Abstract
Cardiovascular disease (CVD) is a significant cause of morbidity and mortality across the world. A large proportion of CVD deaths are secondary to coronary artery disease (CAD) and myocardial infarction (MI). Even though prevention is the best strategy to reduce risk factors associated with MI, the use of cardioprotective interventions aimed at improving patient outcomes is of great interest. Opioid conditioning has been shown to be effective in reducing myocardial ischemia-reperfusion injury (IRI) and cardiomyocyte death. However, the molecular mechanisms behind these effects are under investigation and could provide the basis for the development of novel therapeutic approaches in the treatment of CVD. Non-coding RNAs (ncRNAs), which are functional RNA molecules that do not translate into proteins, are critical modulators of cardiac gene expression during heart development and disease. Moreover, ncRNAs such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are known to be induced by opioid receptor activation and regulate opioid signaling pathways. Recent advances in experimental and computational tools have accelerated the discovery and functional characterization of ncRNAs. In this study, we review the current understanding of the role of ncRNAs in opioid signaling and opioid-induced cardioprotection.
1. Introduction
According to data from the Global, Regional, and National Burden of Cardiovascular Diseases study, more than 17 million people died globally from cardiovascular disease (CVD) in 2015 and a large percentage of these deaths were secondary to coronary artery disease (CAD) [1]. The presence of cholesterol plaques and inflammation in the coronary arteries that supply the heart with blood, oxygen, and nutrients is characteristic of CAD pathophysiology. Plaque growth progressively restricts blood flow in the arteries, which can result in ischemia-reperfusion injury (IRI) and myocardial infarction (MI) [2].
Even though prevention is the best strategy to reduce risk factors associated with MI, interventions such as thrombolytic therapy and primary percutaneous coronary intervention are currently being used to reduce myocardial ischemic injury and limit infarct size in patients with MI [3]. However, the development of other cardioprotective strategies able to further improve patient outcomes is of major interest. Opioid conditioning is an effective intervention to ameliorate myocardial damage in animal models and human subjects [4,5]. It involves the activation of opioid receptor signaling pathways and mimics some of the protective effects achieved by ischemic preconditioning (IPC) [6,7,8].
In recent years, non-coding RNAs (ncRNAs) such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) have gained attention for their remarkable role in cellular function and disease development. The non-coding transcriptome is numerous, which is shown by the latest version of the human lncRNA collection NONCODE that contains 96,308 lncRNA gene loci and exceeds the 22,210 human genes annotated as protein-coding across the Ensembl/GENCODE, RefSeq, and UniProtKB databases [9,10]. Non-coding RNAs participate in DNA synthesis, maintain genome stability, and promote epigenetic modifications [11,12,13]. Most ncRNAs assemble RNA-protein complexes or exploit their intrinsic ability of base pairing to selectively bind and, thus, modulate proteins and other nucleic acids. Multiple studies have identified several ncRNAs implicated in cardiac gene expression and cardiomyocyte behavior during heart development and disease [14]. Furthermore, miRNAs and lncRNAs are known to be induced by opioid receptor activation and can regulate opioid functions in a variety of conditions [15,16]. Thus, polymorphisms affecting the function of ncRNAs involved in opioid signaling could also have a profound impact on drug behavior and efficacy. In this paper, we will summarize the current understanding of the role of miRNAs and lncRNAs in opioid signaling and opioid-induced cardioprotection.
2. The Opioid System
Opioids have potent analgesic effects and are routinely used as medications for managing peri-operative and post-operative pain [17]. Opioid receptor activation mediates the effects of endogenous and exogenous opioid ligands. The distribution of these receptors is broad and includes the central nervous system (CNS), peripheral sensory and autonomic nerves, and multiple cell types including cardiac cells. Activation of opioid receptors at pain modulating sites as well as in limbic, midbrain, and cortical structures prevents spinal cord pain transmission through neuron inhibition [18]. Additionally, the existence of opioid receptors throughout the body contributes to the regulation of complex behaviors and functions such as stress responses, reward processing, mood, respiration, endocrine changes, and immunity [19,20,21].
Opioid receptors belong to the G protein-coupled receptor (GPCR) superfamily. This family includes classical opioid receptors sensitive to naloxone and classified as delta (δ, DOR), kappa (k, KOR), and mu (μ, MOR) as well as the non-classical, naloxone insensitive nociceptin/orphanin receptor [22]. In 1973, three different research groups discovered the existence of opioid receptors in brain homogenates using opioid radio ligand binding [23,24,25]. The Oprm1, Oprd1, and Oprk1 genes were first cloned in the early 1990s and encode MOR, DOR, and KOR, respectively [26,27,28,29]. Both exogenous alkaloids and endogenous peptides such as enkephalins, dynorphins, and β-endorphins can bind with variable affinity and activate opioid receptors. Morphine was the first opiate to be synthesized and is considered a classic MOR agonist [22]. The use of morphine and MOR-selective synthetic opiates called 4-anilidopiperidines, which include fentanyl, remifentanil, sufentanil, alfentanil, lofentanil, ohmefentanil, and carfentanil is common in clinical practice. The potency of these synthetic opiates is between 100-fold and 10,000-fold higher than morphine [30].
Opioid receptors belong to the G protein-coupled receptor (GPCR) superfamily, which is a class of receptors that signal through intracellular heterotrimeric G proteins [18]. Upon activation with opioid agonists, guanosine diphosphate (GDP) is exchanged for guanosine triphosphate (GTP) on the α subunit of heterotrimeric G proteins, which is followed by the dissociation of α and βγ subunits. This dissociation leads to the subsequent interaction of α and βγ subunits with multiple downstream effectors including adenylyl cyclase, phospholipase C, protein kinase C (PKC), and ion channels. All four opioid receptors couple to pertussis toxin-sensitive G proteins like Gαi to inhibit adenylyl cyclase and cyclic adenosine monophosphate (cAMP) formation. Gα subunits can also interact directly with inward rectifying potassium (K+) channels considered essential regulators of electrical excitability in cardiomyocytes [31]. Opioid receptors are also known to interact with and modulate calcium (Ca2+) channels. Thus, when activated they reduce Ca2+ currents by closing P/Q-type, N-type, and L-type voltage-gated Ca2+ channels [32].
Additionally, opioid receptor activation initiates a non-canonical signaling pathway dependent on GPCR kinases (GRKs) and β-arrestins to induce receptor internalization and desensitization [33]. Recycling or degradation of opioid receptors occurs when GRKs trigger signaling termination through opioid receptor phosphorylation, which is followed by recruitment of β-arrestins and formation of receptor complexes able to internalize via clathrin-coated pits [34]. B-arrestins can also act as adaptors to recruit a wide range of signaling molecules to opioid receptor complexes including Src and activate downstream effectors such as mitogen-activated protein kinases (MAPKs) [35].
Opioid signaling via GPCRs can be agonist-selective and, as such, agonists that bind to the same receptor can induce different physiological responses by activating specific downstream signaling pathways [36]. Agonist-selective opioid signaling differentially modulates gene expression, which partially explains some of the conflicting results obtained with different agonists regarding cardioprotection and might have significant implications for opioid tolerance [37,38,39]. During opioid signaling, β-arrestin plays a significant role in the intracellular trafficking of GPCRs and β-arrestins bound to GPCRs also function as signal transducers of MAPKs and the serine/threonine kinase Akt [40]. Opioid agonists can induce extracellular signal-regulated kinase (ERK) phosphorylation through PKC or the β-arrestin pathway [41,42,43]. Morphine and fentanyl can both activate MOR but they differentially phosphorylate ERK [38]. ERK phosphorylation by morphine requires PKC activation and, in this case, ERK remains cytosolic and activates p90 ribosomal S6 kinase [37]. However, morphine ability to induce receptor phosphorylation, β-arrestin recruitment, and receptor internalization is limited in comparison to fentanyl [44]. ERK phosphorylation in response to fentanyl occurs in a β-arrestin dependent manner and leads to ERK translocation into the nucleus where it activates the transcription factor E26 transformation specific ETS domain-containing protein (ELK1) [38,44]. Thus, the differential modulation of opioid receptor trafficking and desensitization is due to the different ability of morphine and fentanyl to activate the β-arrestin pathway.
3. Mechanisms of Opioid Conditioning in the Heart
In CAD, blood flow disruption to the myocardium blocks the oxygen supply, which causes a rapid decline in ATP levels and an increase in AMP/ATP ratios [45]. The cardiac action potential shortens and within minutes of the occlusion extracellular accumulation of K+ occurs partly due to the opening of ATP-dependent K+ channels (KATP) that generate an outward K+ current [46]. Since oxidative phosphorylation supplies most of the energy required for cardiomyocyte function, a lack of oxygen and nutrients unleashes profound metabolic changes within the affected cells. These changes include the arrest of oxidative phosphorylation, membrane depolarization, ATP depletion, 3Na+-2K+ ATPase dysfunction, intracellular sodium (Na+) overload, inhibition of contractile function, and a switch to anaerobic glycolysis [3,47]. Consequently, the cells experience lactate buildup, intracellular pH reduction, activation of the Na+-H+ ion exchanger, and Ca2+ overload. Thus, the ischemic death of cardiomyocytes begins in the subendocardial myocardium and, when the ischemia is prolonged, it extends transmurally across the ventricular wall and towards the epicardium, which causes irreversible damage [48]. There are reports of cardiomyocytes suffering from apoptosis, necrosis, and autophagy in response to ischemic insults [49]. Although reperfusion of acutely ischemic myocardium restores blood flow and normoxia, the events that accompany reperfusion can independently induce cardiomyocyte death, arrhythmias, myocardial stunning, and microvascular obstruction [50,51].
In 1986, Murry, Jennings, and Reimer described how several brief periods of coronary artery occlusion and reperfusion before a prolonged ischemic insult reduce ATP depletion and tissue damage [52]. This intervention is known as IPC. Several endogenous and exogenous mediators such as adenosine, bradykinin, acetylcholine, angiotensin II, endothelin, and opioids activate membrane-bound GPCRs on cardiac cells to mimic IPC [8,53,54,55]. Administration of morphine, remifentanil, methadone or U50488H, a KOR agonist, before ischemia or during reperfusion protects from myocardial damage in animal models [8,56,57]. Furthermore, evidence from multiple studies demonstrates that opioid receptor antagonists block IPC. Naloxone, which is a non-selective opioid receptor antagonist, as well as selective antagonists such as DOR and KOR prevent IPC in multiple species [58,59]. Moreover, δ1-opioid receptor/KATP channel signaling pathway mediates the cardioprotective effects of morphine and PKCδ is an essential mediator of infarct size reduction after δ1-opioid receptor stimulation [60]. Multiple signaling pathways activated in cardiomyocytes downstream of opioid receptors are known to be involved in opioid-induced cardioprotection. Some of these pathways include PKC, MAPKs (ERK1/2, p38, and JNK1/2), mammalian target of rapamycin, cyclooxygenase 2, inducible nitric oxide synthase, KATP channels, and reactive oxygen species [60,61,62,63,64,65,66,67]. Additionally, opioid receptor agonists such as piritramide, pethidine, fentanyl, sufentanil, carfentanil, and buprenorphine elicit arrhythmogenic protection in animal models of acute coronary artery occlusion [68,69,70]. Opioid-induced tolerance to arrhythmia during myocardial ischemia appears to be mediated by the activation of peripheral δ2-opioid and κ1-opioid receptors [71].
