; Tarui, H.; Agata, K.; Takeichi, M.; Nakagawa, S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J. Cell Sci. 2007, 120, 2498–2506. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Tsuiji, H.; Yoshimoto, R.; Hasegawa, Y.; Furuno, M.; Yoshida, M.; Nakagawa, S. Competition between a noncoding exon and introns: Gomafu contains tandem uacuaac repeats and associates with splicing factor-1. Genes Cells 2011, 16, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, A.; Hasegawa, Y.; Ishida, K.; Yanaka, K.; Nakagawa, S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes Cells 2014, 19, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Lipska, B.K.; Hyde, T.M.; Ye, T.Z.; Newburn, E.N.; Morita, Y.; Vakkalanka, R.; Barenboim, M.; Sei, Y.; Weinberger, D.R.; et al. DISC1 splice variants are upregulated in schizophrenia and associated with risk polymorphisms. Proc. Natl. Acad. Sci. USA 2009, 106, 15873–15878. [Google Scholar] [CrossRef]
- Law, A.J.; Kleinman, J.E.; Weinberger, D.R.; Weickert, C.S. Disease-associated intronic variants in the ErbB4 gene are related to altered ErbB4 splice-variant expression in the brain in schizophrenia. Hum. Mol. Genet. 2007, 16, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Gianfrancesco, O.; Warburton, A.; Collier, D.A.; Bubb, V.J.; Quinn, J.P. Novel brain expressed RNA identified at the MIR137 schizophrenia-associated locus. Schizophr. Res. 2017, 184, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Laga, R.; Sedlacek, O. Therapeutic targeting of non-coding RNAs in cancer. Biochem. J. 2017, 474, 4219–4251. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.S.; Rait, A.; Garrido-Sanabria, E.R.; Pirollo, K.F.; Harford, J.B.; Chang, E.H. Nanotherapeutics for gene modulation that prevents apoptosis in the brain and fatal neuroinflammation. Mol. Ther. 2018, 26, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.C.; Gao, J.Q. Exosomes as novel bio-carriers for gene and drug delivery. Int. J. Pharm. 2017, 521, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, H.X.; He, C.P.; Fan, S.; Zhu, Y.L.; Qi, C.; Huang, N.P.; Xiao, Z.D.; Lu, Z.H.; Tannous, B.A.; et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials 2018, 150, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, D.M.; Liu, B.; Duncan, C.E.; Beveridge, N.J.; Tooney, P.A.; Schofield, P.R.; Cairns, M.J. Gene–microRNA interactions associated with antipsychotic mechanisms and the metabolic side effects of olanzapine. Psychopharmacology 2013, 227, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Swathy, B.; Saradalekshmi, K.R.; Nair, I.V.; Nair, C.; Banerjee, M. Pharmacoepigenomic responses of antipsychotic drugs on pharmacogenes are likely to be modulated by miRNAs. Epigenomics 2017, 9, 811–821. [Google Scholar] [CrossRef] [PubMed]
- Song, H.T.; Sun, X.Y.; Zhang, L.; Zhao, L.; Guo, Z.M.; Fan, H.M.; Zhong, A.F.; Niu, W.; Dai, Y.H.; Zhang, L.Y.; et al. A preliminary analysis of association between the down-regulation of microRNA-181b expression and symptomatology improvement in schizophrenia patients before and after antipsychotic treatment. J. Psychiatry Res. 2014, 54, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.D.; Sun, X.Y.; Niu, W.; Kong, L.M.; He, M.J.; Fan, H.M.; Li, W.S.; Zhong, A.F.; Zhang, L.Y.; Lu, J. A preliminary analysis of microRNA-21 expression alteration after antipsychotic treatment in patients with schizophrenia. Psychiatry Res. 2016, 244, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Fraguas, D.; Merchan-Naranjo, J.; del Rey-Mejias, A.; Castro-Fornieles, J.; Gonzalez-Pinto, A.; Rapado-Castro, M.; Pina-Camacho, L.; Diaz-Caneja, C.M.; Graell, M.; Otero, S.; et al. A longitudinal study on the relationship between duration of untreated psychosis and executive function in early-onset first-episode psychosis. Schizophr. Res. 2014, 158, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Subotnik, K.L.; Ventura, J.; Gretchen-Doorly, D.; Hellemann, G.S.; Agee, E.R.; Casaus, L.R.; Luo, J.S.; Villa, K.F.; Nuechterlein, K.H. The impact of second-generation antipsychotic adherence on positive and negative symptoms in recent-onset schizophrenia. Schizophr. Res. 2014, 159, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Scarr, E.; Millan, M.J.; Bahn, S.; Bertolino, A.; Turck, C.W.; Kapur, S.; Möller, H.J.; Dean, B. Biomarkers for psychiatry: The journey from fantasy to fact, a report of the 2013 CINP think tank. Int. J. Neuropsychopharmacol. 