Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application
Abstract
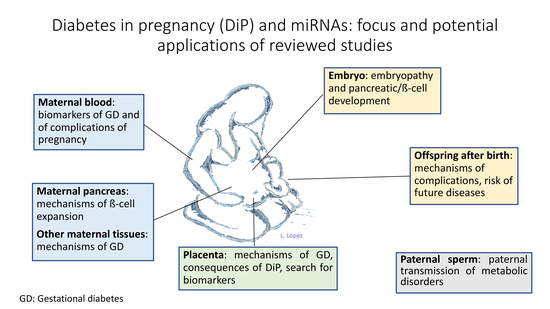
:1. Introduction
1.1. Diabetes in Pregnancy: Classification
1.2. Clinical Consequences of Diabetes in Pregnancy
1.3. Epigenetics and Intrauterine Programming
2. MiRNAs
2.1. Pregnancy and miRNAs: The Role of the Placenta
2.2. MiRNAs and the β-Cell
2.3. Gestational Diabetes and miRNAs
2.3.1. Studies in Maternal Blood
2.3.2. Studies in Placenta
- Some miRNAs could potentially be used as predictors of gestational diabetes and perinatal outcomes.
- The most promising markers of gestational diabetes are: miR-29a, miR-222, miR-16-5p, miR-17-5p and miR-20a-5p.
- Some studies propose a role for miRNAs in the pathogenesis of gestational diabetes and its complications but no conclusive information is available yet. Other non-coding RNAs may also play a role in the pathogenesis and consequences of gestational diabetes.
- Sample source: although some overlap has been described in the results obtained in placental and peripheral blood samples, this is not the rule. Indeed, regarding blood samples, some studies use serum or plasma, whereas others use lysed, whole blood or leukocytes (see Table 2).
- miRNA expression varies with gestational age. Thus, control groups need to be matched to the study group by this variable.
- Other factors, such as mode of delivery, offspring sex and BMI could also add to the risk of bias.
- The diagnostic criteria used to define gestational diabetes also vary among studies, although this is probably a minor, if any, source of bias. All definitions have in common mild hyperglycaemia diagnosed, for the first time, during pregnancy.
- Gestational age should be reported and matched between the study and control groups.
- When placenta samples are obtained, given the mixed nature of this organ, the procedure needs to be standardized to minimize bias. Furthermore, the mode of delivery needs to be reported, as well as offspring sex
- When used as biomarkers of gestational diabetes or its outcomes, the performance of miRNAs should be compared with that of classical, easy-to-assess risk factors.
- The criteria for selecting a certain miRNA or group of miRNAs for assessment or validation should be clear, as well as the choice for endogenous controls.
2.4. Pre-Gestational Diabetes and miRNAs
miRNA and Diabetic Embryopathy
2.5. miRNA and Macrosomia
2.6. Paternal Effects on Offspring and miRNAs
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lawrence, J.M.; Contreras, R.; Chen, W.; Sacks, D.A. Trends in the prevalence of preexisting diabetes and gestational diabetes mellitus among a racially/ethnically diverse population of pregnant women, 1999–2005. Diabetes Care 2008, 31, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, A. Increasing prevalence of gestational diabetes mellitus: A public health perspective. Diabetes Care 2007, 30 (Suppl. 2), S141–S146. [Google Scholar] [CrossRef] [PubMed]
- Catalano, P.M. Trying to understand gestational diabetes. Diabet. Med. 2014, 31, 273–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powe, C.E.; Allard, C.; Battista, M.C.; Doyon, M.; Bouchard, L.; Ecker, J.L.; Perron, P.; Florez, J.C.; Thadhani, R.; Hivert, M.F. Heterogeneous contribution of insulin sensitivity and secretion defects to gestational diabetes mellitus. Diabetes Care 2016, 29, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Committee on Practice Bulletins-Obstetrics. ACOG Practice Bulletin No 190: Gestational Diabetes Mellitus. Obstet. Gynecol. 2018, 131, e49–e64. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, M.W.; Coustan, D.R. Criteria for screening tests for gestational diabetes. Am. J. Obstet. Gynecol. 1982, 144, 768–773. [Google Scholar] [CrossRef]
- National Diabetes Data Group. Classification and diagnosis of diabetes mellitus and other categories of glucose intolerance. Diabetes 1979, 28, 1039–1057. [Google Scholar]
- International Association of Diabetes and Pregnancy Study Groups Consensus Panel; Metzger, B.E.; Gabbe, S.G.; Persson, B.; Buchanan, T.A.; Catalano, P.A.; Damm, P.; Dyer, A.R.; Leiva Ad Hod, M.; Kitzmiler, J.L.; et al. International association of diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010, 33, 676–682. [Google Scholar] [CrossRef] [PubMed]
- American Diabetes Association. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes. Diabetes Care 2018, 41 (Suppl. 1), S13–S27. [Google Scholar] [CrossRef] [PubMed]
- Vandorsten, J.P.; Dodson, W.C.; Espeland, M.A.; Grobman, W.A.; Guise, J.M.; Mercer, B.M.; Minkoff, H.L.; Poindexter, B.; Prosser, L.A.; Sawaya, G.F.; et al. NIH consensus development conference: Diagnosing gestational diabetes mellitus. NIH Consens. State Sci. Statements 2013, 29, 1–31. [Google Scholar] [PubMed]
- Farrar, D.; Duley, L.; Dowswell, T.; Lawlor, D.A. Different strategies for diagnosing gestational diabetes to improve maternal and infant health. Cochrane Database Syst. Rev. 2017, 8, CD007122. [Google Scholar] [CrossRef] [PubMed]
- Crowther, C.A.; Hiller, J.E.; Moss, J.R.; McPhee, A.J.; Jeffries, W.S.; Robinson, J.S.; Australian Carbohydrate Intolerance Study in Pregnant Women (ACHOIS) Trial Group. Effect of treatment of gestational diabetes mellitus on pregnancy outcomes. N. Engl. J. Med. 2005, 352, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Wahabi, H.A.; Alzeidan, R.A.; Esmaeil, S.A. Pre-pregnancy care for women with pre-gestational diabetes mellitus: A systematic review and meta-analysis. BMC Public Health 2012, 12, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Jarvela, I.Y.; Kulmala, P.; Juutinen, K.; Knip, M.; Tapanainen, J.S. Gestational diabetes identifies women at risk for permanent type 1 and type 2 diabetes in fertile age. Diabetes Care 2006, 29, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Persson, M.; Norman, M.; Hanson, U. Obstetric and perinatal outcomes in type 1 diabetic pregnancies: A large, population-based study. Diabetes Care 2009, 32, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Balsells, M.; García-Patterson, A.; Gich, I.; Corcoy, R. Maternal and fetal outcomes in women with type 2 versus type 1 diabetes mellitus: A systematic review and meta-analysis. J. Clin. Endocrinol. Metab. 2009, 94, 4284–4291. [Google Scholar] [CrossRef] [PubMed]