Research on opioid receptor activation in the adult myocardium has focused mainly on DOR and KOR while the contributions of MOR have mainly been ignored due to its low expression in healthy cardiac tissue [72,73,74]. However, MOR expression and its ligand-binding activity increase after ligation of the coronary artery in animal models and in isolated failing hearts [75,76]. Morphine or remifentanil administration before myocardial IRI induces MOR-mediated protection in failing hearts but not in healthy ones [76]. These protective effects appear to be dependent on phosphorylation of ERK1/2 and glycogen synthase kinase 3-βdownstream of MOR activation. Thus, MOR is a potential therapeutic target for opioid-induced cardioprotection during heart failure.
4. Non-Coding RNAs
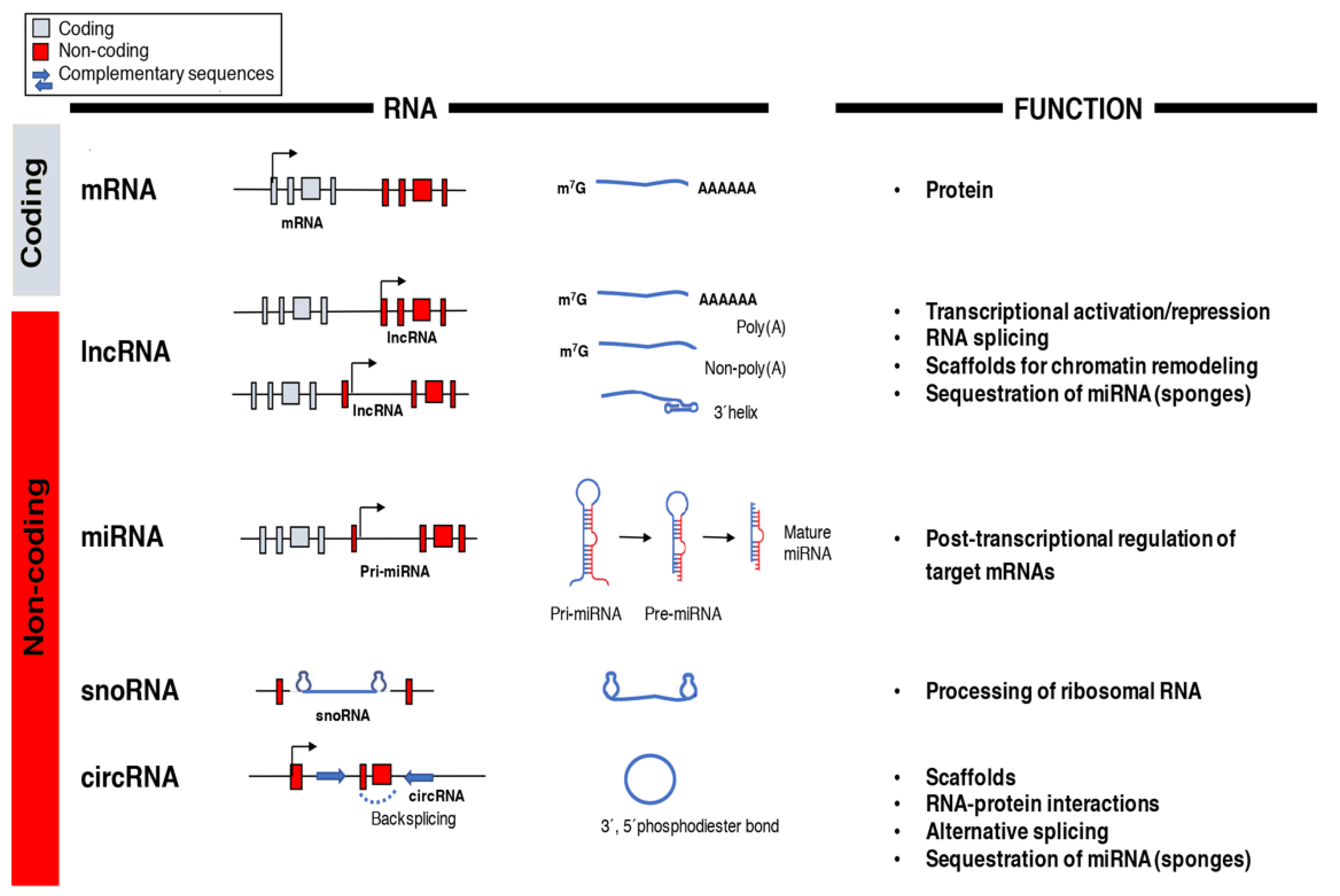
The central dogma of molecular biology established a directional flow of genetic information from DNA to RNA to protein and was still a prevailing idea in the 1970s [77]. It was only after the human genome was decoded in the post-genomic era that the real potential of RNA molecules other than messenger RNA (mRNA) as functional regulators of gene expression and cellular functions began to be appreciated [11]. The arrival of next-generation sequencing led to the discovery of multiple RNA species like miRNAs, lncRNAs, small nucleolar RNAs (snoRNAs), and circular RNAs (circRNAs) with unique biochemical and structural features whose functions are still not entirely understood (Figure 1) [78]. However, recent data highlights essential roles of the non-coding transcriptome in cell biology and human disease particularly in the regulation of gene networks through epigenetic modifications, direct transcriptional regulation, RNA processing, and inhibition of translation [79,80].
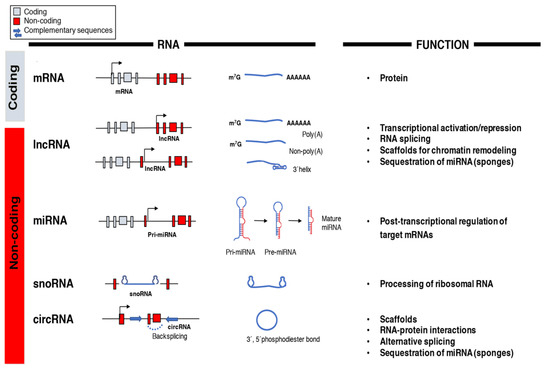
Figure 1.
Classification and function of coding and non-coding RNAs (ncRNAs). The array of RNA molecules is diverse in structure and function across species. Messenger RNAs (mRNAs) are coding transcripts that undergo extensive post-transcriptional processing, 5′capping (m7G) and 3′polyadenylation (poly(A)), and can be translated into proteins. Long non-coding RNAs (lncRNAs) possess little, if any, coding potential and, after their transcription from genic and intergenic regions, they can suffer different types of post-transcriptional processing. MicroRNAs (miRNAs) are transcribed into primary miRNA transcripts (pri-miRNA) by Pol II or are generated through alternative splicing. They are further processed into pre-miRNA and mature miRNA before being incorporated into the RNA-induced silencing complex (RISC)to silence their target genes. Small nucleolar RNAs (snoRNAs) are usually transcribed by Pol II and belong to the translational machinery of the cell and process ribosomal RNAs. Circular RNAs (circRNAs) are produced from back splicing of exons and possess unique features given by the formation of a 3′,5′phosphodiester bond.
The first miRNA genes known as lin-4 and let-7 and their role as post-transcriptional regulators of gene expression were described in Caenorhabditis elegans during the early 1990s [81,82]. Micro RNAs are small, ncRNA molecules of ~22 nucleotides in length that use short regions of complementarity of around six to eight nucleotides of their target mRNAs to repress gene expression [83,84]. Endogenous miRNA-complementary sites are located within the 3′ and 5′ untranslated regions (3′ and 5′ UTRs) of target mRNAs as well as in the coding sequence [85,86,87]. Moreover, combinatory interactions between miRNA and both 3′ and 5′ UTRs of an mRNA have also been reported [88]. Biogenesis of miRNAs begins in the nucleus where RNA polymerases (Pol) II or III transcribe miRNA genes into primary miRNA transcripts (pri-miRNA) [89,90]. Then a nuclear microprocessor complex comprised of the nuclear RNase III enzyme Drosha and DiGeorge critical region 8 protein cleave pri-miRNAs to generate a precursor miRNA (pre-miRNA) [91,92]. Drosha-mediated processing of pri-miRNAs into pre-miRNAs is not required when miRNAs are derived from introns that have been released from their transcripts by alternative splicing [93]. Pre-miRNAs exported into the cytoplasm by exportin 5 are then processed by a RNAse III enzyme, which is called Dicer to generate a short RNA duplex that will give origin to a mature, single-stranded miRNA of around 20 to 30 nucleotides in length [94,95,96,97]. Two different mature miRNAs can arise from an miRNA duplex (-3p and -5p) but usually only one strand is incorporated into the RNA-induced silencing complex to guide the repression of its target mRNAs [98]. When the base pairing between miRNAs and their mRNA targets is fully complementary, it leads to direct cleavage and degradation of the target mRNA through a process involving the machinery of RNA interference while, if miRNAs are only partially complementary to their mRNA targets, the stability of the mRNA target is maintained but the result is inhibition of protein synthesis [89,90].
LncRNAs are a diverse group of long RNA transcripts (>200 nucleotides) without a protein-coding role and, unlike mRNAs, exhibit poor primary sequence conservation, tend to be shorter, and are usually expressed at relatively low levels [99]. This classification separates lncRNAs from short ncRNAs such as tRNAs, miRNAs, and snoRNAs and from longer, protein-coding transcripts. RNA polymerase II transcribes lncRNAs from different DNA elements (promoters, enhancers, and intergenic regions) [100,101]. LncRNAs usually suffer extensive post-transcriptional processing to reach their mature forms. This processing usually involves 5′-capping, splicing, and polyadenylation even though long primary transcripts can also generate mature lncRNAs without 5′ cap structures or 3′ polyadenylated tails (Figure 1) [101]. Some of the non-canonical mechanisms of lncRNAs processing include ribonuclease P cleavage to generate mature 3′ ends, which caps at their ends by snoRNA–protein complexes or circular structure formation [79,101]. Although lncRNAs are considered non-coding transcripts, some contain cryptic open reading frames [102]. LncRNAs act as scaffolds by forming RNA–protein complexes, perform decoy functions by interacting with DNA-binding proteins like transcription factors, work as enhancers in cis to activate or repress genes, and exert epigenetic regulation when they recruit chromatin-modifying proteins to specific genomic sites [103,104]. Additionally, lncRNAs exhibit a diverse repertoire across species and possess nuclear roles as well as cytoplasmic functions [105]. Competing endogenous RNAs (ceRNAs), which contain miRNA-binding sites and act as miRNA sponges, are an example of lncRNAs with cytoplasmic functions [106,107].
