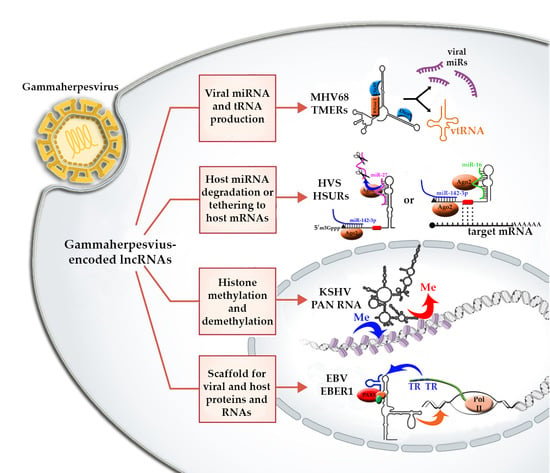
The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis
Abstract
:1. Introduction: Defining Long Non-Coding RNAs
2. The Role of lncRNAs in Gammaherpesviruses
3. Kaposi’s Sarcoma Herpesvirus (KSHV)-Encoded lncRNAs
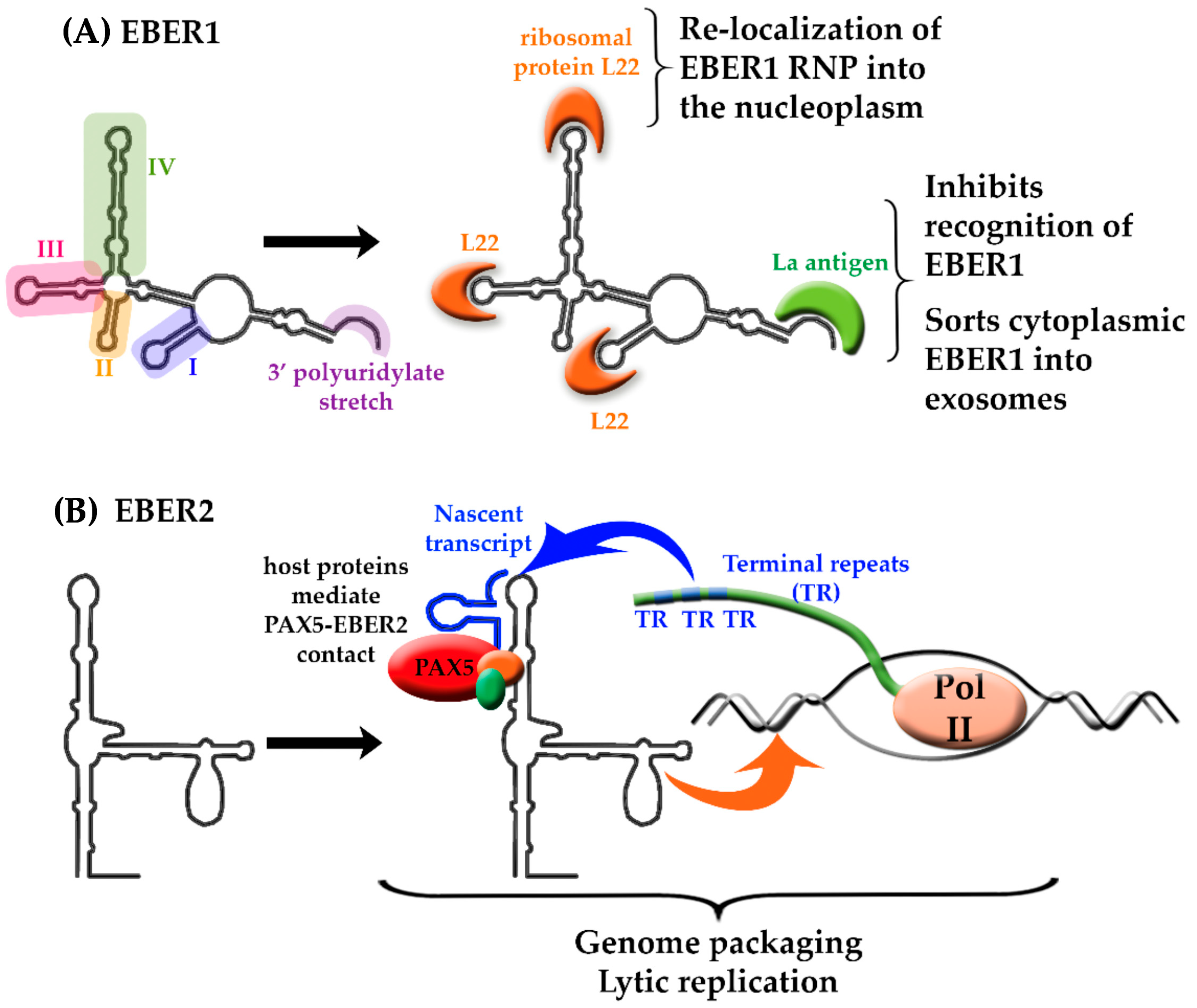
4. Epstein-Barr Virus-Encoded lncRNAs
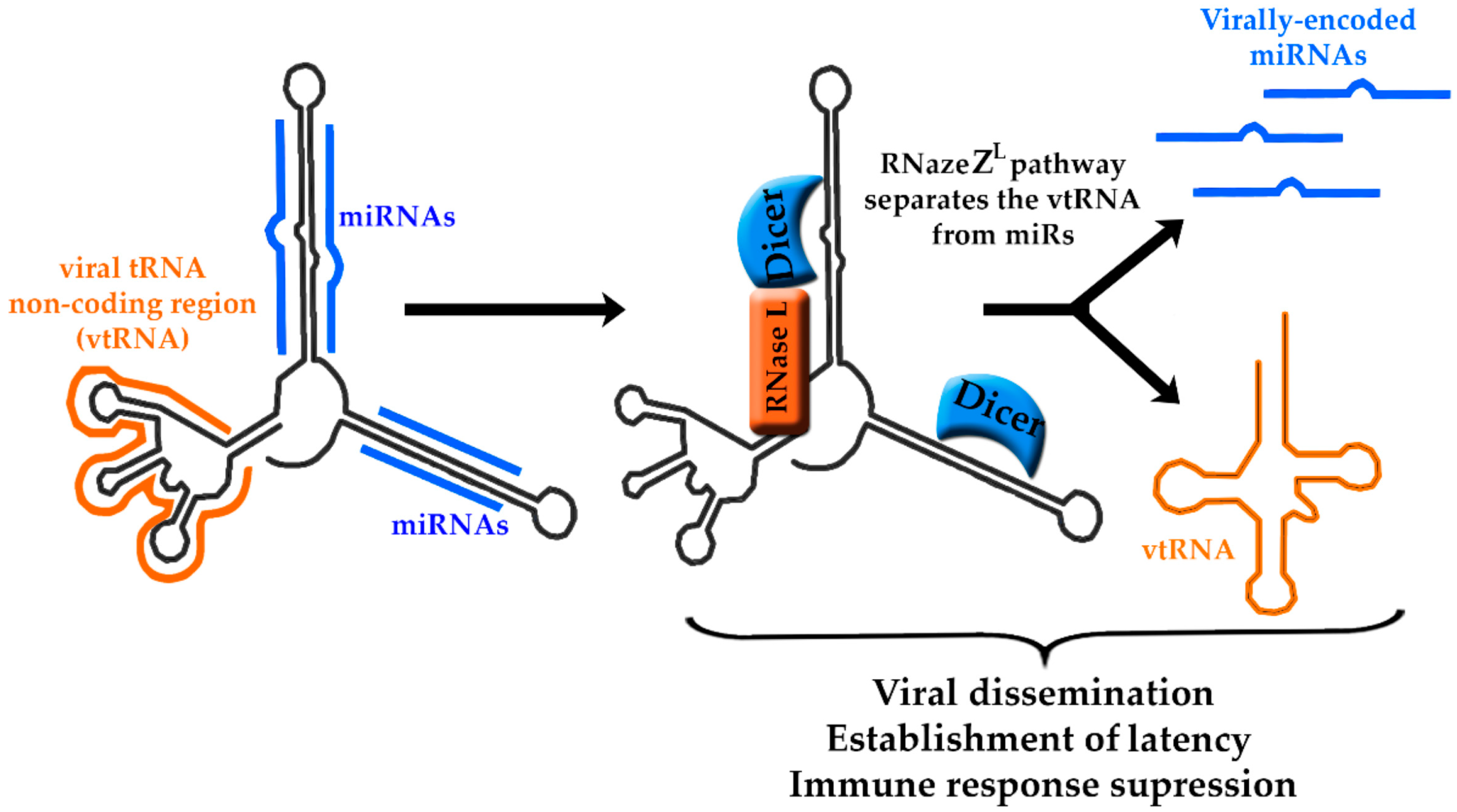
5. Murine Herpesvirus-68-Encoded lncRNAs
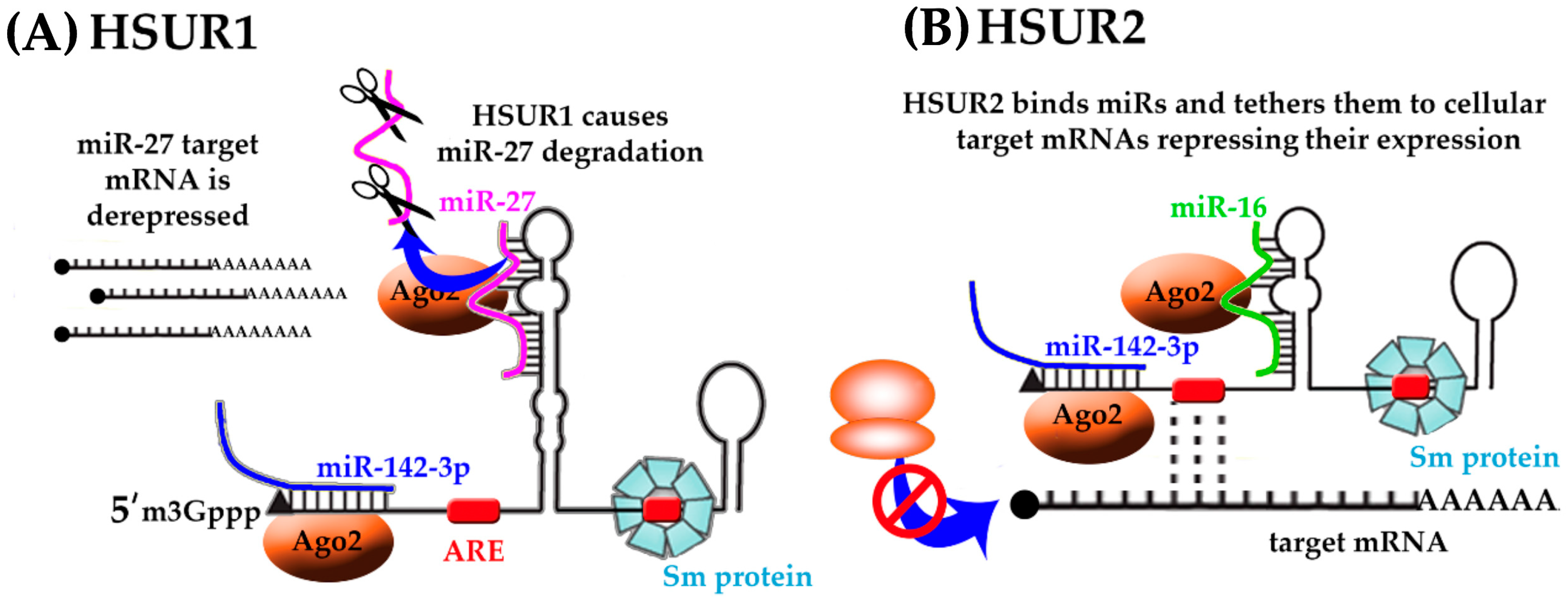
6. Herpesvirus Saimiri-Encoded lncRNAs
7. Gammaherpesvirus-Encoded lncRNAs as Therapeutic Targets
8. Gammaherpesvirus-Encoded lncRNAs: Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Uszczynska-Ratajczak, B.; Lagarde, J.; Frankish, A.; Guigó, R.; Johnson, R. Towards a complete map of the human long non-coding RNA transcriptome. Nat. Rev. Genet. 2018, 19, 535–548. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The diversity of long noncoding RNAs and their generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Galindo, M.I.; Pueyo, J.I.; Fouix, S.; Bishop, S.A.; Couso, J.P. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007, 5, 1052–1062. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.; Weisburd, B.; Stern-Ginossar, N.; Mercier, A.; Madrid, A.S.; Bellare, P.; Holdorf, M.; Weissman, J.S.; Ganem, D. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Dinger, M.E.; Gascoigne, D.K.; Mattick, J.S. The evolution of RNAs with multiple functions. Biochimie 2011, 93, 2013–2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, S.; Moss, W.; O’Grady, T.; Concha, M.; Strong, M.J.; Wang, X.; Yu, Y.; Baddoo, M.; Zhang, K.; Fewell, C. New noncoding lytic transcripts derived from the Epstein-Barr virus latency origin of replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J. Virol. 2015, 89, 7120–7132. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Kim, K.Y.; Chang, P.; Huerta, S.; Shevchenko, B.; Wang, D.; Izumiya, C.; Kung, H.; Izumiya, Y. A lytic viral long noncoding RNA modulates the function of a latent protein. J. Virol. 2014, 88, 1843–1848. [Google Scholar] [CrossRef] [PubMed]
- Krug, L.T. Complexities of gammaherpesvirus transcription revealed by microarrays and RNAseq. Curr. Opin. Virol. 2013, 3, 276–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rennekamp, A.J.; Lieberman, P.M. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA–DNA hybrid molecule at OriLyt. J. Virol. 2011, 85, 2837–2850. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Novikova, I.V.; Hennelly, S.P.; Sanbonmatsu, K.Y. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012, 40, 5034–5051. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bulkley, D.; Wang, J.; Valenstein, M.L.; Yario, T.A.; Steitz, T.A.; Steitz, J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014, 21, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Fields, B.N.; Knipe, D.M.; Howley, P.M. Fields Virology; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007. [Google Scholar]
- Campbell, M.; Kung, H.-J.; Izumiya, Y. Long non-coding RNA and epigenetic gene regulation of KSHV. Viruses 2014, 6, 4165–4177. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, P.; Dabral, P.; Gupta, N.; Sarkar, R.; Verma, S.C. KSHV genome replication and maintenance. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Tsurumi, T.; Fujita, M.; Kudoh, A. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 2005, 15, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Aneja, K.K.; Yuan, Y. Reactivation and lytic replication of Kaposi’s sarcoma-associated herpesvirus: An update. Front. Microbiol. 2017, 8, 613. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.; Sattler, C.; Adler, H. Herpesviruses and their host cells: A successful liaison. Trends Microbiol. 