2015, 18. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.T.; Du, J.L.; Qi, Y.H.; Liang, G.F.; Wang, T.Y.; Li, S.C.; Xie, S.P.; Zeshan, B.; Xiao, Z.D. Aberrant expression of serum miRNAs in schizophrenia. J. Psychiatry Res. 2012, 46, 198–204. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.M.; Sun, X.Y.; Niu, W.; Zhao, L.; Zhang, Q.L.; Li, W.S.; Zhong, A.F.; Zhang, L.Y.; Lu, J. Altered microRNA expression in peripheral blood mononuclear cells from young patients with schizophrenia. J. Mol. Neurosci. 2015, 56, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.C.; Wu, J.; Zhang, H.X.; Zhang, G.L.; Sui, J.; Tong, W.W.; Zhang, X.Y.; Nie, L.L.; Duan, J.H.; Zhang, L.R.; et al. Alterations of miR-132 are novel diagnostic biomarkers in peripheral blood of schizophrenia patients. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2015, 63, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, F.; Shugart, Y.Y.; Yang, L.; Li, X.; Liu, Z.; Sun, N.; Yang, C.; Guo, X.; Shi, J.; et al. The early growth response protein 1-miR-30a-5p-neurogenic differentiation factor 1 axis as a novel biomarker for schizophrenia diagnosis and treatment monitoring. Transl. Psychiatry 2017, 7, e998. [Google Scholar] [CrossRef] [PubMed]
- Camkurt, M.A.; Karababa, F.; Erdal, M.E.; Bayazit, H.; Kandemir, S.B.; Ay, M.E.; Kandemir, H.; Ay, O.I.; Cicek, E.; Selek, S.; et al. Investigation of dysregulation of several microRNAs in peripheral blood of schizophrenia patients. Clin. Psychopharmacol. Neurosci. 2016, 14, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Zhang, J.; Niu, W.; Guo, W.; Song, H.T.; Li, H.Y.; Fan, H.M.; Zhao, L.; Zhong, A.F.; Dai, Y.H.; et al. A preliminary analysis of microRNA as potential clinical biomarker for schizophrenia. Am. J. Med. Genet. B 2015, 168, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Lacro, J.P.; Dunn, L.B.; Dolder, C.R.; Leckband, S.G.; Jeste, D.V. Prevalence of and risk factors for medication nonadherence in patients with schizophrenia: A comprehensive review of recent literature. J. Clin. Psychiatry 2002, 63, 892–909. [Google Scholar] [CrossRef] [PubMed]
- DiBonaventura, M.; Gabriel, S.; Dupclay, L.; Gupta, S.; Kim, E. A patient perspective of the impact of medication side effects on adherence: Results of a cross-sectional nationwide survey of patients with schizophrenia. BMC Psychiatry 2012, 12. [Google Scholar] [CrossRef] [PubMed]
- John, J.; Bhatia, T.; Kukshal, P.; Chandna, P.; Nimgaonkar, V.L.; Deshpande, S.N.; Thelma, B.K. Association study of MiRSNPs with schizophrenia, tardive dyskinesia and cognition. Schizophr. Res. 2016, 174, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, H.Y. Treatment-resistant schizophrenia—The role of clozapine. Curr. Med. Res. Opin. 1997, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Alacam, H.; Akgun, S.; Akca, H.; Ozturk, O.; Kabukcu, B.B.; Herken, H. MiR-181b-5p, miR-195-5p and miR-301a-3p are related with treatment resistance in schizophrenia. Psychiatry Res. 2016, 245, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.Y.; Lu, J.; Zhang, L.; Song, H.T.; Zhao, L.; Fan, H.M.; Zhong, A.F.; Niu, W.; Guo, Z.M.; Dai, Y.H.; et al. Aberrant microRNA expression in peripheral plasma and mononuclear cells as specific blood-based biomarkers in schizophrenia patients. J. Clin. Neurosci. 2015, 22, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.J.; Zhou, Y.; Xu, G.H.; Geng, B.; Cui, Q.H. Transcriptome analysis reveals non-identical microRNA profiles between arterial and venous plasma. Oncotarget 2017, 8, 28471–28480. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.; Olsen, L.; Lindow, M.; Jakobsen, K.D.; Ullum, H.; Jonsson, E.; Andreassen, O.A.; Djurovic, S.; Melle, I.; Agartz, I.; et al. Brain expressed microRNAs implicated in schizophrenia etiology. PLoS ONE 2007, 2, e873. [Google Scholar] [CrossRef] [PubMed]