5. miRNAs in Control of Opioid Signaling
Several ncRNAs are known to be under the control of opioid receptors and can act as fine tuners of opioid signaling pathways by working as downstream targets of GPCRs and associated signaling molecules [108]. Evidence suggests that miRNAs can have profound effects on opioid receptor expression and, thus, in opioid receptor-mediated physiological and pathological processes. A genome-wide association study in human monocyte-derived macrophages treated with morphine identified 26 differentially expressed miRNAs (Table 1) of which miR-15b-5p and miR-181b-5p participate in inflammatory and oxidative stress processes responsible for human immunodeficiency virus 1 (HIV1) viral reservoir expansion in the CNS [109]. According to these results, opiates promote HIV1 propagation in immune cells while simultaneously suppressing immune functions. In a different study, oral administration of hydromorphone and oxycodone induced differential changes in plasma profiles of 179 miRNAs [15]. These opioids upregulate 9 miRNAs and downregulate 17 others (Table 1). Many of these miRNAs especially the let-7 family of miRNAs, miR-23b-5p, and miR-16-5p, have been associated with MOR signaling regulation elsewhere [110,111,112].

Table 1.
Effect of opioids on miRNA expression in a variety of cells and tissues.
The 3′ UTRs of MOR, KOR, and DOR mRNAs contain multiple regulatory elements that are able to modulate their translational efficiency, mRNA stability, and transport [118,119]. Evidence indicates that opioids affect MOR expression through alternative splicing and changes in translational efficiency via miRNAs rather than altering its transcription [120,121]. The let-7 family of miRNAs, which include miR-23b-5p, miR-212/132, miR-16-5p, miR-339-3p, and miR-103/107, have all been identified as miRNAs that are able to bind to MOR 3′ UTR to post-transcriptionally repress its expression.
The let-7 family of miRNAs is abundant across species and its members share a ‘seed sequence’ responsible for mRNA target recognition by the RNA-induced silencing complex (RISC), which suggests that their functions are evolutionarily conserved and highly redundant [122,123]. Twelve different genomic loci in the human genome with some clustered together encode the let-7 family of miRNAs [124].
Although the expression of let-7 miRNAs is ubiquitous and appears to be context-dependent, it is clear that they are essential regulators of proliferation, stem-cell renewal, and terminal differentiation [125,126,127,128]. Interestingly, the expression of let-7 miRNAs increases in response to morphine treatment in neuroblastoma derived SH-SY5Y cells and these miRNAs modulate opioid tolerance by post-transcriptionally regulating MOR expression [110]. Thus, MOR downregulation represents one of many mechanisms contributing to opioid tolerance. In multiple studies, MOR mRNA expression remains unaltered by opioid agonists, which suggests that agonist-selective MOR downregulation occurs post-transcriptionally [129,130,131,132,133]. Let-7 miRNAs can downregulate MOR without affecting its mRNA expression through a mechanism that interferes with translation initiation [134]. Translational repression occurs when cytosolic let-7 miRNAs get incorporated into RISC and its interaction with MOR 3′ UTR leads to MOR mRNA recruitment to P-bodies where it becomes degraded by de-capping enzymes and exonucleases [110,134]. Other mechanisms are likely involved in opioid tolerance besides let-7 downregulation of MOR since knocking down let-7 in the brain only partially reduces morphine antinociceptive tolerance.
Wu Q et al. first described miR-23b-5p as a repressor of MOR translation through its interaction with a K box motif located in MOR 3′ UTR [113]. miR-23b-5p inhibits the association of MOR mRNA with polysomes and is responsible for suppressing MOR translation in mouse neuronal N2A-MOR cells. The same group later demonstrated that long-term morphine treatment upregulates miR-23b-5p in a dose-dependent and time-dependent manner, which describes miR-23b-5p as a post-transcriptional negative feedback mechanism between MOR activation and MOR expression [111].
Expression of the miR-212/132 cluster is required for adequate development, maturation, and function of neurons [135]. Morphine regulates the miR-212/132 cluster via MOR activation and its expression actively represses MOR mRNA translation, which highlights the close relationship that exists between these miRNAs and opioid signaling [114]. Transcription and processing of miR-212/132 occur through the activation of the MAPK MEK1/2, calmodulin, calmodulin kinases II and IV, and protein kinase A that converge in phosphorylation of cAMP Response Element Binding (CREB) protein. Additionally, the miR-212/132 cluster promoter is regulated by the methyl CpG binding protein 2, which forms an epigenetic protein complex and acts as a transcriptional repressor on methylated DNA after sustained morphine administration [114,136]. Another miRNA that post-transcriptionally targets MOR 3′ UTR is miR-134-5p. In a model of Freund’s adjuvant-induced chronic inflammatory pain downregulation of miR-134-5p inversely correlates with MOR expression in rat dorsal root ganglia and functional experiments revealed that MOR is a target of miR-134-5p [137]. Similarly, miR-339-3p expression increases in hippocampus isolated from mice chronically treated with opioid agonists [115]. Furthermore, morphine and fentanyl differentially modulate miR-339-3p expression levels through increased transcription of its pri-miRNA. Thus, the consistent differences between fentanyl and morphine in their ability to modulate miR-339-3p expression hint to molecular mechanisms underlying their distinct pharmacological properties.
The ubiquitous and highly conserved miR-16-5p is a miRNA linked to tumorigenesis, cell cycle regulation, proliferation, and apoptosis [138]. miR-16-5p also exerts miRNA-mediated post-transcriptional MOR regulation. Morphine inhibits miR-16-5p expression in lymphocytes, which is an effect that can be reversed by the antagonist naloxone [112]. Interestingly, miR-16-5p suppresses MOR expression when it binds to a site in its 3′ UTR located between 8699 and 8719 nucleotides away from the stop codon [112]. Thus, downregulation of miR-16-5p is a post-transcriptional mechanism through which morphine increases MOR receptor levels by stabilizing its mRNA.
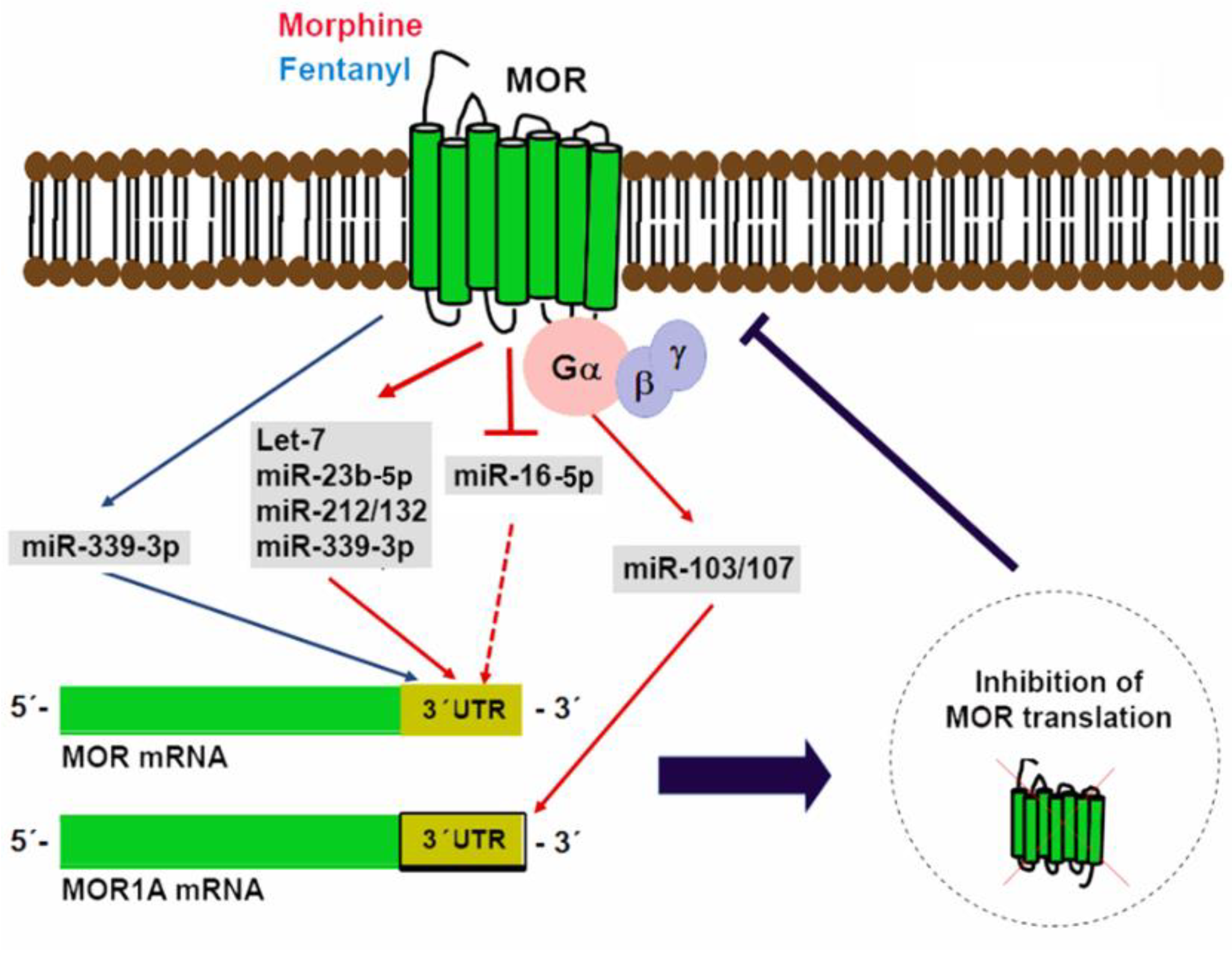
The Oprm1 gene suffers considerable alternative splicing that results in the generation of multiple splice variants including a carboxyl-terminal splice variant that retains exon 3b and is known as MOR1A [116,139]. A conserved miR-103/107 targeting site in the 3′ UTR of both mouse and human MOR1A was identified through computational modeling and was further verified as a functional binding site for miR-103/107 in HEK-293 cells and a neuroblastoma cell line that endogenously expresses human MOR1A [116]. miR-103/107 is a pair of miRNAs with almost identical sequences except for a nucleotide located at the 3′-end and overlapping targets [140,141]. In a wide variety of tissues, the transcription of these miRNAs occurs from introns that map to three different pantothenate kinase family genes that encode regulatory enzymes necessary for coenzyme A biosynthesis [142,143]. In a neuroblastoma cell line and the striatum of a morphine-tolerant mouse, chronic morphine treatment increases miR-103/107 levels, which, in turn, reduce polyribosome-associated MOR1A mRNA [116]. Evidence that miR-103/107 regulate an Oprm1 splice variant suggests that miRNAs could differentially regulate other opioid receptor variants with diverse 3′-UTRs. Figure 2 shows a graphical depiction of miRNAs in control of MOR translation.