2017, 25, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Baglio, S.R.; van Eijndhoven, M.A.J.; Koppers-Lalic, D.; Berenguer, J.; Lougheed, S.M.; Gibbs, S.; Léveillé, N.; Rinkel, R.N.P.M.; Hopmans, E.S.; Swaminathan, S.; et al. Sensing of latent EBV infection through exosomal transfer of 5′pppRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E587–E596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossetto, C.C.; Pari, G.S. Kaposi’s sarcoma-associated herpesvirus noncoding polyadenylated nuclear RNA interacts with virus- and host cell-encoded proteins and suppresses expression of genes involved in immune modulation. J. Virol. 2011, 85, 13290–13297. [Google Scholar] [CrossRef] [PubMed]
- Chandriani, S.; Xu, Y.; Ganem, D. The lytic transcriptome of Kaposi’s sarcoma-associated herpesvirus reveals extensive transcription of noncoding regions, including regions antisense to important genes. J. Virol. 2010, 84, 7934–7942. [Google Scholar] [CrossRef] [PubMed]
- Dresang, L.R.; Teuton, J.R.; Feng, H.; Jacobs, J.M.; Camp, D.G.; Purvine, S.O.; Gritsenko, M.A.; Li, Z.; Smith, R.D.; Sugden, B.; et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: Insights on the detection and discovery of viral genes. BMC Genom. 2011, 12, 625. [Google Scholar] [CrossRef] [PubMed]
- Schifano, J.M.; Corcoran, K.; Kelkar, H.; Dittmer, D.P. Expression of the antisense-to-latency transcript long noncoding RNA in Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2017, 91, e01698-16. [Google Scholar] [CrossRef] [PubMed]
- Majerciak, V.; Ni, T.; Yang, W.; Meng, B.; Zhu, J.; Zheng, Z.M. A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Lerner, M.R.; Andrews, N.C.; Miller, G.; Steitz, J.A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1981, 78, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Toczyski, D.P.; Matera, A.G.; Ward, D.C.; Steitz, J.A. The Epstein-Barr virus (EBV) small RNA EBER1 binds and relocalizes ribosomal protein L22 in EBV-infected human B lymphocytes. Proc. Natl. Acad. Sci. USA 1994, 91, 3463–3467. [Google Scholar] [CrossRef] [PubMed]
- Fok, V.; Mitton-Fry, R.M.; Grech, A.; Steitz, J.A. Multiple domains of EBER 1, an Epstein-Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA 2006, 12, 872–882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moss, W.N.; Lee, N.; Pimienta, G.; Steitz, J.A. RNA families in Epstein-Barr virus. RNA Biol. 2014, 11, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: More surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, H.; Nishikura, K. A new function for the RNA-editing enzyme ADAR1. Nat. Immunol. 2009, 10, 16–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, T.V.; Schwemmle, M.; Jeffrey, I.; Laing, K.; Mellor, H.; Proud, C.G.; Hilse, K.; Clemens, M.J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993, 21, 4483–4490. [Google Scholar] [CrossRef] [PubMed]
- Jeang, K.T.; Hayward, S.D. Organization of the Epstein-Barr virus DNA molecule. III. Location of the P3HR-1 deletion junction and characterization of the NotI repeat units that form part of the template for an abundant 12-O-tetradecanoylphorbol-13-acetate-induced mRNA transcript. J. Virol. 1983, 48, 135–148. [Google Scholar] [PubMed]
- Feldman, E.R.; Kara, M.; Coleman, C.B.; Grau, K.R.; Oko, L.M.; Krueger, B.J.; Renne, R.; van Dyk, L.F.; Tibbetts, S.A. Virus-encoded microRNAs facilitate gammaherpesvirus latency and pathogenesis in vivo. MBio 2014, 5, e00981-14. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.I.; Murthy, S.C.S.; Trimble, J.J.; Desrosiers, R.C.; Steitz, J.A. Four novel U RNAs are encoded by a herpesvirus. Cell 1988, 54, 599–607. [Google Scholar] [CrossRef]
- Myer, V.E.E.; Lee, S.I.I.; Steitz, J.A.A. Viral small nuclear ribonucleoproteins bind a protein implicated in messenger RNA destabilization. Proc. Natl. Acad. Sci. USA 1992, 89, 1296–1300. [Google Scholar] [CrossRef] [PubMed]