- Hillier, T.; Pedula, K.L.; Schimdt, M.M.; Mullen, J.D.; Charles, M.A.; Pettitt, D.J. Childhood obesity and metabolic imprinting: The ongoing effects of maternal hyperglycemia. Diabetes Care 2007, 30, 2287–2292. [Google Scholar] [CrossRef] [PubMed]
- Tam, W.H.; Ma, R.C.W.; Ozaki, R.; Li, A.M.; Chan, M.H.M.; Yuen, L.Y.; Lao, T.T.H.; Yang, X.; Ho, C.S.; Tutino, G.E.; et al. In utero exposure to maternal hyperglycemia increases childhood cardiometabolic risk in offspring. Diabetes Care 2017, 40, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.D.; Mathiesen, E.R.; Hansen, T.; Pedersen, O.; Jensen, D.M.; Lauenborg, J.; Damm, P. High prevalence of type 2 diabetes and pre-diabetes in adult offspring of women with gestational diabetes mellitus or type 1 diabetes: The role of intrauterine hyperglycemia. Diabetes Care 2008, 31, 340–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, T.P.; Watkins, A.J.; Velazquez, M.A.; Mathers, J.C.; Prentice, A.M.; Stephenson, J.; Barker, M.; Saffery, R.; Yajnik, C.S.; Eckert, J.J.; et al. Origins of lifetime health around the time of conception: Causes and consequences. Lancet 2018, 391, 1842–1852. [Google Scholar] [CrossRef]
- Saffery, R.; Novakovic, B. Epigenetics as the mediator of fetal programming of adult onset disease: What is the evidence? Acta Obstet. Gynecol. Scand. 2014, 93, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perception of epigenetics. Nature 2007, 7143, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Ling, C. DNA methylation links genetics, fetal environment, and an unhealthy lifestyle to the development of type 2 diabetes. Clin. Epigenet. 2017, 9, 105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Z.-J.; Zhang, C.L.; Schatten, H.; Sun, Q.-Y. Maternal diabetes mellitus and the origin of non-communicable diseases in offspring: The role of epigenetics. Biol. Reprod. 2014, 90, 139. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, S.J.; Tellam, R.L.; Morrison, J.L.; Muhlhausler, B.S.; Mollo, P.L. Recent developments on the role of epigenetics in obesity and metabolic disease. Clin. Epigenet. 2015, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Cantone, I.; Fisher, A.G. Epigenetic programming and reprogramming during development. Nat. Struct. Mol. Biol. 2013, 20, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Smallwood, S.A.; Kelsay, G. De novo DNA methylation: A germ cell perspective. Trends Genet. 2012, 281, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Roccaro, A.M.; Sacco, A.; Jia, X.; Azab, A.K.; Maiso, P.; Ngo, H.T.; Azab, F.; Runnels, J.; Quang, P.; Ghobrial, I.M. MicroRNA—Dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood 2010, 116, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Denis, H.; Ndlovu, M.N.; Fuks, F. Regulation of mammalian DNA methyltransferases: A route to new mechanisms. EMBO Rep. 2011, 12, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Napso, T.; Yong, H.E.J.; Lopez-Tello, J.; Sferruzzi-Perri, A.N. The Role of Placental Hormones in Mediating Maternal Adaptations to Support Pregnancy and Lactation. Front. Physiol. 2018, 9, 1091. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.F.; Ouyang, Y.; Coyne, C.B.; Sadovsky, Y. MicroRNAs in placental health and disease. Am. J. Obstet. Gynecol. 2015, 213 (Suppl. 4), S163–S172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, S.S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Morales-Prieto, D.M.; Chaiwangyen, W.; Ospina-Prieto, S.; Schneider, U.; Herrmann, J.; Gruhn, B.; Markert, U.R. MicroRNA expression profiles of trophoblastic cells. Placenta 2012, 33, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Kolloru, G.K.; Ahmed, A. Small molecule, big prospects: Micro RNA in pregnancy and its complications. J. Pregnancy 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 which suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Lu, C.; Ji, X.; Miao, Z.; Long, W.; Ding, H.; Lv, M. Roles of microRNAs in preeclampsia. J. Cell. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bounds, K.R.; Chiasson, V.L.; Pan, L.J.; Gupta, S.; Chatterjee, P. MicroRNAs: New Players in the Pathobiology of Preeclampsia. Front. Cardiovasc. Med. 2017, 4, 60. [Google Scholar] [CrossRef] [PubMed]
- Huynh, J.; Dawson, D.; Roberts, D.; Bentley-Lewis, R. A systematic review of placental pathology in maternal diabetes mellitus. Placenta 2015, 36, 101–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, S.M.; Coan, P.M.; Burton, G.J.; Lindsay, R.S. Placental structure in type 1 diabetes. Relation to fetal insulin, leptin and IGF-I. Diabetes 2009, 58, 2634–2641. [Google Scholar] [CrossRef] [PubMed]
- Starikov, R.; Inman, K.; Chen, K.; Lopes, V.; Coviello, E.; Pinar, H.; He, M. Comparison of placental findings in type 1 and type 2 diabetic pregnancies. Placenta 2014, 35, 1001–1006. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, T.; Varastehpour, A.; Catalano, P.; Hauguel-de Mouzon, S. Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 2003, 52, 2951–2958. [Google Scholar] [CrossRef] [PubMed]