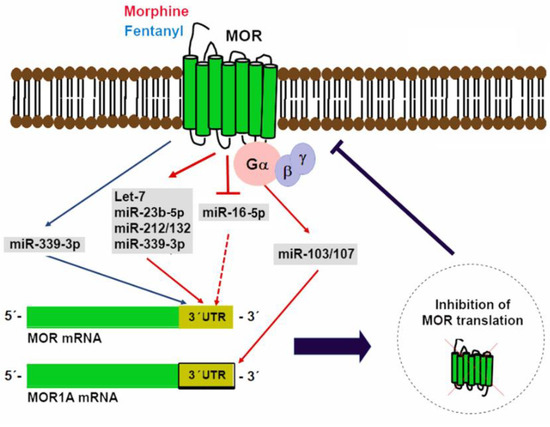
Figure 2.
miRNAs in control of mu opioid receptor (MOR) translation. Multiple miRNAs able to bind to regulatory elements located in MOR 3′ UTR have been identified. Expression of some of these miRNAs (miR-339-3p, let-7, miR-23b-5p, miR-212/132, and miR-103/107) is induced by opioid agonists as a negative feedback mechanism. In contrast, morphine can also reduce miR-16-5p expression to increase MOR translation.
Although fewer studies of DOR and KOR post-transcriptional regulation exist, DOR is a direct target of miR-874-5p in the context of hepatocellular carcinoma [144]. In this context, miR-874-5p expression inversely correlates with DOR expression and miR-874-5p downregulation is associated with a larger tumor size, increased vascular invasion, poor tumor differentiation, and worse outcomes.
In addition to miRNAs that regulate opioid receptor expression, miR-365-5p is a regulator of β-arrestin 2 able to reverse morphine anti-nociceptive tolerance in a rat model [117]. Neurons of the spinal cord express miR-365-5p and its levels decrease after chronic morphine administration. Thus, miR-365-5p plays an essential regulatory role in the development of morphine tolerance by targeting β-arrestin 2, which is a key molecule in opioid signaling and receptor trafficking [117,145].
6. Non-Coding RNAs in Opioid-Induced Cardioprotection
DOR activation protects against ischemic injury in the heart. Novel evidence demonstrates that DOR signaling affects miRNA expression in the heart and some of these miRNAs could be involved in mediating its protective effects (Table 2). Administration of the DOR agonist UFP-512 in rats maintained under normoxic conditions increases cardiac expression of miR-107-3p, miR-141-3p, and miR-350-5p while DOR activation under hypoxic conditions increases miR-7a/b, mi-107-3p, miR-200b-5p, miR-376a-3p, and miR134-5p levels [146]. Even though the exact contribution of these miRNAs and their targets to DOR-mediated cardioprotection is unknown, some of them are key modulators of apoptosis, angiogenesis, and tissue repair [147,148,149]. miR-107-3p targets hypoxia-inducible factor 1 βand prevents endothelial progenitor cell differentiation during hypoxia. This demonstrates the role of this miRNA in tissue repair during ischemic heart disease [147]. miR-200b-5p promotes angiogenesis by inhibiting its target ETS1 and miR-7a/b expression acts as a protective mechanism against IRI-induced apoptosis through poly(ADP-ribose) polymerase repression [148,149].

Table 2.
Effect of opioids on miRNA expression in the heart and cardiac cells.
Additionally, morphine preconditioning modifies the miRNA expression profile in isolated cardiomyocytes while decreasing cell death and lactate dehydrogenase levels in models of hypoxia/reoxygenation in vitro [150,151]. A total of 39 miRNAs are differentially expressed in adult rat ventricular myocytes after morphine conditioning (Table 2) of which miR-133b-5p appears to have a preponderant role in morphine signaling and cardioprotection [150].
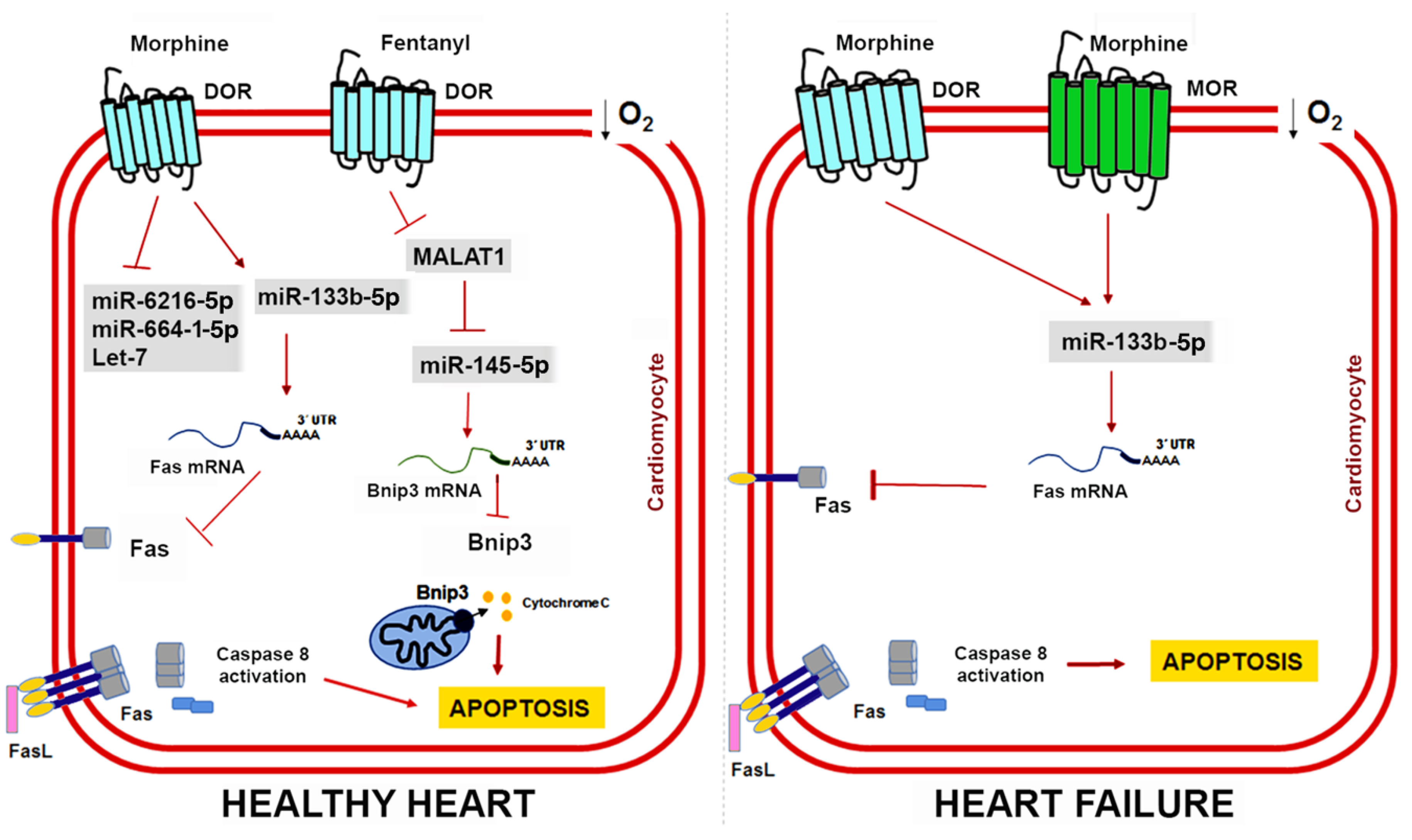
Initially defined as a muscle-specific miRNA, miR-133b belongs to a family of myomiRs that also includes miR-133a, miR-133b, miR-1, and miR-206. These miRNAs share tissue expression specificity but their mature sequences present nucleotide variants and, as such, their functions could be different [153]. Morphine administration in cardiac muscle cells upregulates miR-133b-5p expression and its knockdown blocks the protective effects of morphine preconditioning by promoting Fas expression, which is a death receptor that induces apoptosis by interacting with the Fas ligand [150]. Interestingly, some studies have described opioidergic conditioning interventions as being less effective in pathological conditions such as hypertrophy and heart failure [154,155,156]. To address this issue, Zhu HJ et al. further compared the expression profile of miRNAs in a rat model of heart failure induced by chronic doxorubicin injection [151]. They found 12 miRNAs whose expression pattern was significantly altered by hypoxia and morphine preconditioning (Table 2) and validated miR-133b-5p, miR-6216-5p, miR-664-1-5p, and let-7e as cardioprotective miRNAs in vitro. In cardiomyocytes derived from failing hearts, miR-133b-5p is severely downregulated, but morphine administration restores its expression [151]. Moreover, miR-133b-5p also mediates the protective effect of morphine in cardiomyocytes derived from failing hearts through its target Fas. However, the protective role of other miR-133b-5p targets remains to be determined. Morphine regulates the dopaminergic neuron differentiation in zebrafish via miR-133b-5p and its target Pitx3 and miR-133b-5p also targets the small GTPase RhoA to promote neurite outgrowth and spinal cord regeneration [157,158,159]. Figure 3 shows the cardioprotective mechanisms mediated by opioids and ncRNAs in healthy hearts and damaged hearts.
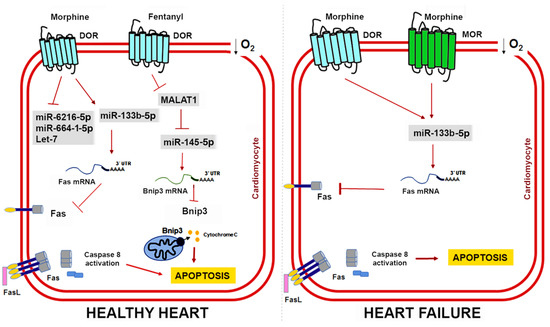
Figure 3.
ncRNAs and opioid-induced cardioprotection in healthy and failing hearts. Experimental evidence shows that opioids can reduce cardiomyocyte apoptosis caused by myocardial IRI through miR-133b-5p and its target Fas in both healthy and failing hearts. In healthy hearts, fentanyl activation of DOR inhibits MALAT1 expression and a ceRNA for miR-145-5p, which allows miR-145-5p to target Bnip3 and reduce apoptosis. Interestingly, heart failure induces expression of MOR in cardiomyocytes, which could have important implications for opioid-mediated cardioprotection in patients with MI.