- Cook, H.L.; Mischo, H.E.; Steitz, J.A. The Herpesvirus saimiri small nuclear RNAs recruit AU-rich element-binding proteins but do not alter host au-rich element-containing mRNA levels in virally transformed T cells. Mol. Cell. Biol. 2004, 24, 4522–4533. [Google Scholar] [CrossRef] [PubMed]
- Cazalla, D.; Yario, T.; Steitz, J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science 2010, 328, 1563–1566. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, J.C.; Nicholas, J.; Biller, D.; Cameron, K.R.; Biesinger, B.; Newman, C.; Wittmann, S.; Craxton, M.A.; Coleman, H.; Fleckenstein, B. Primary structure of the Herpesvirus saimiri genome. J. Virol. 1992, 66, 5047–5058. [Google Scholar] [PubMed]
- Oksenhendler, E.; Boulanger, E.; Galicier, L.; Du, M.Q.; Dupin, N.; Diss, T.C.; Hamoudi, R.; Daniel, M.T.; Agbalika, F.; Boshoff, C.; et al. High incidence of Kaposi sarcoma-associated herpesvirus-related non-Hodgkin lymphoma in patients with HIV infection and multicentric Castleman disease. Blood 2002, 99, 2331–2336. [Google Scholar] [CrossRef] [PubMed]
- Dupin, N.; Diss, T.L.; Kellam, P.; Tulliez, M.; Du, M.Q.; Sicard, D.; Weiss, R.A.; Isaacson, P.G.; Boshoff, C. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 2000, 95, 1406–1412. [Google Scholar] [PubMed]
- Du, M.Q.; Liu, H.; Diss, T.C.; Ye, H.; Hamoudi, R.A.; Dupin, N.; Meignin, V.; Oksenhendler, E.; Boshoff, C.; Isaacson, P.G. Kaposi sarcoma-associated herpesvirus infects monotypic (IgMλ) but polyclonal naive B cells in Castleman disease and associated lymphoproliferative disorders. Blood 2001, 97, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ganem, D. Making sense of antisense: Seemingly noncoding RNAs antisense to the master regulator of Kaposi’s sarcoma-associated herpesvirus lytic replication do not regulate that transcript but serve as mRNAs encoding small peptides. J. Virol. 2010, 84, 5465–5475. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, J.; Grundhoff, A.; Ganem, D. RNAs in the virion of Kaposi’s sarcoma-associated herpesvirus. J. Virol. 2005, 79, 10138–10146. [Google Scholar] [CrossRef] [PubMed]
- Bruce, A.G.; Barcy, S.; DiMaio, T.; Gan, E.; Garrigues, H.J.; Lagunoff, M.; Rose, T.M. Quantitative analysis of the KSHV transcriptome following primary infection of blood and lymphatic endothelial cells. Pathogens 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Sztuba-Solinska, J.; Rausch, J.W.; Smith, R.; Miller, J.T.; Whitby, D.; le Grice, S.F.J. Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA: A structural scaffold for nuclear, cytoplasmic and viral proteins. Nucleic Acids Res. 2017, 45, 6805–6821. [Google Scholar] [CrossRef] [PubMed]
- Massimelli, M.J.; Kang, J.G.; Majerciak, V.; Le, S.Y.; Liewehr, D.J.; Steinberg, S.M.; Zheng, Z.M. Stability of a long noncoding viral RNA depends on a 9-nt core element at the RNA 5’ end to interact with viral ORF57 and cellular PABPC1. Int. J. Biol. Sci. 2011, 7, 1145–1160. [Google Scholar] [CrossRef] [PubMed]
- Conrad, N.K.; Steitz, J.A. A Kaposi’s sarcoma virus RNA element that increases the nuclear abundance of intronless transcripts. EMBO J. 2005, 24, 1831–1841. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Pari, G.S. PAN’s Labyrinth: Molecular biology of Kaposi’s sarcoma-associated herpesvirus (KSHV) PAN RNA, a multifunctional long noncoding RNA. Viruses 2014, 6, 4212–4226. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi’s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef] [PubMed]