- Pickard, B. Progress in defining the biological causes of schizophrenia. Expert Rev. Mol. Med. 2011, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Gogos, A.; Sbisa, A.M.; Sun, J.; Gibbons, A.; Udawela, M.; Dean, B. A role for estrogen in schizophrenia: Clinical and preclinical findings. Int. J. Endocrinol. 2015, 2015, 615356. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Galdzicka, M.; Ginns, E.; Baker, S.P.; Rogaev, E.; Xu, J.; Akbarian, S. Gender-specific reduction of estrogen-sensitive small RNA, miR-30b, in subjects with schizophrenia. Schizophr. Bull. 2012, 38, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Huang, X.F.; Chen, D.C.; Xiu, M.H.; Hui, L.; Liu, H.B.; Kosten, T.R.; Zhang, X.Y. Gender differences in cognitive function of patients with chronic schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2012, 39, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Fugger, H.N.; Foster, T.C.; Gustafsson, J.A.; Rissman, E.F. Novel effects of estradiol and estrogen receptor α and β on cognitive function. Brain Res. 2000, 883, 258–264. [Google Scholar] [CrossRef]
- Foster, T.C.; Rani, A.; Kumar, A.; Cui, L.; Semple-Rowland, S.L. Viral vector-mediated delivery of estrogen receptor-α to the hippocampus improves spatial learning in estrogen receptor-α knockout mice. Mol. Ther. 2008, 16, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Huo, Y.; Huang, C.J.; Chen, C.L.; Ye, J. MicroRNA-30b promotes axon outgrowth of retinal ganglion cells by inhibiting Semaphorin3A expression. Brain Res. 2015, 1611, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Gao, Y.; Meng, Z.Y.; Zhang, C.; Qi, Q.D. Regulatory role of microRNA-30b and plasminogen activator inhibitor-1 in the pathogenesis of cognitive impairment. Exp. Ther. Med. 2016, 11, 1993–1998. [Google Scholar] [CrossRef] [PubMed]
- Arnedo, J.; Svrakic, D.M.; del Val, C.; Romero-Zaliz, R.; Hernandez-Cuervo, H.; Fanous, A.H.; Pato, M.T.; Pato, C.N.; de Erausquin, G.A.; Cloninger, C.R.; et al. Uncovering the hidden risk architecture of the schizophrenias: Confirmation in three independent genome-wide association studies. Am. J. Psychiatry 2015, 172, 139–153. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, A.S.; Scarr, E.; Boer, S.; Money, T.; Jeon, W.J.; Felder, C.; Dean, B. Widespread decreases in cortical muscarinic receptors in a subset of people with schizophrenia. Int. J. Neuropsychopharmacol. 2013, 16, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Scarr, E.; Cowie, T.F.; Kanellakis, S.; Sundram, S.; Pantelis, C.; Dean, B. Decreased cortical muscarinic receptors define a subgroup of subjects with schizophrenia. Mol Psychiatry 2009, 14, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- Scarr, E.; Craig, J.M.; Cairns, M.J.; Seo, M.S.; Galati, J.C.; Beveridge, N.J.; Gibbons, A.; Juzva, S.; Weinrich, B.; Parkinson-Bates, M.; et al. Decreased cortical muscarinic M1 receptors in schizophrenia are associated with changes in gene promoter methylation, mRNA and gene targeting microRNA. Transl. Psychiatry 2013, 3, e230. [Google Scholar] [CrossRef] [PubMed]
- Tylee, D.S.; Kawaguchi, D.M.; Glatt, S.J. On the outside, looking in: A review and evaluation of the comparability of blood and brain “-omes”. Am. J. Med. Genet. B 2013, 162, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Lee, S.Y.; Scarr, E.; Yu, Y.H.; Lin, Y.T.; Liu, C.M.; Hwang, T.J.; Hsieh, M.H.; Liu, C.C.; Chien, Y.L.; et al. Aberrant expression of microRNAs as biomarker for schizophrenia: From acute state to partial remission, and from peripheral blood to cortical tissue. Transl. Psychiatry 2016, 6, e717. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Yu, S.L.; Hsieh, M.H.; Chen, C.H.; Chen, H.Y.; Wen, C.C.; Huang, Y.H.; Hsiao, P.C.; Hsiao, C.K.; Liu, C.M.; et al. MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PLoS ONE 2011, 6, e21635. [Google Scholar] [CrossRef] [PubMed]