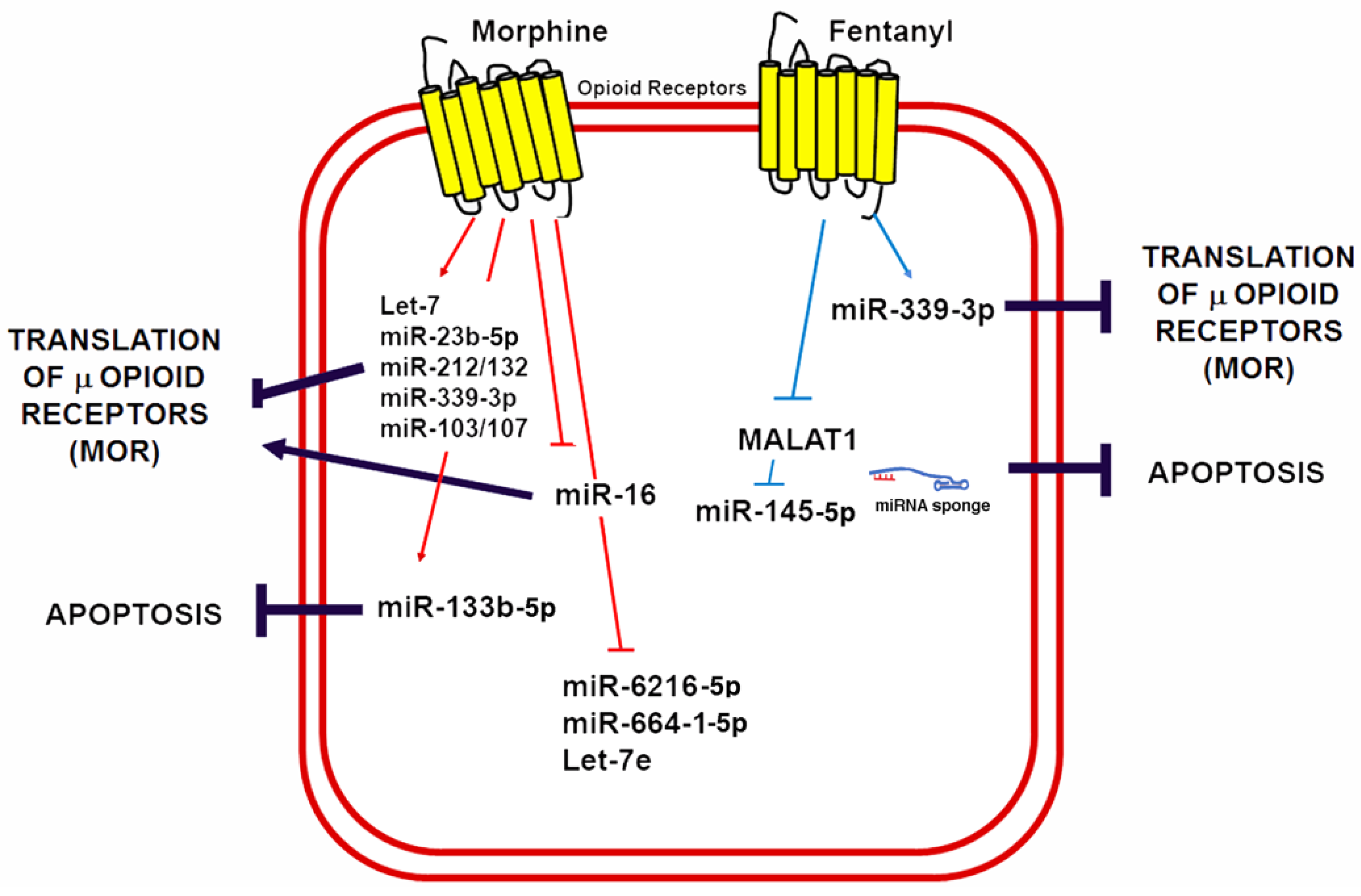
Knowledge regarding the role of lncRNAs in opioid signaling and cardioprotection is still in its early days. Nonetheless, recent studies have demonstrated that lncRNAs could be a therapeutic target in myocardial IRI [160,161]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA upregulated in response to hypoxia and myocardial ischemia [162,163]. Increased MALAT1 associates with cardiomyocyte apoptosis and its downregulation improves the ventricular function after myocardial IRI [164]. Moreover, fentanyl reduces hypoxia/reoxygenation injury through the MALAT1/miR-145-5p/Bcl-2 interacting protein 3 (BNIP3) axis [152]. MALAT1 promotes apoptosis by acting as a ceRNA for miR-145-5p, which allows the expression of miR-145 target BNIP3 (a pro-apoptotic protein associated with autophagy) [165]. Fentanyl exerts its protective effects through MALAT1 downregulation and negatively regulates the miR-145-5p/BNIP3 pathway in vitro and in mice subjected to myocardial IRI [152]. Figure 4 summarizes the role of ncRNAs in opioid signaling and opioid-induced cardioprotection.
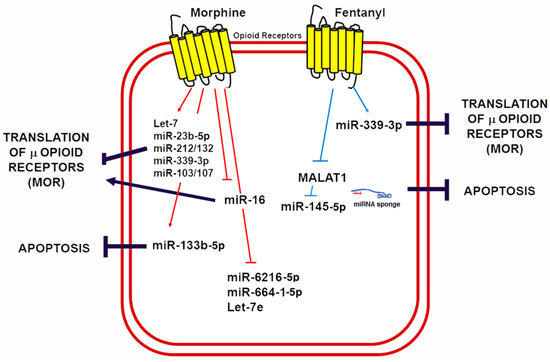
Figure 4.
Morphine and fentanyl induction of ncRNAs involved in opioid signaling and cardioprotection. A schematic summary of miRNAs known to be induced by opioids in a variety of cells and animal models as well as its mechanisms of opioid signaling regulation and its potential implications for cardioprotection by reducing apoptosis.
7. Future Perspectives
Non-coding RNAs are essential modulators of opioid signaling and gene expression in physiological processes and CVD. A better understanding of the molecular networks regulated by miRNAs and lncRNAs in response to the activation of opioid signaling pathways in the heart may be exploited in the future to develop effective interventions to reduce myocardial ischemic injury and limit infarct size in patients with MI.
Dysregulation of the non-coding transcriptome (miRNAs, lncRNAs, and circRNAs) has been reported across various heart diseases, which suggests that ncRNAs are critical modulators of cardiac pathophysiology [166]. Our current knowledge regarding the origin and function of ncRNAs in the heart is still in its infancy. However, some of these ncRNAs are known to be released into the circulation and their concentration levels in blood, serum, urine, saliva, or microvesicles could allow us to discriminate between heathy and diseased subjects. For this reason, ncRNAs have gained recognition as potential non-invasive biomarkers to improve diagnosis, prognosis, and risk assessment of cardiac injury [167]. Moreover, their biochemical characteristics make them more stable, sensitive, and specific than other types of circulating biomarkers [168]. The miRNAs miR-1-5p, miR-126-5p, miR-197-5p, and miR-208a-5p have been proposed as diagnostic markers of acute MI while additional panels of ncRNAs useful in tracking complications derived from myocardial ischemia have also been reported [169,170,171,172,173]. Unfortunately, studies evaluating the true sensitivity and specificity of these ncRNAs as biomarkers in different populations are still lacking. Additionally, mimicking opioid cardioprotection through in vitro delivery of ncRNA as drug targets represents a promising therapeutic strategy for a myriad of cardiac conditions [174]. However, there is still not enough evidence on the effectiveness of these new approaches in CVD treatment and many challenges remain such as specific delivery, poor cellular uptake, off-target effects, and immunogenicity [175].
Opioids are drugs with a narrow therapeutic index that are commonly used to treat pain. Multiple genetic factors regulate opioid pharmacokinetics and pharmacodynamics [176,177]. The pharmacokinetic genes coding for phase I and II enzymes involved in opioid metabolism are highly polymorphic [176,177]. Thus, the presence of single-nucleotide polymorphisms, copy number variations, or differences in expression levels of pharmacokinetic genes can manifest as variations of opioid-induced responses among different individuals. The field of pharmacogenomics has mostly focused on drug targets and its relationship with metabolism or transport genes. Since ncRNAs can potentially regulate the expression of pharmacogenomic-related genes and processes, they could have profound implications on personalized responses to opioid analgesics.