- Borah, S.; Darricarrère, N.; Darnell, A.; Myoung, J.; Steitz, J.A. A viral nuclear noncoding RNA binds re-localized poly(A) binding protein and is required for late KSHV gene expression. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Forero, A.; McCormick, K.D.; Jenkins, F.J.; Sarkar, S.N. Downregulation of IRF4 induces lytic reactivation of KSHV in primary effusion lymphoma cells. Virology 2014, 458–459, 4–10. [Google Scholar] [CrossRef] [PubMed]
- McDowell, M.E.; Purushothaman, P.; Rossetto, C.C.; Pari, G.S.; Verma, S.C. Phosphorylation of Kaposi’s sarcoma-associated herpesvirus processivity factor ORF59 by a viral kinase modulates its ability to associate with RTA and oriLyt. J. Virol. 2013, 87, 8038–8052. [Google Scholar] [CrossRef] [PubMed]
- Henle, G.; Henle, W.; Diebel, K.W.; Smith, A.L.; van Dyk, L.F.; Efstathiou, S.; Davis, A.J.; Simas, J.P.; Bowden, R.J.; Campbell, M.; et al. Viral lncRNA: A regulatory molecule for controlling virus life cycle. J. Virol. 2016, 11, 5894–5904. [Google Scholar]
- Johansson, B.; Klein, G.; Henle, W.; Henle, G. Epstein-Barr virus (EBV)-associated antibody patterns in malignant lymphoma and leukemia. I. Hodgkin’s disease. Int. J. Cancer 1970, 6, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Young, L.S.; Dawson, C.W. Epstein-Barr virus and nasopharyngeal carcinoma. Chin. J. Cancer 2014, 33, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Fernandez, S.G.; Somberg, J.J.; Keck, K.M.; Miranda, J.J.L. Epstein–Barr virus latency type and spontaneous reactivation predict lytic induction levels. Biochem. Biophys. Res. Commun. 2016, 474, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Maruo, S.; Yajima, M.; Kanda, T.; Takada, K. Epstein-Barr virus (EBV)-encoded RNA 2 (EBER2) but not EBER1 plays a critical role in EBV-induced B-cell growth transformation. J. Virol. 2007, 81, 11236–11245. [Google Scholar] [CrossRef] [PubMed]
- Clarke, P.A.; Sharp, N.A.; Clemens, M.J. Expression of genes for the Epstein—Barr virus small RNAs EBER-1 and EBER-2 in Daudi Burkitt’s lymphoma cells: Effects of interferon treatment. J. Gen. Virol. 1992, 73, 3169–3175. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D. Epstein-Barr Virus-encoded RNAs: Key molecules in viral pathogenesis. Cancers 2014, 6, 1615–1630. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Joo, E.H.; Song, K.A.; Choi, B.; Kim, M.; Kim, S.H.; Kim, S.J.; Kang, M.S. Effects of lymphocyte profile on development of EBV-induced lymphoma subtypes in humanized mice. Proc. Natl. Acad. Sci. USA 2015, 112, 13081–13086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aromseree, S.; Middeldorp, J.M.; Pientong, C.; van Eijndhoven, M.; Ramayanti, O.; Lougheed, S.M.; Pegtel, D.M.; Steenbergen, R.D.M.; Ekalaksananan, T. High levels of EBV-Encoded RNA 1 (EBER1) trigger interferon and inflammation-related genes in keratinocytes expressing HPV16 E6/E7. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Boelens, M.C.; Wu, T.J.; Nabet, B.Y.; Xu, B.; Qiu, Y.; Yoon, T.; Azzam, D.J.; Twyman-Saint Victor, C.; Wiemann, B.Z.; Ishwaran, H.; et al. Exosome transfer from stromal to breast cancer cells regulates therapy resistance pathways. Cell 2014, 159, 499–513. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Pimienta, G.; Steitz, J.A. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein-Barr virus. RNA 2012, 18, 2073–2082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gouble, A.; Grazide, S.; Meggetto, F.; Mercier, P.; Delsol, G.; Morello, D. A New player in oncogenesis: AUF1/hnRNPD overexpression leads to tumorigenesis in transgenic mice 1. Cancer Res. 2002, 62, 1489–1495. [Google Scholar] [PubMed]
- Lee, N.; Yario, T.A.; Gao, J.S.; Steitz, J.A. EBV noncoding RNA EBER2 interacts with host RNA-binding proteins to regulate viral gene expression. Proc. Natl. Acad. Sci. USA 2016, 113, 3221–3226. [Google Scholar] [CrossRef] [PubMed]