- Enquobahrie, D.A.; Williams, M.A.; Qiu, C.; Meller, M.; Sorensen, T.K. Global placental gene expression in gestational diabetes mellitus. Am. J. Obstet. Gynecol. 2009, 200, 206.e1–206.e13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.H.; Wang, D.P.; Zhang, L.L.; Zhang, F.; Wang, D.M.; Zhang, W.Y. Genomic expression profiles of blood and placenta reveal significant immune-related pathways and categories in Chinese women with gestational diabetes mellitus. Diabet. Med. 2011, 28, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, T.; Lepercq, J.; Varastehpour, A.; Basu, S.; Catalano, P.M.; Hauguel-De Mouzon, S. Differential regulation of genes for fetoplacental lipid pathways in pregnancy with gestational and type 1 diabetes mellitus. Am. J. Obstet. Gynecol. 2009, 201, 209.e1–209.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bari, M.F.; Ngo, S.; Bastie, C.C.; Sheppard, A.M.; Vatish, M. Gestational diabetic transcriptomic profiling of microdissected human trophoblast. J. Endocrinol. 2016, 229, 47–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martino, J.; Sebert, S.; Segura, M.T.; García-Valdés, L.; Florido, J.; Padilla, M.C.; Marcos, A.; Rueda, R.; McArdle, H.J.; Budge, H.; et al. Maternal Body Weight and Gestational Diabetes Differentially Influence Placental and Pregnancy Outcomes. J. Clin. Endocrinol. Metab. 2016, 101, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, J.H.; Haase, T.N.; Jaksch, C.; Nalla, A.; Søstrup, B.; Nalla, A.A.; Larsen, L.; Rasmussen, M.; Dalgaard, L.T.; Gaarn, L.W.; et al. Impact of fetal and neonatal environment on beta cell function and development of diabetes. Acta Obstet. Gynecol. Scand. 2014, 93, 1109–1122. [Google Scholar] [CrossRef] [PubMed]
- Jacovetti, C.; Abderrahmani, A.; Parnaud, G.; Jonas, J.C.; Peyot, M.L.; Cornu, M.; Laybutt, R.; Meugnier, E.; Rome, S.; Thorens, B.; et al. MicroRNAs contribute to compensatory β-cell expansion during pregnancy and obesity. J. Clin. Investig. 2012, 122, 3541–3551. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Valverde, S.L.; Taft, R.J.; Mattick, J.S. MicroRNAs in B-cell biology, insulin resistance, diabetes and its complications. Diabetes 2011, 60, 1825–1830. [Google Scholar] [CrossRef] [PubMed]
- Rosero, S.; Bravo-Egana, V.; Jiang, Z.; Khuri, S.; Tsinoremas, N.; Klein, D.; Sabates, E.; Correa-Medina, M.; Ricordi, C.; Domínguez-Bendala, J.; et al. MicroRNA signature of the human developing pancreas. BMC Genom. 2010, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, M.V.; Joglekar, V.M.; Hardikar, A.A. Expression of islet-specific miRNAs during human pancreatic development. Gene Expr. Patterns 2009, 9, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Dotta, F.; Ventriglia, G.; Snowhite, I.V.; Pugliese, A. MicroRNAs: Markers of β-cell stress and autoimmunity. Curr. Opin. Endocrinol. Diabetes Obes. 2018, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Assmann, T.S.; Recamonde-Mendoza, M.; De Souza, B.M.; Crispim, D. MicroRNA expression profiles and type 1 diabetes mellitus: Systematic review and bioinformatic analysis. Endocr. Connect. 2017, 6, 773–790. [Google Scholar] [CrossRef] [PubMed]
- Dumortier, O.; Hinault, C.; Gautier, N.; Patoureaux, N.; Casamento, V.; Van Obberghen, E. Maternal protein restriction leads to pancreatic failure in offspring: Role of missexpressed microRNA-375. Diabetes 2014, 63, 3416–3427. [Google Scholar] [CrossRef] [PubMed]
- Moen, G.H.; Sommer, C.; Prasad, R.B.; Sletner, L.; Groop, L.; Qvigstad, E.; Birkeland, K.I. Mechanisms in endocrinology: Epigenetic modifications and gestational diabetes: A systematic review of published literature. Eur. J. Endocrinol. 2017, 176, R247–R267. [Google Scholar] [CrossRef] [PubMed]
- Poirier, C.; Desgagne, V.; Guerin, R.; Bouchard, L. MicroRNAs in pregnancy and gestational diabetes: Emerging role in maternal metabolic regulation. Curr. Diabetes Rep. 2017, 17, 35. [Google Scholar] [CrossRef] [PubMed]
- Guarino, E.; Delli Poggi, C.; Grieco, G.E.; Cenci, V.; Ceccarelli, E.; Crisci, I.; Sebastiani, G.; Dotta, F. Circulating microRNAs as biomarkers of gestational diabetes mellitus: Updates and perspectives. Int. J. Endocrinol. 2018, 12, 6380463. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.L.; Jia, Y.J.; Xing, B.H.; Shi, D.D.; Dong, X.J. Plasma microRNA-16-5p, -17-5p and -20a-5p: Novel diagnostic biomarkers for gestational diabetes mellitus. J. Obstet. Gynaecol. Res. 2017, 43, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.L.; Zhang, L.; Li, J.; Tian, S.; Lv, X.D.; Wang, X.Q.; Su, X.; Li, Y.; Hu, Y.; Ma, X.; et al. Up-regulation of miR-98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci. Rep. 2016, 6, 32268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Tian, F.; Li, H.; Zhou, Y.; Lu, J.; Ge, Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynaecol. Obstet. 2015, 130, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Collares, C.V.; Evangelista, A.F.; Xavier, D.J.; Rassi, D.M.; Arns, T.; Foss-Freitas, M.C.; Foss, M.C.; Puthier, D.; Sakamoto-Hojo, E.T.; Passos, G.A.; et al. Identifying common and specific microRNAs expressed in peripheral blood mononuclear cell of type 1, type 2, and gestational diabetes mellitus patients. BMC Res. Notes 2013, 6, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Floris, I.; Descamps, B.; Vardeu, A.; Mitić, T.; Posadino, A.M.; Shantikumar, S.; Sala-Newby, G.; Capobianco, G.; Mangialardi, G.; Howard, L.; et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 664–674. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Bai, J.; Liu, P.; Dong, J.; Tang, Y.; Zhou, J.; Han, P.; Xing, J.; Chen, Y.; Yu, X. miR-494 protects pancreatic β-cell function by targeting PTEN in gestational diabetes mellitus. EXCLI J. 2017, 16, 1297–1307. [Google Scholar] [CrossRef] [PubMed]