Author Contributions
Conceptualization, Z.M. and R.E. Literature Review, C.I., M.d.l.P.G. and R.R. Writing—Original Draft Preparation, R.E. Writing—Review & Editing, Z.M. and C.I.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Theroux, P. Pathophysiology of coronary artery disease. Circulation 2005, 111, 3481–3488. [Google Scholar] [CrossRef] [PubMed]
- Hausenloy, D.J.; Yellon, D.M. Myocardial ischemia-reperfusion injury: A neglected therapeutic target. J. Clin. Investig. 2013, 123, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Headrick, J.P.; See Hoe, L.E.; Du Toit, E.F.; Peart, J.N. Opioid receptors and cardioprotection-‘Opioidergic conditioning’ of the heart. Br. J. Pharmacol. 2015, 172, 2026–2050. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.E.; Gross, G.J. Opioids and cardioprotection. Pharmacol. Ther. 2001, 89, 123–137. [Google Scholar] [CrossRef]
- Schultz, J.E.; Hsu, A.K.; Gross, G.J. Ischemic preconditioning in the intact rat heart is mediated by delta1- but not mu- or kappa-opioid receptors. Circulation 1998, 97, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.J.; Hsu, A.K.; Gross, G.J. Ischemic preconditioning and morphine-induced cardioprotection involve the delta (δ)-opioid receptor in the intact rat heart. J. Mol. Cell. Cardiol. 1997, 29, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.E.; Hsu, A.K.; Gross, G.J. Morphine mimics the cardioprotective effect of ischemic preconditioning via a glibenclamide-sensitive mechanism in the rat heart. Circ. Res. 1996, 78, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Abascal, F.; Juan, D.; Jungreis, I.; Martinez, L.; Rigau, M.; Rodriguez, J.M.; Vazquez, J.; Tress, M.L. Loose ends: Almost one in five human genes still have unresolved coding status. Nucleic Acids Res. 2018, 46, 7070–7084. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012, 337, 1159–1161. [Google Scholar] [CrossRef] [PubMed]
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Frías-Lasserre, D.; Villagra, C.A. The importance of ncRNAs as epigenetic mechanisms in phenotypic variation and organic evolution. Front. Microbiol. 2017, 8, 2483. [Google Scholar] [CrossRef] [PubMed]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Heart J. 2017. [Google Scholar] [CrossRef] [PubMed]
- Toyama, K.; Kiyosawa, N.; Watanabe, K.; Ishizuka, H. Identification of circulating miRNAs differentially regulated by opioid treatment. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Law, P.Y.; Loh, H.H. Non-coding RNAs regulating morphine function: With emphasis on the in vivo and in vitro functions of miR-190. Front. Genet. 2012, 3, 113. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.; Benveniste, H.; McLellan, A.T. Use and misuse of opioids in chronic pain. Annu. Rev. Med. 2018, 69, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Al-Hasani, R.; Bruchas, M.R. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 2011, 115, 1363–1381. [Google Scholar] [CrossRef] [PubMed]
- Bodnar, R.J. Endogenous opiates and behavior: 2009. Peptides 2010, 31, 2325–2359. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, O.; Hester, J. Opioids and endocrine dysfunction. Br. J. Pain 2012, 6, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Le Merrer, J.; Becker, J.A.; Befort, K.; Kieffer, B.L. Reward processing by the opioid system in the brain. Physiol. Rev. 2009, 89, 1379–1412. [Google Scholar] [CrossRef] [PubMed]
- Pathan, H.; Williams, J. Basic opioid pharmacology: An update. Br. J. Pain 2012, 6, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Pert, C.B.; Snyder, S.H. Properties of opiate-receptor binding in rat brain. Proc. Natl. Acad. Sci. USA 1973, 70, 2243–2247. [Google Scholar] [CrossRef] [PubMed]
- Simon, E.J.; Hiller, J.M.; Edelman, I. Stereospecific binding of the potent narcotic analgesic (3H) Etorphine to rat-brain homogenate. Proc. Natl. Acad. Sci. USA 1973, 70, 1947–1949. [Google Scholar] [CrossRef] [PubMed]
- Terenius, L. Stereospecific interaction between narcotic analgesics and a synaptic plasm a membrane fraction of rat cerebral cortex. Acta Pharmacol. Toxicol. 1973, 32, 317–320. [Google Scholar] [CrossRef]
- Evans, C.J.; Keith, D.E.; Morrison, H.; Magendzo, K.; Edwards, R.H. Cloning of a delta opioid receptor by functional expression. Science 1992, 258, 1952–1955. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, B.L.; Befort, K.; Gaveriaux-Ruff, C.; Hirth, C.G. The delta-opioid receptor: Isolation of a cDNA by expression cloning and pharmacological characterization. Proc. Natl. Acad. Sci. USA 1992, 89, 12048–12052. [Google Scholar] [CrossRef] [PubMed]
- Minami, M.; Toya, T.; Katao, Y.; Maekawa, K.; Nakamura, S.; Onogi, T.; Kaneko, S.; Satoh, M. Cloning and expression of a cDNA for the rat kappa-opioid receptor. FEBS Lett. 1993, 329, 291–295. [Google Scholar] [CrossRef]
- Chen, Y.; Mestek, A.; Liu, J.; Hurley, J.A.; Yu, L. Molecular cloning and functional expression of a mu-opioid receptor from rat brain. Mol. Pharmacol. 1993, 44, 8–12. [Google Scholar] [CrossRef]
- Bagley, J.R.; Kudzma, L.V.; Lalinde, N.L.; Colapret, J.A.; Huang, B.S.; Lin, B.S.; Jerussi, T.P.; Benvenga, M.J.; Doorley, B.M.; Ossipov, M.H. Evolution of the 4-anilidopiperidine class of opioid analgesics. Med. Res. Rev. 1991, 11, 403–436. [Google Scholar] [CrossRef] [PubMed]
- Zylbergold, P.; Ramakrishnan, N.; Hebert, T. The role of G proteins in assembly and function of Kir3 inwardly rectifying potassium channels. Channels 2010, 4, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Rusin, K.I.; Giovannucci, D.R.; Stuenkel, E.L.; Moises, H.C. Kappa-opioid receptor activation modulates Ca2+ currents and secretion in isolated neuroendocrine nerve terminals. J. Neurosci. 1997, 17, 6565–6574. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Loh, H.H.; Law, P.Y. A novel noncanonical signaling pathway for the μ-opioid receptor. Mol. Pharmacol. 2013, 84, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, R.J.; Shenoy, S.K. Transduction of receptor signals by beta-arrestins. Science 2005, 308, 512–517. [Google Scholar] [CrossRef] [PubMed]
- Luttrell, L.M.; Daaka, Y.; Lefkowitz, R.J. Regulation of tyrosine kinase cascades by G-protein-coupled receptors. Curr. Opin. Cell Biol. 1999, 11, 177–183. [Google Scholar] [CrossRef]
- Urban, J.D.; Clarke, W.P.; von Zastrow, M.; Nichols, D.E.; Kobilka, B.; Weinstein, H.; Javitch, J.A.; Roth, B.L.; Christopoulos, A.; Sexton, P.M.; et al. Functional selectivity and classical concepts of quantitative pharmacology. J. Pharmacol. Exp. Ther. 2007, 320, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zeng, Y.; Zhang, X.; Chu, J.; Loh, H.H.; Law, P.Y. mu-opioid receptor agonists differentially regulate the expression of miR-190 and NeuroD. Mol. Pharmacol. 2010, 77, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Loh, H.H.; Law, P.Y. Beta-arrestin-dependent mu-opioid receptor-activated extracellular signal-regulated kinases (ERKs) translocate to nucleus in contrast to G protein-dependent ERK activation. Mol. Pharmacol. 2008, 73, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Chu, J.; Qiu, Y.; Loh, H.H.; Law, P.Y. Agonist-selective signaling is determined by the receptor location within the membrane domains. Proc. Natl. Acad. Sci. USA 2008, 105, 9421–9426. [Google Scholar] [CrossRef] [PubMed]
- Takenouchi, O.; Yoshimura, H.; Ozawa, T. Unique roles of β-arrestin in GPCR trafficking revealed by photoinducible dimerizers. Sci. Rep. 2018, 8, 677. [Google Scholar] [CrossRef] [PubMed]
- DeWire, S.M.; Ahn, S.; Lefkowitz, R.J.; Shenoy, S.K. Beta-arrestins and cell signaling. Annu. Rev. Physiol. 2007, 69, 483–510. [Google Scholar] [CrossRef] [PubMed]
- Violin, J.D.; Lefkowitz, R.J. Beta-arrestin-biased ligands at seven-transmembrane receptors. Trends Pharmacol. Sci. 2007, 28, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Gesty-Palmer, D.; Chen, M.; Reiter, E.; Ahn, S.; Nelson, C.D.; Wang, S.; Eckhardt, A.E.; Cowan, C.L.; Spurney, R.F.; Luttrell, L.M.; et al. Distinct beta-arrestin- and G protein-dependent pathways for parathyroid hormone receptor-stimulated ERK1/2 activation. J. Biol. Chem. 2006, 281, 10856–10864. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Chu, J.; Zhang, Y.; Loh, H.H.; Law, P.Y. Modulating micro-opioid receptor phosphorylation switches agonist-dependent signaling as reflected in PKCepsilon activation and dendritic spine stability. J. Biol. Chem. 2011, 286, 12724–12733. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated protein kinase: The guardian of cardiac energy status. J. Clin. Investig. 2004, 114, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Gross, G.J.; Auchampach, J.A. Role of ATP dependent potassium channels in myocardial ischaemia. Cardiovasc. Res. 1992, 26, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Turer, A.T.; Hill, J.A. Pathogenesis of myocardial ischemia-reperfusion injury and rationale for therapy. Am. J. Cardiol. 2010, 106, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Reimer, K.A.; Lowe, J.E.; Rasmussen, M.M.; Jennings, R.B. The wavefront phenomenon of ischemic cell death. 1. Myocardial infarct size vs duration of coronary occlusion in dogs. Circulation 1977, 56, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Chiong, M.; Wang, Z.V.; Pedrozo, Z.; Cao, D.J.; Troncoso, R.; Ibacache, M.; Criollo, A.; Nemchenko, A.; Hill, J.A.; Lavandero, S. Cardiomyocyte death: Mechanisms and translational implications. Cell Death Dis. 2011, 2, e244. [Google Scholar] [CrossRef] [PubMed]
- Yellon, D.M.; Hausenloy, D.J. Myocardial reperfusion injury. N. Engl. J. Med. 2007, 357, 1121–1135. [Google Scholar] [CrossRef] [PubMed]
- Braunwald, E.; Kloner, R.A. Myocardial reperfusion: A double-edged sword? J. Clin. Investig. 1985, 76, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Murry, C.E.; Jennings, R.B.; Reimer, K.A. Preconditioning with ischemia: A delay of lethal cell injury in ischemic myocardium. Circulation 1986, 74, 1124–1136. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.N.; Patel, H.H.; Gross, G.J. Delta-opioid receptor activation mimics ischemic preconditioning in the canine heart. J. Cardiovasc. Pharmacol. 2003, 42, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Mayfield, K.P.; D’Alecy, L.G. Role of endogenous opioid peptides in the acute adaptation to hypoxia. Brain Res. 1992, 582, 226–231. [Google Scholar] [CrossRef]