- Ruf, I.K.; Lackey, K.A.; Warudkar, S.; Sample, J.T. Protection from interferon-induced apoptosis by Epstein-Barr virus small RNAs is not mediated by inhibition of PKR. J. Virol. 2005, 79, 14562–14569. [Google Scholar] [CrossRef] [PubMed]
- McKenna, S.A.; Lindhout, D.A.; Shimoike, T.; Aitken, C.E.; Puglisi, J.D. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J. Mol. Biol. 2007, 372, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y.; Zhang, Y. Viral lncRNA: A regulatory molecule for controlling virus life cycle. Non-Coding RNA Res. 2017, 2, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Arvey, A.; Tempera, I.; Tsai, K.; Chen, H.S.; Tikhmyanova, N.; Klichinsky, M.; Leslie, C.; Lieberman, P.M. An atlas of the Epstein-Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe 2012, 12, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Moss, W.N.; Steitz, J.A. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genom. 2013, 14, 543. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, V.S.; Valverde, D.P.; Moss, W.N. Human regulatory proteins associate with non-coding RNAs from the EBV IR1 region. BMC Res. Notes 2018, 11, 139. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Moss, W.N.; Yario, T.A.; Steitz, J.A. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell 2015, 160, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Aligo, J.; Walker, M.; Bugelski, P.; Weinstock, D. Is murine gammaherpesvirus-68 (MHV-68) a suitable immunotoxicological model for examining immunomodulatory drug-associated viral recrudescence? J. Immunotoxicol. 2015, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.R.; Kara, M.; Oko, L.M.; Grau, K.R.; Krueger, B.J.; Zhang, J.; Feng, P.; van Dyk, L.F.; Renne, R.; Tibbetts, S.A. A gammaherpesvirus noncoding RNA is essential for hematogenous dissemination and establishment of peripheral latency. mSphere 2016, 1, e00105–e00115. [Google Scholar] [CrossRef] [PubMed]
- Steer, B.; Strehle, M.; Sattler, C.; Bund, D.; Flach, B.; Stoeger, T.; Haas, J.G.; Adler, H. The small noncoding RNAs (sncRNAs) of murine gammaherpesvirus 68 (MHV-68) are involved in regulating the latent-to-lytic switch in vivo. Sci. Rep. 2016, 6, 32128. [Google Scholar] [CrossRef] [PubMed]
- Diebel, K.W.; Claypool, D.J.; van Dyk, L.F. A conserved RNA polymerase III promoter required for gammaherpesvirus TMER transcription and microRNA processing. Gene 2014, 544, 8–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, L.J.; Frank, R.; Rossi, J.J. Optimization and characterization of tRNA-shRNA expression constructs. Nucleic Acids Res. 2007, 35, 2620–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diebel, K.W.; Oko, L.M.; Medina, E.M.; Niemeyer, B.F.; Warren, C.J.; Claypool, D.J.; Tibbetts, S.A.; Cool, C.D.; Clambey, E.T.; van Dyk, L.F. Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo. MBio 2015, 6, e01670-14. [Google Scholar] [CrossRef] [PubMed]
- Ensser, A.; Fleckenstein, B. T-cell transformation and oncogenesis by γ2-herpesviruses. Adv. Cancer Res. 2005, 93, 91–128. [Google Scholar] [PubMed]
- Chen, C.Y.A.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995, 20, 465–470. [Google Scholar] [CrossRef]
- Fan, X.C.; Myer, V.E.; Steitz, J.A. AU-rich elements target small nuclear RNAs as well as mRNAS for rapid degradation. Genes Dev. 1997, 11, 2557–2568. [Google Scholar] [CrossRef] [PubMed]
- Guttilla, I.K.; White, B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ami, R.; Rodríguez-Baño, J.; Arslan, H.; Pitout, J.D.D.; Quentin, C.; Calbo, E.S.; Azap, O.K.; Arpin, C.; Pascual, A.; Livermore, D.M.; et al. A multinational survey of risk factors for infection with extended-spectrum β-lactamase-producing enterobacteriaceae in nonhospitalized patients. Clin. Infect. Dis. 2009, 49, 682–690. [Google Scholar] [CrossRef] [PubMed]