- Houshmand-Oeregaard, A.; Schrölkamp, M.; Kelstrup, L.; Hansen, N.S.; Hjort, L.; Thuesen, A.C.B.; Damm, P. Increased expression of microRNA-15a and microRNA-15b in skeletal muscle from adult offspring of women with diabetes in pregnancy. Hum. Mol. Genet. 2018, 27, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Lamadrid-Romero, M.; Solís, K.H.; Cruz-Reséndiz, M.S.; Pérez, J.E.; Díaz, N.F.; Flores-Herrera, H.; García-López, G.; Perichart, O.; Reyes-Muñoz, E.; Arenas-Huertero, F.; et al. Central nervous system development-related microRNAs levels increase in the serum of gestational diabetic women during the first trimester of pregnancy. Neurosci. Res. 2018, 130, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, L.; Zhou, L.; Wu, J.; Sheng, C.; Chen, H.; Liu, Y.; Gao, S.; Huang, W. A MicroRNA Signature in Gestational Diabetes Mellitus Associated with Risk of Macrosomia. Cell. Physiol. Biochem. 2015, 37, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Muralimanoharan, S.; Maloyan, A.; Myatt, L. Mitochondrial function and glucose metabolism in the placenta with gestational diabetes mellitus: Role of miR-143. Clin. Sci. (Lond.) 2016, 130, 931–941. [Google Scholar] [CrossRef] [PubMed]
- Pheiffer, C.; Dias, S.; Rheeder, P.; Adam, S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018, 22, 345–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahimi, G.; Jafari, N.; Khodabakhsh, M.; Shirzad, Z.; Dogaheh, H.P. Upregulation of microRNA processing enzymes Drosha and Dicer in gestational diabetes mellitus. Gynecol. Endocrinol. 2015, 31, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, G.; Guarino, E.; Grieco, G.E.; Formichi, C.; Delli Poggi, C.; Ceccarelli, E.; Dotta, F. Circulating microRNA (miRNA) Expression Profiling in Plasma of Patients with Gestational Diabetes Mellitus Reveals Upregulation of miRNA miR-330-3p. Front. Endocrinol. 2017, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, C.; Guo, X.; Ding, H.; Cui, Y.; Shen, R.; Liu, J. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERα expression in estrogen-induced insulin resistance. Endocrinology 2014, 155, 1982–1990. [Google Scholar] [CrossRef] [PubMed]
- Stirm, L.; Huypens, P.; Sass, S.; Batra, R.; Fritsche, L.; Brucker, S.; Abele, H.; Hennige, A.M.; Theis, F.; Beckers, J.; et al. Maternal whole blood cell miRNA-340 is elevated in gestational diabetes and inversely regulated by glucose and insulin. Sci. Rep. 2018, 8, 1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagoma, A.; Alnek, K.; Kirss, A.; Uibo, R.; Haller-Kikkatalo, K. MicroRNA profiling of second trimester maternal plasma shows upregulation of miR-195-5p in patients with gestational diabetes. Gene 2018, 672, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Peng, Q.J.; Chen, L.C. MiR-95, -548am and -1246 expression in placenta tissue of gestationl diabetes mellitus as well as their relationship with adipocytokines and glucose transporters. J. Hainan Med. Univ. 2016, 22, 5–8. [Google Scholar]
- Tryggestad, J.B.; Vishwanath, A.; Jiang, S.; Mallappa, A.; Teague, A.M.; Takahashi, Y.; Thompson, D.M.; Chernausek, S.D. Influence of gestational diabetes mellitus on human umbilical vein endothelial cell miRNA. Clin. Sci. (Lond.) 2016, 130, 1955–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wander, P.L.; Boyko, E.J.; Hevner, K.; Parikh, V.J.; Tadesse, M.G.; Sorensen, T.K.; Williams, M.A.; Enquobahrie, D.A. Circulating early- and mid-pregnancy microRNAs and risk of gestational diabetes. Diabetes Res. Clin. Pract. 2017, 132, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Bian, D.; Hao, L.; Huang, F.; Xu, M.; Qin, J.; Liu, Y. microRNA-503 contribute to pancreatic beta cell dysfunction by targeting the mTOR pathway in gestational diabetes mellitus. EXCLI J. 2017, 16, 1177–1187. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Feng, J.; Cheng, F.; Cui, X.; Gao, L.; Chen, Y.; Wang, F.; Zhong, T.; Li, Y.; Liu, L. Circular RNA expression profiles in placental villi from women with gestational diabetes mellitus. Biochem. Biophys. Res. Commun. 2018, 498, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Dong, J.; Jiang, T.; Shi, Z.; Yu, B.; Zhu, Y.; Chen, D.; Xu, J.; Huo, R.; Dai, J.; et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS ONE 2011, 6, e23925. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, T.; Shi, Z.; Ding, H.; Ling, X. MicroRNA-518d regulates PPARα protein expression in the placentas of females with gestational diabetes mellitus. Mol. Med. Rep. 2014, 9, 2085–2090. [Google Scholar] [CrossRef] [PubMed]
- Crossland, R.E.; Norden, J.; Bibby, L.A.; Davis, J.; Dickinson, A.M. Evaluation of optimal extracellular vesicle small RNA isolation and qRT-PCR normalisation for serum and urine. J. Immunol. Methods 2016, 429, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- Wägner, A.M.; González García-Cano, D.; Vega-Guedes, B.; Lorente-Arencibia, P.; Figueras-Falcón, T.; Brito-Casillas, Y.; Armas-Roca, M.; Pérez-Matos, C.; Fleitas-Ojeda, C.; Correa-González, S.; et al. Epigenetic effects of maternal type 1 diabetes. Placental miRNAs, methylations and their effects on gene expression. Diabetologia 2015, 58 (Suppl. 1), A120. [Google Scholar]
- Ibarra, A.; Vega, B.; Armas, M.; Gonzalez, D.; Perera, S.; Horres, R.; Valls, R.; Wiebe, J.C.; Wägner, A.M. Placental microRNA expression patterns in pregestational diabetes and identification of specific potential biomarkers. Diabetologia 2018, 61 (Suppl. 1), A169. [Google Scholar]