- Korthuis, R.J. Filling GAPs in the understanding of cardioprotection induced by GPCR activation: RGS proteins modulate ischaemic injury. Cardiovasc. Res. 2011, 91, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.R.; Hsu, A.K.; Gross, G.J. Acute methadone treatment reduces myocardial infarct size via the delta-opioid receptor in rats during reperfusion. Anesth. Analg. 2009, 109, 1395–1402. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Sun, Y.; Li, J.; Xing, W.; Zhang, S.; Gu, X.; Feng, N.; Zhao, L.; Fan, R.; Wang, Y.; et al. Quaternary ammonium salt of U50488H, a new κ-opioid receptor agonist, protects rat heart against ischemia/reperfusion injury. Eur. J. Pharmacol. 2014, 737, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.E.; Rose, E.; Yao, Z.; Gross, G.J. Evidence for involvement of opioid receptors in ischemic preconditioning in rat hearts. Am. J. Physiol. 1995, 268, H2157–H2161. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Cohen, M.V.; Downey, J.M. Opioid receptor contributes to ischemic preconditioning through protein kinase C activation in rabbits. Mol. Cell. Biochem. 1998, 186, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Fryer, R.M.; Wang, Y.; Hsu, A.K.; Gross, G.J. Essential activation of PKC-delta in opioid-initiated cardioprotection. Am. J. Physiol. Heart Circ. Physiol. 2001, 280, H1346–H1353. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Liu, F.; Ji, F.; Liang, J.; Liu, L.; Wu, Q.; Wang, T. Effect of c-Jun N-terminal kinase (JNK)/p38 mitogen-activated protein kinase (p38 MAPK) in morphine-induced tau protein hyperphosphorylation. Behav. Brain Res. 2013, 237, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Fryer, R.M.; Hsu, A.K.; Gross, G.J. ERK and p38 MAP kinase activation are components of opioid-induced delayed cardioprotection. Basic. Res. Cardiol. 2001, 96, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.R.; Hsu, A.K.; Gross, G.J. The JAK/STAT pathway is essential for opioid-induced cardioprotection: JAK2 as a mediator of STAT3, Akt, and GSK-3 beta. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H827–H834. [Google Scholar] [CrossRef] [PubMed]
- Gross, E.R.; Hsu, A.K.; Gross, G.J. Opioid-induced cardioprotection occurs via glycogen synthase kinase beta inhibition during reperfusion in intact rat hearts. Circ. Res. 2004, 94, 960–966. [Google Scholar] [CrossRef] [PubMed]
- Kodani, E.; Xuan, Y.T.; Shinmura, K.; Takano, H.; Tang, X.L.; Bolli, R. Delta-opioid receptor-induced late preconditioning is mediated by cyclooxygenase-2 in conscious rabbits. Am. J. Physiol. Heart Circ. Physiol. 2002, 283, H1943–H1957. [Google Scholar] [CrossRef] [PubMed]
- Peart, J.N.; Gross, G.J. Adenosine and opioid receptor-mediated cardioprotection in the rat: Evidence for cross-talk between receptors. Am. J. Physiol. Heart Circ. Physiol. 2003, 285, H81–H89. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.V.; Yang, X.M.; Liu, G.S.; Heusch, G.; Downey, J.M. Acetylcholine, bradykinin, opioids, and phenylephrine, but not adenosine, trigger preconditioning by generating free radicals and opening mitochondrial K(ATP) channels. Circ. Res. 2001, 89, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Fagbemi, O.; Kane, K.A.; Lepran, I. Anti-arrhythmic actions of meptazinol, a partial agonist at opiate receptors, in acute myocardial ischemia. Br. J. Pharmacol. 1983, 78, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Saini, V.; Carr, D.B.; Verrier, R.L. Comparative effects of the opioids fentanyl and buprenorphine on ventricular vulnerability during acute coronary artery occlusion. Cardiovasc. Res. 1989, 23, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Hess, L.; Vrana, M.; Vranova, Z.; Fejfar, Z. The antifibrillatory effect of fentanyl, sufentanil and carfentanil in acute phase of local myocardial ischemia in the dog. Acta Cardiol. 1989, 44, 303–311. [Google Scholar] [PubMed]
- Maslov, L.N.; Oeltgen, P.R.; Lishmanov, Y.B.; Brown, S.A.; Barzakh, E.I.; Krylatov, A.V.; Pei, J.M. Activation of peripheral delta opioid receptors increases cardiac tolerance to arrhythmogenic effect of ischemia/reperfusion. Acad. Emerg. Med. 2014, 21, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Wittert, G.; Hope, P.; Pyle, D. Tissue distribution of opioid receptor gene expression in the rat. Biochem. Biophys. Res. Commun. 1996, 218, 877–881. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, L.O.; Bergh, N.; Li, L.; Bissessar, E.; Bobrova, I.; Gross, G.J.; Akyürek, L.M.; Grip, L. Dose-dependent cardioprotection of enkephalin analogue Eribis peptide 94 and cardiac expression of opioid receptors in a porcine model of ischaemia and reperfusion. Eur. J. Pharmacol. 2012, 674, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Theisen, M.M.; Schlottmann, S.; August, C.; Herzog, C.; Theilmeier, G.; Maas, M.; Blumenstiel, J.M.; Weber, T.P.; Van Aken, H.K.; Kaerlein, K.T. Detection and distribution of opioid peptide receptors in porcine myocardial tissue. Pharmacol. Res. 2014, 84, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Head, B.P.; Patel, H.H.; Roth, D.M.; Lai, N.C.; Niesman, I.R.; Farquhar, M.G.; Insel, P.A. G-protein-coupled receptor signaling components localize in both sarcolemmal and intracellular caveolin-3-associated microdomains in adult cardiac myocytes. J. Biol. Chem. 2005, 280, 31036–31044. [Google Scholar] [CrossRef] [PubMed]
- He, S.F.; Jin, S.Y.; Yang, W.; Pan, Y.L.; Huang, J.; Zhang, S.J.; Zhang, L.; Zhang, Y. Cardiac μ-opioid receptor contributes to opioid-induced cardioprotection in chronic heart failure. Br. J. Anaesth. 2018, 121, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Saleembhasha, A.; Mishra, S. Novel molecules lncRNAs, tRFs and circRNAs deciphered from next-generation sequencing/RNA sequencing: Computational databases and tools. Brief. Funct. Genom. 2018, 17, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef] [PubMed]
- DiStefano, J.K. The emerging role of long noncoding RNAs in human disease. Methods Mol. Biol. 2018, 1706, 91–110. [Google Scholar] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Kim, D.; Chang, H.R.; Baek, D. Rules for functional microRNA targeting. BMB Rep. 2017, 50, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Xu, Y.; Xie, X.; Wang, T.; Ko, J.H.; Zhou, T. The role of RNA structure at 5’ untranslated region in microRNA-mediated gene regulation. RNA 2014, 20, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Syed, A.P.; Bilen, B.; Zavolan, M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Res. 2013, 23, 604–615. [Google Scholar] [CrossRef] [PubMed]
- Lytle, J.R.; Yario, T.A.; Steitz, J.A. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc. Natl. Acad. Sci. USA 2007, 104, 9667–9672. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Schirle, N.T.; Sheu-Gruttadauria, J.; MacRae, I.J. Structural basis for microRNA targeting. Science 2014, 31, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Hutvágner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar] [CrossRef]
- Goff, L.A.; Rinn, J.L. Linking RNA biology to lncRNAs. Genome Res. 2015, 25, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim. Biophys. Acta 2016, 1859, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.E.; Alvarez-Dominguez, J.R.; Kline, N.; Huynh, N.J.; Geisler, S.; Hu, W.; Coller, J.; Baker, K.E. Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep. 2014, 7, 1858–1866. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Kutter, C.; Watt, S.; Stefflova, K.; Wilson, M.D.; Goncalves, A.; Ponting, C.P.; Odom, D.T.; Marques, A.C. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.; Chang, H.Y. Uncovering the role of genomic “dark matter” in human disease. J. Clin. Investig. 2012, 122, 1589–1595. [Google Scholar] [CrossRef] [PubMed]
- Barbierato, M.; Zusso, M.; Skaper, S.D.; Giusti, P. MicroRNAs: Emerging role in the endogenous μ opioid system. CNS Neurol. Disord. Drug. Targets 2015, 14, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Dave, R.S.; Khalili, K. Morphine treatment of human monocyte-derived macrophages induces differential miRNA and protein expression: Impact on inflammation and oxidative stress in the central nervous system. J. Cell Biochem. 2010, 110, 834–845. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, C.; Kirkmire, C.M.; Wang, Z.J. Regulation of opioid tolerance by let-7 family microRNA targeting the mu opioid receptor. J. Neurosci. 2010, 30, 10251–10258. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, L.; Law, P.Y.; Wei, L.N.; Loh, H.H. Long-term morphine treatment decreases the association of mu-opioid receptor (MOR1) mRNA with polysomes through miRNA23b. Mol. Pharmacol. 2009, 75, 744–750. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S.; Bhattacharyya, S.N.; Artus, C.G.; Zoller, T.; Cougot, N.; Basyuk, E.; Bertrand, E.; Filipowicz, W. Inhibition of translational initiation by let-7 MicroRNA in human cells. Science 2005, 309, 1573–1576. [Google Scholar] [CrossRef] [PubMed]
- Wanet, A.; Tacheny, A.; Arnould, T.; Renard, P. miR-212/132 expression and functions: Within and beyond the neuronal compartment. Nucleic Acids Res. 2012, 40, 4742–4753. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Gao, Y.; Gong, S.; Guo, S.; Hisamitsu, T.; Jiang, X. Regulation of μ-opioid type 1 receptors by microRNA134 in dorsal root ganglion neurons following peripheral inflammation. Eur. J. Pain 2013, 17, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, Y.; Li, X.; Liang, X.; Wang, L.; Song, J.; Zhang, X.; Zhang, C.; Niu, J.; Zhang, P.; et al. microRNA-874 suppresses tumor proliferation and metastasis in hepatocellular carcinoma by targeting the DOR/EGFR/ERK pathway. Cell Death Dis. 2018, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Li, H.; Jiang, W.; Zhang, C.; McNutt, M.A.; Li, G. Simian immunodeficiency virus impacts microRNA-16 mediated post-transcriptional regulation of mu opioid receptor in CEM × 174 cells. J. Cell. Biochem. 2016, 117, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Ide, S.; Han, W.; Kasai, S.; Hata, H.; Sora, I.; Ikeda, K. Characterization of the 3’ untranslated region of the human mu-opioid receptor (MOR-1) mRNA. Gene 2005, 364, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Kasai, S.; Hata, H.; Takahashi, T.; Takamatsu, Y.; Yamamoto, H.; Uhl, G.R.; Sora, I.; Ikeda, K. Intracisternal A-particle element in the 3’ noncoding region of the mu-opioid receptor gene in CXBK mice: A new genetic mechanism underlying differences in opioid sensitivity. Pharm. Genom. 2006, 16, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Regan, P.M.; Sariyer, I.K.; Langford, T.D.; Datta, P.K.; Khalili, K. Morphine-induced MOR-1X and ASF/SF2 expressions are independent of transcriptional regulation: Implications for MOR-1X signaling. J. Cell. Physiol. 2016, 231, 1542–1553. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, M.; Elliott, K.; Hynansky, A.; Inturrisi, C.E. CNS levels of mu opioid receptor (MOR-1) mRNA during chronic treatment with morphine or naltrexone. Brain Res. Bull. 1995, 38, 135–141. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016, 7, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Chafin, C.B.; Regna, N.L.; Caudell, D.L.; Reilly, C.M. MicroRNA-let-7a promotes E2F-mediated cell proliferation and NFκB activation in vitro. Cell. Mol. Immunol. 2014, 11, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Büssing, I.; Slack, F.J.; Grosshans, H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Klee, W.A.; Streaty, R.A. Narcotic receptor sites in morphine-dependent rats. Nature 1974, 248, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Holaday, J.W.; Hitzemann, R.J.; Curell, J.; Tortella, F.C.; Belenky, G.L. Repeated electroconvulsive shock or chronic morphine treatment increases the number of 3H-D-Ala2, D-Leu5-enkephalin binding sites in rat brain membranes. Life Sci. 1982, 31, 2359–2362. [Google Scholar] [CrossRef]