- Zheutlin, A.B.; Jeffries, C.D.; Perkins, D.O.; Chung, Y.; Chekroud, A.M.; Addington, J.; Bearden, C.E.; Cadenhead, K.S.; Cornblatt, B.A.; Mathalon, D.H.; et al. The role of microRNA expression in cortical development during conversion to psychosis. Neuropsychopharmacology 2017, 42, 2188–2195. [Google Scholar] [CrossRef] [PubMed]
- Gurwitz, D. Exosomal microRNAs in tissue crosstalk. Drug Dev. Res. 2015, 76, 259–262. [Google Scholar] [CrossRef] [PubMed]
- Street, J.M.; Barran, P.E.; Mackay, C.L.; Weidt, S.; Balmforth, C.; Walsh, T.S.; Chalmers, R.T.A.; Webb, D.J.; Dear, J.W. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J. Transl. Med. 2012, 10. [Google Scholar] [CrossRef] [PubMed]
- Shankar, G.M.; Balaj, L.; Stott, S.L.; Nahed, B.; Carter, B.S. Liquid biopsy for brain tumors. Expert Rev. Mol. Diagn. 2017, 17, 943–947. [Google Scholar] [CrossRef] [PubMed]


| Study | Tissue Source | Altered RNA Expression in Schizophrenia vs. Control |
|---|---|---|
| Alacam et al., 2016 [136] | Plasma | Increased in treatment-resistant schizophrenia: miR-181b, miR-195, miR-301 Decreased in treatment-responsive schizophrenia: miR-181b, miR-195, miR-301 |
| Banigan et al., 2013 [49] | Prefrontal cortex exosomes | Increased: miR-497 Not changed: miR-15b, miR-29c, miR-31, miR-149, miR-219 |
| Beveridge et al., 2008 [50] | Superior temporal gyrus | Increased: miR-181b Not changed: Let7g |
| Beveridge et al., 2010 [11] | Prefrontal cortex | Increased: Let7d, miR-128, miR-16, miR-181a, miR-181b, miR-20a, miR-219, miR-27a, miR-29c, miR-7 |
| Superior temporal gyrus | Increased: miR-107; miR-15a, miR-15b, miR-195, miR-181b, Let7e, miR-20a, miR-26b Not changed: miR-16, miR-19a | |
| Burmistrova et al., 2007 [40] | Superior parietal lobule | Not altered: miR-130b |
| Guella et al., 2013 [65] | Prefrontal cortex | Not changed: miR-137; decreased in rs1625579-T/T control but not schizophrenia cases compared to G/T & G/G genotype |
| Kim et al., 2010 [55] | Prefrontal cortex | Increased: miR-34a, miR-132, miR-212, miR-544, miR-7, miR-154 |
| Lai et al., 2016 [153] | Peripheral blood mononuclear cells | Increased: miR-34a, miR-449a, miR-564, miR-548d |
| Prefrontal cortex | Not changed: miR-34a | |
| Stiatum | Not changed: miR-34a | |
| Melios et al., 2012 [142] | Prefrontal cortex | Decreased: miR-30b in females only |
| Perkins et al., 2007 [35] | Prefrontal cortex | Decreased: miR-30b, miR-26b, miR-92, miR-24, miR-30e Not changed: miR-29b, miR-195, miR-7 |
| Santarelli et al., 2011 [13] | Prefrontal cortex | Increased: mi-R17, miR-107, miR-134, miR-328, miR-382, miR-652 Not changed: miR-150, miR-199a, miR-25, miR-487a |
| Shi et al., 2012 [126] | Serum | Increased: miR-181b, miR-219, miR-1308, Let7g, miR-346, Decreased: miR-195 Not changed: miR-103 |
| Sun et al., 2015 [131] | Plasma | Increased: miR-30e, miR-181b, miR-34a, miR-346, miR-7 |
| Sun et al., 2015 [137] | Plasma | Increased: miR-132, miR-195, miR-30e, miR-7 |
| Peripheral blood mononuclear cells | Increased: miR-212, miR-34a, miR-30e | |
| Wei et al., 2015 [41] | Plasma | Increased: miR-130b, miR-193a Not changed: miR-122, miR-130a, miR-13b, miR-193a, miR-502, miR-652, miR-886 |
| Yu et al., 2015 [128] | Peripheral blood mononuclear cells | Increased: miR-132, miR-134, miR-1271, miR-664, miR-200c, miR-432 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibbons, A.; Udawela, M.; Dean, B. Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia. Non-Coding RNA 2018, 4, 11. https://doi.org/10.3390/ncrna4020011
Gibbons A, Udawela M, Dean B. Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia. Non-Coding RNA. 2018; 4(2):11. https://doi.org/10.3390/ncrna4020011
Chicago/Turabian StyleGibbons, Andrew, Madhara Udawela, and Brian Dean. 2018. "Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia" Non-Coding RNA 4, no. 2: 11. https://doi.org/10.3390/ncrna4020011
APA StyleGibbons, A., Udawela, M., & Dean, B. (2018). Non-Coding RNA as Novel Players in the Pathophysiology of Schizophrenia. Non-Coding RNA, 4(2), 11. https://doi.org/10.3390/ncrna4020011