- Dong, D.; Zhang, Y.; Reece, E.A.; Wang, L.; Harman, C.R.; Yang, P. MicroRNA expression profiling and functional anotation analysis of their targets modulated by oxidative sress during embryonic heart development in diabetic mice. Reprod. Toxicol. 2016, 65, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Fu, N.; Yang, P. MiR-17 downregulation by high glucose stabilizes thioredoxin-interacting protein and removes thioredoxin inhibitio on ASK1 leading to apoptosis. Toxicol. Sci. 2016, 150, 84–86. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Teague, A.M.; Tryggestad, J.B.; Chernausek, S.D. Role of microRNA-130b in placental PGC-1a/TFAM mitochondrial biogenesis pathway. Biochem. Biophys. Res. Commun. 2017, 487, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Ramya, S.; Shyamasundar, S.; Huat Bay, B.; Thameem Dheen, S. Maternal diabetes alters expression of microRNAs that reulate genes critical for neural tube development. Front. Mol. Neurosci. 2017, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Zhao, L.; Cai, W.; Wei, M.; Zhou, M.; Yang, G.; Yuan, L. Maternal exosomes in diabetes contribute to the cardiac development deficiency. Biochem. Byophys. Res. Commun. 2017, 483, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Shyamasundar, S.; Jadhav, S.P.; Bay, B.H.; Tay, S.S.; Kumar, S.D.; Rangasamy, D.; Dheen, S.T. Analysis of epigenetic factors in mouse embryonic neural stem cells exposed to hyperglycemia. PLoS ONE 2013, 8, e65945. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Xu, C.; Reece, E.A.; Li, X.; Wu, Y.; Harman, C.; Yu, J.; Dong, D.; Wang, C.; Yang, P.; et al. Protein kinase C-α suppresses autophagy and induces neural tube defects via miR-129-2 in diabetic pregnancy. Nat. Commun. 2017, 8, 15182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Hakvoort, T.B.M.; Jongejan, A.; Ruijter, J.M.; van Kampen, A.H.C.; Lamers, W.H. Unexpected regulation of miRNA abundance during adaptation of early-somite mouse embryos to diabetic pregnancy. Biochem. Biophys. Res. Commun. 2017, 482, 1013–1018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, D.; Reece, E.A.; Wang, A.R.; Yang, P. Oxidative stress-induced miR-27a targets the redox gene nuclear factor erythroid 2-related factor 2 in dibaetic embryopaty. Am. J. Obstet. Gynecol. 2018, 218, 136.e1–136.e10. [Google Scholar] [CrossRef] [PubMed]
- Erickson, U.J.; Wentzel, P. The status of diabetic embryopathy. Upsala J. Med. Sci. 2015, 2, 96–112. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, A.; Zhang, J.; Dagvadorj, A.; Hirayama, F.; Shibuya, K.; Souza, J.P.; Gulmezoglu, A.M. Macrosomia in 23 developing countries: An analysis of a multicountry, facility-based, cross-sectional survey. Lancet 2013, 381, 476–483. [Google Scholar] [CrossRef]
- Pasek, R.C.; Gannon, M. Advancements and challenges in generating accurate animal models of gestational diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1327–E1338. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Han, J.; Zheng, F.; Ma, H.; Chen, J.; Jiang, Y.; Jiang, H. Early second-trimester serum microRNAs as potential biomarker for nondiabetic macrosomia. Biomed. Res. Int. 2014, 2014, 394125. [Google Scholar] [CrossRef] [PubMed]
- Merzouk, H.; Khan, N.A. Implication of lipids in macrosomia of diabetic pregnancy: Can n-3 polyunsaturated fatty acids exert beneficial effects? Clin. Sci. (Lond.) 2003, 105, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Kc, K.; Shakya, S.; Zhang, H. Gestational diabetes mellitus and macrosomia: A literature review. Ann. Nutr. Metab. 2015, 66 (Suppl. 2), 14–20. [Google Scholar] [CrossRef] [PubMed]
- Silveira, P.P.; Portella, A.K.; Goldani, M.Z.; Barbieri, M.A. Developmental origins of health and disease (DOHaD). J. Pediatr. 2007, 83, 494–504. [Google Scholar] [CrossRef]
- Jiang, H.; Wen, Y.; Hu, L.; Miao, T.; Zhang, M.; Dong, J. Serum MicroRNAs as Diagnostic Biomarkers for Macrosomia. Reprod. Sci. 2015, 22, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Maccani, M.A.; Marsit, C.J. Exposure and fetal growth-associated miRNA alterations in the human placenta. Clin. Epigenet. 2011, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wu, W.; Zhang, M.; Li, J.; Peng, Y.; Miao, T.T.; Zhu, H.; Xu, G. Aberrant upregulation of miR-21 in placental tissues of macrosomia. J. Perinatol. 2014, 34, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Tang, Q.; Wu, W.; Gu, H.; Liu, L.; Wu, J.; Jiang, H.; Ding, H.; Xia, Y.; et al. The role, mechanism and potentially novel biomarker of microRNA-17-92 cluster in macrosomia. Sci. Rep. 2015, 5, 17212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.T.; Cai, Q.Y.; Ji, S.S.; Zhang, H.X.; Wang, Y.H.; Yan, H.T.; Yang, X.J. Decreased miR-143 and increased miR-21 placental expression levels are associated with macrosomia. Mol. Med. Rep. 2016, 13, 3273–3280. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhu, Y.; Li, H.; Tian, F.; Xie, X.; Bai, Y. Differential expression of circulating miRNAs in maternal plasma in pregnancies with fetal macrosomia. Int. J. Mol. Med. 2015, 35, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhao, C.; Long, W.; Ding, H.; Shen, R. Microarray Expression Profile Analysis of Long Non-Coding RNAs in Umbilical Cord Plasma Reveals their Potential Role in Gestational Diabetes-Induced Macrosomia. Cell. Physiol. Biochem. 2015, 36, 542–554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodil-Garcia, P.; Arellanes-Licea, E.D.C.; Montoya-Contreras, A.; Salazar-Olivo, L.A. Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards. Int. J. Mol. Sci. 2017, 18, E2552. [Google Scholar] [CrossRef] [PubMed]