- Lewis, J.W.; Lewis, M.E.; Loomus, D.J.; Akil, H. Acute systemic administration of morphine selectively increases mu opioid receptor binding in the rat brain. Neuropeptides 1984, 5, 117–120. [Google Scholar] [CrossRef]
- Rothman, R.B.; Danks, J.A.; Jacobson, A.E.; Burke, T.R.; Rice, K.C.; Tortella, F.C.; Holaday, J.W. Morphine tolerance increases mu-noncompetitive delta binding sites. Eur. J. Pharmacol. 1986, 124, 113–119. [Google Scholar] [CrossRef]
- Koch, T.; Höllt, V. Role of receptor internalization in opioid tolerance and dependence. Pharmacol. Ther. 2008, 117, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Law, P.Y.; Wei, L.N.; Loh, H.H. Post-transcriptional regulation of mouse mu opioid receptor (MOR1) via its 3’ untranslated region: A role for microRNA23b. FASEB J. 2008, 22, 4085–4095. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Concejo, A.; Jimenez-Gonzalez, A.; Rodríguez, R.E. μ opioid receptor expression after morphine administration is regulated by miR-212/132 cluster. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.E.; Lioy, D.T.; Ma, L.; Impey, S.; Mandel, G.; Goodman, R.H. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 2007, 10, 1513–1514. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Hwang, C.K.; Zheng, H.; Wagley, Y.; Lin, H.Y.; Kim, D.K.; Law, P.Y.; Loh, H.H.; Wei, L.N. MicroRNA 339 down-regulates μ-opioid receptor at the post-transcriptional level in response to opioid treatment. FASEB J. 2013, 27, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, J.; Xu, M.; Pasternak, G.W.; Pan, Y.X. Morphine regulates expression of μ-opioid receptor MOR-1A, an intron-retention carboxyl terminal splice variant of the μ-opioid receptor (OPRM1) gene via miR-103/miR-107. Mol. Pharmacol. 2014, 85, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Bare, L.A.; Mansson, E.; Yang, D. Expression of two variants of the human mu opioid receptor mRNA in SK-N-SH cells and human brain. FEBS Lett. 1994, 354, 213–216. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Surapureddi, S.; Coulter, S.; Ferguson, S.S.; Goldstein, J.A. Human CYP2C8 is post-transcriptionally regulated by microRNAs 103 and 107 in human liver. Mol. Pharmacol. 2012, 82, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Polster, B.J.; Westaway, S.K.; Nguyen, T.M.; Yoon, M.Y.; Hayflick, S.J. Discordant expression of miR-103/7 and pantothenate kinase host genes in mouse. Mol. Genet. Metab. 2010, 101, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Wilfred, B.R.; Wang, W.X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, W.; Zhong, T.; Song, Z.; Zou, Y.; Ding, Z.; Guo, Q.; Dong, X.; Zou, W. miR-365 targets β-arrestin 2 to reverse morphine tolerance in rats. Sci. Rep. 2016, 6, 38285. [Google Scholar] [CrossRef] [PubMed]
- Bohn, L.M.; Gainetdinov, R.R.; Lin, F.T.; Lefkowitz, R.J.; Caron, M.G. Mu-opioid receptor desensitization by beta-arrestin-2 determines morphine tolerance but not dependence. Nature 2000, 408, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Xue, L.; Shao, N.; Deng, D.; Kang, X.; Chao, D.; Xu, Y.; Wang, R.; Yang, Y.; Xia, Y. δ-Opioid receptor activation and microRNA expression in the rat heart under prolonged hypoxia. Cell Physiol. Biochem. 2016, 39, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Cao, J.; Wang, L.; Zhou, Q.; Li, Y.; Shen, C.; Zhang, X.; Wang, C. MicroRNA 107 partly inhibits endothelial progenitor cells differentiation via HIF-1β. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.C.; Khanna, S.; Roy, S.; Sen, C.K. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J. Biol. Chem. 2011, 286, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, R.; Zhang, C.; Bian, H.J.; Wang, F.; Xiao, J.; Liu, S.W.; Yi, W.; Zhang, M.X.; Wang, S.X.; et al. MicroRNA-7a/b protects against cardiac myocyte injury in ischemia/reperfusion by targeting poly (ADP-ribose) polymerase. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- He, S.F.; Zhu, H.J.; Han, Z.Y.; Wu, H.; Jin, S.Y.; Irwin, M.G.; Zhang, Y. MicroRNA-133b-5p is involved in cardioprotection of morphine preconditioning in rat cardiomyocytes by targeting fas. Can. J. Cardiol. 2016, 32, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Gu, H.; Xu, W.; Zhou, X. Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis and improves left ventricular function in diabetic rats. Int. J. Cardiol. 2016, 203, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Han, Z.Y.; He, S.F.; Jin, S.Y.; Xu, S.J.; Fang, X.D.; Zhang, Y. Specific MicroRNAs comparisons in hypoxia and morphine preconditioning against hypoxia-reoxgenation injury with and without heart failure. Life Sci. 2017, 170, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xia, L.; Chen, M.; Lin, C.; Wu, H.; Zhang, Y.; Pan, S.; Li, X. miR-133b, a particular member of myomiRs, coming into playing its unique pathological role in human cancer. Oncotarget 2017, 8, 50193–50208. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Miura, T.; Tanno, M.; Sakamoto, J.; Kuno, A.; Genda, S.; Matsumoto, T.; Ichikawa, Y.; Shimamoto, K. Interruption of signal transduction between G protein and PKC-epsilon underlies the impaired myocardial response to ischemic preconditioning in postinfarct remodeled hearts. Mol. Cell. Biochem. 2003, 247, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, S.Y.; Kwak, Y.L.; Hwang, K.C.; Shim, Y.H. Hyperglycemia attenuates myocardial preconditioning of remifentanil. J. Surg. Res. 2012, 174, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Andersen, A.; Povlsen, J.A.; Bøtker, H.E.; Nielsen-Kudsk, J.E. Right ventricular hypertrophy and failure abolish cardioprotection by ischaemic pre-conditioning. Eur. J. Heart Fail. 2013, 15, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Simon, F.M.; Zhang, X.X.; Loh, H.H.; Law, P.Y.; Rodriguez, R.E. Morphine regulates dopaminergic neuron differentiation via miR-133b. Mol. Pharmacol. 2010, 78, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.C.; Zheng, J.Y.; Tang, L.J.; Huang, B.S.; Li, K.; Tao, Y.; Yu, W.; Zhu, R.L.; Li, S.; Li, L.X. miR-133b promotes neurite outgrowth by targeting RhoA expression. Cell. Physiol. Biochem. 2015, 35, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.M.; Gibbs, K.M.; Davila, J.; Campbell, N.; Sung, S.; Todorova, T.I.; Otsuka, S.; Sabaawy, H.E.; Hart, R.P.; Schachner, M. MicroRNA miR-133b is essential for functional recovery after spinal cord injury in adult zebrafish. Eur. J. Neurosci. 2011, 33, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wu, Z.; Tian, L.; Li, D.; Wang, X.; He, Y.; Jin, W.; Li, M.; Zhu, Q.; Shang, T.; et al. Long noncoding RNA AK12348 is involved in the regulation of myocardial ischaemia-reperfusion injury by targeting PARP and caspase-3. Heart Lung Circ. 2018, 27, e51–e58. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.B.; Katwadi, K.; Kwek, X.Y.; Ismail, N.I.; Chinda, K.; Ong, S.G.; Hausenloy, D.J. Non-coding RNAs as therapeutic targets for preventing myocardial ischemia-reperfusion injury. Expert Opin. Ther. Targets 2018, 22, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Q.; Yuan, Z.; Chen, A.; Liu, Z.; Wang, Z.; Li, H. LncRNA-MALAT1 promotes CPC proliferation and migration in hypoxia by up-regulation of JMJD6 via sponging miR-125. Biochem. Biophys. Res. Commun. 2018, 499, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.Y.; Dong, B.; Tang, L.; Zhou, S.H. LncRNA MALAT1 sponges miR-133 to promote NLRP3 inflammasome expression in ischemia-reperfusion injured heart. Int. J. Cardiol. 2018, 254. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.H.; Hao, W.; Meng, Q.T.; Du, X.B.; Lei, S.Q.; Xia, Z.Y. Long non-coding RNA MALAT1 functions as a mediator in cardioprotective effects of fentanyl in myocardial ischemia-reperfusion injury. Cell. Biol. Int. 2017, 41, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Li, J.; Xu, T.; Zhou, D.D.; Zhang, L.; Wang, X. MicroRNA-145 induces apoptosis of glioma cells by targeting BNIP3 and Notch signaling. Oncotarget 2017, 8, 61510–61527. [Google Scholar] [CrossRef] [PubMed]
- Viereck, J.; Thum, T. Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 2017, 120, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Huang, W. MicroRNAs: Biomarkers, diagnostics, and therapeutics. Methods Mol. Biol. 2017, 1617, 57–67. [Google Scholar] [PubMed]
- E, S.; Costa, M.C.; Kurc, S.; Drożdż, A.; Cortez-Dias, N.; Enguita, F.J. The circulating non-coding RNA landscape for biomarker research: Lessons and prospects from cardiovascular diseases. Acta Pharmacol. Sin. 2018, 39, 1085–1099. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Willeit, P.; Tilling, L.; Drozdov, I.; Prokopi, M.; Renard, J.M.; Mayr, A.; Weger, S.; Schett, G.; Shah, A.; et al. Prospective study on circulating microRNAs and risk of myocardial infarction. J. Am. Coll. Cardiol. 2012, 60, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.K.; Zhu, J.Q.; Zhang, J.T.; Li, Q.; Li, Y.; He, J.; Qin, Y.W.; Jing, Q. Circulating microRNA: A novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J. 2010, 31, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Devaux, Y.; Vausort, M.; McCann, G.P.; Kelly, D.; Collignon, O.; Ng, L.L.; Wagner, D.R.; Squire, I.B. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Feistritzer, H.J.; Klug, G.; Reinstadler, S.J.; Reindl, M.; Mayr, A.; Mair, J.; Metzler, B. Novel biomarkers predicting cardiac function after acute myocardial infarction. Br. Med. Bull. 2016, 119, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Choong, O.K.; Lee, D.S.; Chen, C.Y.; Hsieh, P.C.H. The roles of non-coding RNAs in cardiac regenerative medicine. Noncoding RNA Res. 2017, 2, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J. Therapeutic potentials of noncoding RNAs: Targeted delivery of ncRNAs in cancer cells. Adv. Exp. Med. Biol. 2016, 927, 429–458. [Google Scholar] [PubMed]
- Somogyi, A.A.; Barratt, D.T.; Coller, J.K. Pharmacogenetics of opioids. Clin. Pharmacol. Ther. 2007, 81, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Owusu Obeng, A.; Hamadeh, I.; Smith, M. Review of opioid pharmacogenetics and considerations for pain management. Pharmacotherapy 2017, 37, 1105–1121. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).