- Crist, C.G.; Montarras, D.; Pallafacchina, G.; Rocancourt, D.; Cumano, A.; Conway, S.J.; Buckingham, M. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. USA 2009, 106, 13383–13387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcinowski, L.; Tanguy, M.; Krmpotic, A.; Rädle, B.; Lisnić, V.J.; Tuddenham, L.; Chane-Woon-Ming, B.; Ruzsics, Z.; Erhard, F.; Benkartek, C.; et al. Degradation of cellular miR-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorbea, C.; Mosbruger, T.; Cazalla, D. A viral Sm-class RNA base-pairs with mRNAs & recruits microRNAs to inhibit apoptosis. Nature 2017, 550, 275–279. [Google Scholar] [PubMed]
- Coen, N.; Duraffour, S.; Topalis, D.; Snoeck, R.; Andrei, G. Spectrum of activity and mechanisms of resistance of various nucleoside derivatives against gammaherpesviruses. Antimicrob. Agents Chemother. 2014, 58, 7312–7323. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Bestman-Smith, J.; Boivin, G. Resistance of herpesviruses to antiviral drugs: Clinical impacts and molecular mechanisms. Drug Resist. Updates 2002, 5, 88–114. [Google Scholar] [CrossRef]
- Cao, Y. EBV based cancer prevention and therapy in nasopharyngeal carcinoma. NPJ Precis. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.A.; Woo, J.K.S.; King, A.; Zee, B.C.Y.; Lam, W.K.J.; Chan, S.L.; Chu, S.W.I.; Mak, C.; Tse, I.O.L.; Leung, S.Y.M.; et al. Analysis of plasma Epstein–Barr virus DNA to screen for nasopharyngeal cancer. N. Engl. J. Med. 2017, 378, 973. [Google Scholar] [CrossRef] [PubMed]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Moulec, S.L.E.; Guigay, J.; Hirashima, M.; Guemira, F.; et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Misawa, N.; Nie, C.; Satou, Y.; Iwakiri, D.; Matsuoka, M.; Takahashi, R.; Kuzushima, K.; Ito, M.; Takada, K.; et al. A novel animal model of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood 2011, 117, 5663–5673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, W.; Philip, P.S.; Tariq, S.; Khan, G. Epstein-Barr virus-encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV-transformed cells. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Aga, M.; Bentz, G.L.; Raffa, S.; Torrisi, M.R.; Kondo, S.; Wakisaka, N.; Yoshizaki, T.; Pagano, J.S.; Shackelford, J. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 2014, 33, 4613–4622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meckes, D.G. Exosomal Communication Goes Viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwab, A.; Meyering, S.S.; Lepene, B.; Iordanskiy, S.; van Hoek, M.L.; Hakami, R.M.; Kashanchi, F. Extracellular vesicles from infected cells: Potential for direct pathogenesis. Front. Microbiol. 2015, 6, 1132. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cao, H.; Shen, B.; Feng, J. Tumor-derived exosomes in cancer progression and treatment failure. Oncotarget 2015, 6, 37151–37168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iero, M.; Valenti, R.; Huber, V.; Filipazzi, P.; Parmiani, G.; Fais, S.; Rivoltini, L. Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ. 2008, 15, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T. Small molecules targeting viral RNA. Wiley Interdiscip. Rev. RNA 2016, 7, 726–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Spurgers, K.B.; Sharkey, C.M.; Warfield, K.L.; Bavari, S. Oligonucleotide antiviral therapeutics: Antisense and RNA interference for highly pathogenic RNA viruses. Antivir. Res. 2008, 78, 26–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, X.; Corey, D.R. Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Res. 2018, 46, 1584–1600. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Connelly, C.M.; Moon, M.H.; Schneekloth, J.S. The emerging role of RNA as a therapeutic target for small molecules. Cell Chem. Biol. 2016, 23, 1077–1090. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.; Castaldi, M.P.; Dutta, S.; Dibrov, S.M.; Wyles, D.L.; Hermann, T. Conformational inhibition of the hepatitis C virus internal ribosome entry site RNA. Nat. Chem. Biol. 2009, 5, 823–825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.J.; Kim, Y.G.; Park, H.J. Identification of RNA pseudoknot-binding ligand that inhibits the −1 ribosomal frameshifting of SARS-coronavirus by structure-based virtual screening. J. Am. Chem. Soc. 2011, 133, 10094–10100. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Bottini, A.; Kim, M.; Bardaro, M.F.; Zhang, Z.; Pellecchia, M.; Choi, B.S.; Varani, G. Correction: A novel small-molecule binds to the influenza A virus RNA promoter and inhibits viral replication. Chem. Commun. 2014, 50, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G. HIV: Shock and kill. Nature 2012, 487, 439–440. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; Perrine, S.P.; Williams, R.M.; Faller, D.V. Histone deacetylase inhibitors are potent inducers of gene expression in latent EBV and sensitize lymphoma cells to nucleoside antiviral agents. Blood 2012, 119, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Anderson, J.L.; Lewin, S.R. Getting the ‘Kill’ into ‘Shock and Kill’: Strategies to eliminate latent HIV. Cell Host Microbe 2018, 23, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.H.; Kenney, S.C. Valproic acid enhances the efficacy of chemotherapy in EBV-positive tumors by increasing lytic viral gene expression. Cancer Res. 2006, 66, 8762–8769. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Shimoda, M.; Olney, L.; Lyu, Y.; Tran, K.; Jiang, G.; Nakano, K.; Davis, R.R.; Tepper, C.G.; Maverakis, E.; et al. Oncolytic reactivation of KSHV as a therapeutic approach for primary effusion lymphoma. Mol. Cancer Ther. 2017, 16, 2627–2638. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, M.; Zhou, F.; Ye, F.; Gao, S.-J. Activation of Kaposi’s sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: Identification of sirtuin 1 as a regulator of the KSHV life cycle. J. Virol. 2014, 88, 6355–6367. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, S.; Gay, L.A.; Jain, V.; Haecker, I.; Renne, R. microRNA dependent and independent deregulation of long non-coding RNAs by an oncogenic herpesvirus. PLoS Pathog. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Sethuraman, S.; Thomas, M.; Gay, L.A.; Renne, R. Computational analysis of ribonomics datasets identifies long non-coding RNA targets of γ-herpesviral miRNAs. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Vadis, Q.; Soldin, O.P.; Elin, R.J.; Soldin, S.J. Therapeutic drug monitoring in human immunodeficiency virus/acquired immunodeficiency syndrome. Arch. Pathol. Lab. Med. 2003, 127, 102–105. [Google Scholar]
- Diaz-Toledano, R.; Lozano, G.; Martinez-Salas, E. In-cell SHAPE uncovers dynamic interactions between the untranslated regions of the foot-and-mouth disease virus RNA. Nucleic Acids Res. 2017, 45, 1416–1432. [Google Scholar] [CrossRef] [PubMed]
- Francisco-Velilla, R.; Fernandez-Chamorro, J.; Lozano, G.; Diaz-Toledano, R.; Martínez-Salas, E. RNA-protein interaction methods to study viral IRES elements. Methods 2015, 91, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.; Lander, E.S.; Guttman, M. RNA Antisense Purification (RAP) for Mapping RNA Interactions with Chromatin; Springer: New York, NY, USA, 2015; Volume 1262. [Google Scholar]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D. Capture hybridization analysis of RNA targets (CHART). Curr. Protoc. Mol. Biol. 2013, 101, 21–25. [Google Scholar]