- Pescador, N.; Perez-Barba, M.; Ibarra, J.M.; Corbaton, A.; Martinez-Larrad, M.T.; Serrano-Rios, M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 2013, 8, e77251. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yin, L.; Chen, H.; Yang, S.; Pan, C.; Lu, S.; Miao, M.; Jiao, B. miR-376a suppresses proliferation and induces apoptosis in hepatocellular carcinoma. FEBS Lett. 2012, 586, 2396–2403. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.D.; Kruger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Bronneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Bottger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; Lopez-Bermejo, A.; Fernandez-Real, J.M. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef] [PubMed]
- Mouillet, J.F.; Chu, T.; Hubel, C.A.; Nelson, D.M.; Parks, W.T.; Sadovsky, Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta 2010, 31, 781–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, L.; Zhao, S.; Zhu, M.; Yu, M. Differential expression of microRNAs in porcine placentas on days 30 and 90 of gestation. Reprod. Fertil. Dev. 2010, 22, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Maccani, M.A.; Padbury, J.F.; Marsit, C.J. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS ONE 2011, 6, e21210. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Hwang, S.H.; Cho, H.H.; Shin, K.K.; Bae, Y.C.; Jung, J.S. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J. Cell. Physiol. 2012, 227, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.; Gburcik, V.; Petrovic, N.; Gallagher, I.J.; Nedergaard, J.; Cannon, B.; Timmons, J.A. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr. Disord. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.A. The Jak/STAT pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011205. [Google Scholar] [CrossRef] [PubMed]
- Puff, R.; Dames, P.; Weise, M.; Goke, B.; Parhofer, K.; Lechner, A. No non-redundant function of suppressor of cytokine signaling 2 in insulin producing β-cells. Islets 2010, 2, 252–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasche, A.; Al-Hasani, H.; Herwig, R. Meta-analysis approach identifies candidate genes and associated molecular networks for type-2 diabetes mellitus. BMC Genom. 2008, 9, 310. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Tang, H.; Leng, J.; Jiang, Q. Suppressors of cytokine signaling (SOCS) and type 2 diabetes. Mol. Biol. Rep. 2014, 41, 2265–2274. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Lv, C.; Wu, C.; Chen, F.; Shao, Y.; Wang, Q. Suppressor of cytokine signaling (SOCS) 2 attenuates renal lesions in rats with diabetic nephropathy. Acta Histochem. 2014, 116, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Rico-Bautista, E.; Greenhalgh, C.J.; Tollet-Egnell, P.; Hilton, D.J.; Alexander, W.S.; Norstedt, G.; Flores-Morales, A. Suppressor of cytokine signaling-2 deficiency induces molecular and metabolic changes that partially overlap with growth hormone-dependent effects. Mol. Endocrinol. 2005, 19, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Brito-Casillas, Y.; Aranda-Tavío, H.; Rodrigo-González, L.; Expósito-Montesdeoca, A.B.; Martín-Rodríguez, P.; Guerra, B.; Wägner, A.M.; Fernández-Pérez, L. Socs2-/- mouse as a potential model of macrosomia and gestational diabetes. Diabetologia 2017, 60 (Suppl. 1), 1–608. [Google Scholar]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hou, J.; Ye, L.; Chen, Y.; Cui, J.; Tian, W.; Li, C.; Liu, L. MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci. Rep. 2014, 4, 3819. [Google Scholar] [CrossRef] [PubMed]
- Szabo, S.; Xu, Y.; Romero, R.; Fule, T.; Karaszi, K.; Bhatti, G.; Varkonyi, T.; Varkonyi, I.; Krenacs, T.; Dong, Z.; et al. Changes of placental syndecan-1 expression in preeclampsia and HELLP syndrome. Virchows Arch. 2013, 463, 445–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borralho, P.M.; Simoes, A.E.; Gomes, S.E.; Lima, R.T.; Carvalho, T.; Ferreira, D.M.; Vasconcelos, M.H.; Castro, R.E.; Rodrigues, C.M. miR-143 overexpression impairs growth of human colon carcinoma xenografts in mice with induction of apoptosis and inhibition of proliferation. PLoS ONE 2011, 6, e23787. [Google Scholar] [CrossRef] [PubMed]
- Olive, V.; Jiang, I.; He, L. Mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010, 42, 1348–1354. [Google Scholar] [CrossRef] [PubMed]
- Davalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somel, M.; Liu, X.; Tang, L.; Yan, Z.; Hu, H.; Guo, S.; Jiang, X.; Zhang, X.; Xu, G.; Xie, G.; et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011, 9, e1001214. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Yu, H.R.; Huang, L.T.; Huang, H.C.; Chen, R.F.; Lin, I.C.; Ou, C.Y.; Hsu, T.Y.; Yang, K.D. MiRNA-125b regulates TNF-α production in CD14+ neonatal monocytes via post-transcriptional regulation. J. Leukoc. Biol. 2012, 92, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Kaati, G.; Bygren, L.O.; Edvinsson, S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur. J. Hum. Genet. 2002, 10, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, J.; Heslehurst, N.; Hall, J.; Schoenaker, D.A.J.M.; Hutchinson, J.; Cade, J.E.; Poston, L.; Barrett, G.; Crozier, S.R.; Barker, M.; et al. Before the beninning: Nutrition and lifestyle in the preconception period and its importance for future health. Lancet 2018, 391, 1830–1841. [Google Scholar] [CrossRef]
- Kort, H.I.; Massey, J.B.; Elsner, C.W.; Mitchell-Leef, D.; Shapiro, D.B.; Witt, M.A.; Roudebush, W.E. Impact of body mass index values on sperm quantity and quality. J. Androl. 2006, 27, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Fullston, T.; Ohlsson Teague, E.M.; Palmer, N.O.; DeBlasio, M.J.; Mitchell, M.; Corbett, M.; Print, C.G.; Owens, J.A.; Lane, M. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J. 2013, 27, 4226–4243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakos, H.W.; Henshaw, R.C.; Mitchell, M.; Lane, M. Paternal body mass index is associated with decreased blastocyst development and reduced live birth rates following assisted reproductive technology. Fertil. Steril. 2011, 95, 1700–1704. [Google Scholar] [CrossRef] [PubMed]
- Slyvka, Y.; Zhang, Y.; Nowak, F.V. Epigenetic effects of paternal diet on offspring: Emphasis on obesity. Endocrine 2015, 48, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Carone, B.R.; Fauquier, L.; Habib, N.; Shea, J.M.; Hart, C.E.; Li, R.; Bock, C.; Li, C.; Gu, H.; Zamore, P.D.; et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 2010, 143, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Sjöberg, L.; Pitkäniemi, J.; Haapala, L.; Kaaja, R.; Tuomilehto, J. Fertility in people with childhood-onset type 1 diabetes. Diabetologia 2013, 56, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Wiebe, J.C.; Santana, A.; Medina-Rodríguez, N.; Hernández, M.; Nóvoa, J.; Mauricio, D.; Wägner, A.M. T1DGC. Fertility is reduced in women and in men with type 1 diabetes: Results from the Type 1 Diabetes Genetics Consortium (T1DGC). Diabetologia 2014, 57, 2501–2504. [Google Scholar] [CrossRef] [PubMed]
- Agbaje, I.M.; Rogers, D.A.; McVicar, C.M.; McClure, N.; Atkinson, A.B.; Mallidis, C.; Lewis, S.E. Insulin dependant diabetes mellitus: Implications for male reproductive function. Hum. Reprod. 2007, 22, 1871–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Vignera, S.; Condorelli, R.A.; Di Mauro, M.; Lo Presti, D.; Mongioì, L.M.; Russo, G.; Calogero, A.E. Reproductive function in male patients with type 1 diabetes mellitus. Andrology 2015, 3, 1082–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Yang, C.R.; Wei, Y.P.; Zhao, Z.A.; Hou, Y.; Schatten, H.; Sun, Q.Y. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl. Acad. Sci. USA 2014, 111, 1873–1878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Yan, M.; Cao, Z.; Li, X.; Zhang, Y.; Shi, J.; Feng, G.H.; Peng, H.; Zhang, X.; Zhang, Y.; et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016, 351, 397–400. [Google Scholar] [CrossRef] [PubMed]
| After 100 g Glucose [6] | After 100 g Glucose [7] | After 75 g Glucose [8,9] | |
|---|---|---|---|
| Fasting | 95/5.3 | 105/5.8 | 92/5.1 |
| 1 h | 180/10.0 | 190/10.6 | 180/10.0 |
| 2 h | 155/8.6 | 165/9.2 | 153/8.5 |
| 3 h | 140/7.8 | 145/8.0 | -- |
| Author Year [Reference] | miRNA | Methods/Control | Diagnostic Criteria | GA (wk) | N (GD/C) | Tissue | Results | Comments |
|---|---|---|---|---|---|---|---|---|
| Maternal blood | ||||||||
| Zhao 2011 [81] | miRNA profiling miR-132 miR-29a miR-222 | Array (Applied Biosystems)+qPCR/cel-miR-36 | At 24–28 weeks. Two-step. 75 g 3 h OGTT | 16–19 | 36/36 | Serum | Reduced in the women developing GD | Profiling in two pools of 24 samples. 10 miRNA validated. miR-222, miR-29a validated in 2 external samples (16/group in each centre) |
| Zhu 2015 [62] | miRNA profiling miR-16-5p miR-17-5p miR-19a-3p miR-19b-3p miR-20a-5p | Massive sequencing profiling+qPCR/miR-221 | Two-step. 75 g, 3 h OGTT | 16–19 | 10/10 (one pool each) | Plasma | Upregulated in the women developing GD | Pilot discovery study. The 5 mentioned miRNAs, validated with q-RT-PCR also in the pools, apparently |
| Cao 2017 [61] | miR-16-5p miR-17-5p miR-20a-5p miR-19a-3p miR-19b-3p | qRT-PCR/U6 | Three-hour 75 g OGTT | Serial sampling until week 24–28 | 85/72 | Plasma | miR-16-5p, miR-17-5p, miR-20a-5p up-regulated in GD at diagnosis miR-16-5p and miR-17-5p in earlier pregnancy. AUC ROC 0.92 for 16-5p | Selection based on [56] |
| Wander 2017 [78] | miR-126-3p miR-155-5p miR-21-3p miR-146b-5p miR-210-3p miR-222-3p miR-223-3p miR-517-5p miR-518a-3p miR-29a-3p | qPCR/miR-423-3p | Two-step diagnosis 100 g OGTT [6] | 7–22 | 36/80 | Plasma | miR-155-5p and miR-21-3p up-regulated in GD Differential regulation according to maternal obesity and offspring sex. Analysis adjusted for gestational age (logistic regression) | Ten candidate miRNA selected based on previous association with pregnancy complications |
| Pheiffer 2018 [70] | miR-16-5p miR-17-5p miR-19a-3p miR-19b-3p miR-20a-5p miR-29a-3p miR-132-3p miR-222-3p | qPCR array (Qiagen)/Cel-miR-39 | Two-hour 75 g OGTT at 24–28 weeks of pregnancy [8] | 13–31 | 28/53 | Serum | miR-20a-5p and miR-222-3p down-regulated in GD | miRNAs selected for previous association with GD. Only 20a-5p significant predictor of GD in multivariate logistic regression analysis |
| Tagoma 2018 [75] | Array of 84 miRNA miR-195-5p | miRNA array+qPCR/Cel-miR-39 | Two-hour OGTT with 75 g glucose during the second trimester of pregnancy [8] | 23–31 | 13/9 | Plasma | miR-195 upregulated in GD | Higher gestational age in GD. Of the 84 miRNA array, 15 were upregulated in GD. Top 3 were validated with qRT-PCR. miR195-5p has targets in lipid metabolism |
| Sebastiani 2017 [72] | Array of 384 miRNAs miR-330-3p | TaqMan miRNA Human Array Panel A platform (Life-technologies)+qPCR/miR-320 and miR-374a | Two-hour 75 g OGTT at 16–19 weeks or 24–28 weeks of pregnancy [8] | 24–33 | 25/14$ | Plasma | miR-330-3p upregulated in GD | Bimodal expression observed in GD. Low expression associated with less caesarean sections |
| Collares 2013 [63] | miRNA profiling | Array (Agilent)/Median expression, quantile normalization | GD vs. non-pregnant DM1 and DM2 | 28–37 | 6/7/7 | Blood (PBMC) | The authors conclude that miRNA profiles distinguished types of diabetes | Non-pregnant DM1 and DM2 differed from GD in age and sex distribution, as well as pregnancy state |