- Jacob, R.; Zander, S.; Gutschner, T. The dark side of the epitranscriptome: Chemical modifications in long non-coding RNAs. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhu, P.; Ma, S.; Song, J.; Bai, J.; Sun, F.; Yi, C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 2015, 11, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [PubMed]


| Virus | Name | Alias(es) | Size (Kb) | Associated Proteins | Method of Detection | References |
|---|---|---|---|---|---|---|
| KSHV | K1/ORF4-11 | ALE; K1.3; K1/11-AS | ~17.0 | RNA-seq, ribosomal profiling and tilling microarray | [9] | |
| vIL6/ORF2 | K1.5 | ~1.0 | Genome-tilling microarray | [26] | ||
| vIL6/K4.2 | K2/K4.2-AS | ~5.8 | RNA-seq and ribosomal profiling | [9] | ||
| K4s | K3.5 | ~0.9 | Tilling microarray | [27] | ||
| PAN/K7 | K7.3; anti-PAN | ~1.3 | Tilling microarray | [27] | ||
| K9/ORF62 | ~17.3 | RNA-seq and ribosomal profiling | [9] | |||
| ORF58/59 | K11.5 | ~2.5 | Tilling microarray | [27] | ||
| ORF65/69 | ~7.5 | Tilling microarray | [27] | |||
| vGPCR | ~4.0 | Tilling microarray | [27] | |||
| ORF7 | ~0.8 | Tilling microarray | [27] | |||
| K5/K6 | K4.5; T6.1; K5/6-AS | 6.1 | RNA-seq, ribosomal profiling | [9,28] | ||
| IR | K4.7; T1.5 | 1.2 | RNA-seq, ribosomal profiling | [9,28] | ||
| PAN RNA | nut1; T1.1 | 1.1 | hnRNP C1, PABPC1, LANA, ORF57, PCRC2 | RNA-seq, ribosomal profiling | [9,28,29] | |
| RTA | K7.7; T3.0; 50L | 2.9 | RNA-seq | [9,28] | ||
| RTA | T1.2; 50S | 1 | RNA-seq, ribosomal profiling | [9,28] | ||
| miR/K13/72/LANA | ALT; K12.5 | 10.1; 9.9 | Genome-tiling microarray | [26] | ||
| EBV | EBER1 | 0.167 | La, L22, hnRNPD | RNA-seq | [30,31,32,33,34] | |
| EBER2 | 0.172 | La, nucleolin, PAX5 | RNA-seq | [30,31,34,35,36] | ||
| EBV-sisRNA-2 | 2.971 | Computational modeling and RNA-Seq | [37] | |||
| BHLF1 | 2.5 | Northern blot | [14,37] | |||
| oriPtLs | Leftleftward oriPt | 2.304 | NONO, ADAR | Tilling microarray, RNA-seq, FISH | [11] | |
| oriPtRs | Rightward oirPt | 2.304 | NONO, ADAR | Tilling microarray, RNA-seq, FISH | [11] | |
| MHV-68 | TMER1 | 0.2–0.25 | AGO2, Sm proteins | Plasmid construction and DNA | [38] | |
| TMER2 | 0.2–0.25 | AGO2, Sm proteins | sequencing paired with tRNA search | [38] | ||
| TMER3 | 0.2–0.25 | programs, RT-PCR analysis | [38] | |||
| TMER4 | 0.2–0.25 | [38] | ||||
| TMER5 | 0.2–0.25 | [38] | ||||
| TMER6 | 0.2–0.25 | [38] | ||||
| TMER7 | 0.2–0.25 | [38] | ||||
| TMER8 | 0.2–0.25 | [38] | ||||
| HVS | HSUR1 | 0.114–0.143 | Sm, Ago2, HuR, hnRNP D | RNA-seq | [39,40,41,42] | |
| HSUR2 | 0.114–0.144 | Sm, Ago2, HuR, hnRNP D | RNA-seq | [39,43] | ||
| HSUR5 | 0.114–0.145 | Sm, Ago2, HuR, hnRNP D | RNA-seq | [39,43] | ||
| HSUR3 | 0.075–0.106 | Sm | RNA-seq | [39,43] | ||
| HSUR4 | 0.075–0.106 | Sm | RNA-seq | [39,43] | ||
| HSUR6 | 0.075–0.106 | Sm | RNA-seq | [39,43] | ||
| HSUR7 | 0.075–0.106 | Sm | RNA-seq | [39,43] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chavez-Calvillo, G.; Martin, S.; Hamm, C.; Sztuba-Solinska, J. The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis. Non-Coding RNA 2018, 4, 24. https://doi.org/10.3390/ncrna4040024
Chavez-Calvillo G, Martin S, Hamm C, Sztuba-Solinska J. The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis. Non-Coding RNA. 2018; 4(4):24. https://doi.org/10.3390/ncrna4040024
Chicago/Turabian StyleChavez-Calvillo, Gabriela, Sarah Martin, Chad Hamm, and Joanna Sztuba-Solinska. 2018. "The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis" Non-Coding RNA 4, no. 4: 24. https://doi.org/10.3390/ncrna4040024
APA StyleChavez-Calvillo, G., Martin, S., Hamm, C., & Sztuba-Solinska, J. (2018). The Structure-To-Function Relationships of Gammaherpesvirus-Encoded Long Non-Coding RNAs and Their Contributions to Viral Pathogenesis. Non-Coding RNA, 4(4), 24. https://doi.org/10.3390/ncrna4040024