| He 2017 [65] | miR-494 | RT-qPCR/U6 | NS | NS | 20/20 | Peripheral blood | Down-regulated in GD Overexpression of miR-494 enhanced insulin secretion and increased total insulin content, induced cell proliferation and inhibited cell apoptosis in INS1 cells | From pre-existing database in Chinese women with differential expression in GD. MiR-494 chosen because of relation with apoptosis in other tissues |
| Lamadrid 2018 [67] | miR-125b-5p and 11 other miRNA | RT-qPCR array/Cel-miR-39-3p | American diabetes association 2016 two-step protocol (any trimester) | Sampling in 3 trimesters | 14/27 | Serum | miR-183-5p, miR-200-3p, miR-125b-5p, miR-1290 higher in GD in first trimester | miRNAs selected because of previous association with neural development |
| Rahimi 2014 [71] | Drosha, Dicer, DGCR8 mRNA | qPCR/RPL38 mRNA | NS | Average 32–33 (SD2.7) | 20/20 | Whole blood-lysis | Drosha and Dicer Upregulated and DGCR8 down-regulated in GD | Components of the miRNA machinery are altered in GD |
| Stirm 2018 [74] | miR-340 | miRNA profiling (massive RNA sequencing-Illumina HiSeq. 2500 platform)+qPCR/RNU6B | IADPSG recommendations [8] | 24–32 | 30/30 | Whole blood cells from mothers and offspring, lymphocytes | Upregulated in GD and up-regulated by glucose, down by insulin, in vitro (lymphocytes) | Screening in 8/8, validation in 30/30 and 8/8 offspring. 29 miRNA upregulated in GD, one validated by qRT-PCR. |
| Xu 2017 [79] | miRNA profiling Validation of miR-503 | Array (Agilent)+qPCR/NS | NS | NS | 3/3 25/25 | Placenta Maternal peripheral blood | Upregulated in GD. In vitro inhibition of miR-503 increases insulin content and secretion in INS1 cells | One of the 28 upregulated miRNAs in an array was selected. Array in placentas, validation in blood |
| Placenta | ||||||||
| Cao 2016 [60] | miR-98 | qRT-PCR/U6 snRNA | NS | 39+/−1 | 193/202 | Placenta | Upregulated in GD, increases global methylation | Single miRNA |
| Li 2015 [68] | miR-508-3p miR-27a miR-9 miR-137 miR-92a miR-33a miR-30d miR-362-5p miR-502-5p | Array (Agilent), qPCR/U6 snRNA | Fasting glucose >5.1 mmol/L | Term | 15/15 * | Placenta | 29 differently expressed miRNA in the array, 9 replicated by qPCR. miR-508-3p upregulated and the rest, down-regulated | In silico prediction shows EGFR/PI3K/Akt pathway involvement, which plays a role in foetal growth. EGFR/PI3K/Akt upregulated in GD placentas |
| Muralimanoharan 2016 [69] | mir-143 | qPCR/U18 RNA | NS. Include A1 and A2 | Term (38–39), caesarean section only in GD | 12/6 | Placenta (trophoblasts) | 50% reduction in A2 but not A1 GD | Selected for previous association with metabolic switch between glycolysis and oxidation. Overexpression of miR-143 reduces aerobic glycolysis and rescues mitochondrial complexes in trophoblast cells |
| Tan 2016 [76] | miR-95 miR-548 miR-1246 | qPCR?/U6 | NS | 38+/−4 | 45/40 | Placenta | miR-95 and miR-548 upregulatedmiR-1246 downregulated | Correlated with serum lipids and adipokines, also with placental GLUTs |
| Zhao 2014 [82] | miR-518d | qPCR/snRNA U6 | 2 h, 75 g OGTT: fasting glucose > 5.6 mmol/L or 2 h > 8.6 mmol/L | 37–40 | 40/40 | Placenta | Upregulated in GD. PPAR-α is a predicted and validated target, with inverse placental protein expression | miRNA selected as placental marker |
| Yan 2018 [80] | Circular RNA profiling | NGS (Illumina HiSeq)+qPCR/GAPDH | NS | 38–41 | 30/30 | Placenta | From a total of 48,270 circRNAs, 227 were upregulated and 255 down-regulated | Enrichment of pathways involved in glucose and lipid metabolism |
| Other tissues | ||||||||
| Floris 2015 [64] | miR-101 | qPCR/SnU6B | At 24–28 weeks’ gestation with fasting glycemia >95 mg/dL and >155 mg/dL two hours after a 75 g OGTT | Term (NS) | 18/18 | HUVEC | Increased expression in HUVECs of GD, which affects their survival and functional capabilities | Mir101 selected due to relationship with endothelial function and angiogenesis |
| Tryggestad 2016 [77] | miR-30c-5p miR-452-5p miR-126-3p miR-130b-3p miR-148a-3p miR-let-7a-5p miR-let-7g-5p | Array+qPCR on 32 miRNA/RNU 48 | American Diabetes Association criteria [9] (A1 and A2) | NS | 7/12 | HUVEC | Among 19 detectable by qPCR, 7 upregulated in offspring of GD | Functional studies show decreased expression of AMPKa by transfection of miR-130 and miR-148a. AMPKa is known to stimulate glucose uptake and fatty acid oxidation |
| Shi 2014 [73] | miRNA expression profiling | AFFX miRNA expression chips +qPCR/miR-16 | American Diabetes Association criteria (A1) [9] | 38–39 (Caesarean section) | 13/13 | Omental adipose tissue | miR-222 upregulated in GD | One of 17 differentially expressed miRNAs. |
| Houshmand-Oeregaard 2018 [66] | mirR-15a miR-15b | qPCR/RNU48 | NS. Screening performed in high-risk women | NA | 76/42 | Muscle of adult offspring of women with GD | Both miRNAs upregulated. Expression was correlated with personal and maternal glucose | miRNAs selected based on previous results. Involved in insulin secretion and resistance. |
| Author Year [Reference] | Species | miRNA | Target Gene | Methods | Tissue | Results | Comments |
|---|---|---|---|---|---|---|---|
| Dong 2016 [87] | Mice | 149 miRNAs | 2111 potential target genes | RNA microarrays/RT-qPCR | Heart | Cardiac development-related pathways (STAT3 and IGF-1) and transcription factors associated to altered miRNAs, leading to CHDs | Oxidative stress as responsible for dysregulation of miRNAs |
| Dong D 2016 [88] | Mice | miR-17 downregulated | Thioredoxin interactive protein upregulated | RT-qPCR | Neural stem cells | Proapoptotic hyperglycaemia (via ASK1 pathway) | ASK1 leads to NTDs |
| Ibarra 2018 [85] | Human | miR-125-5p miR-20a-5p | RNA-Seq/qPCR | Placenta | Classifiers composed by 2–3 miRNAs were identified | miR-125-5p and miR-20a-5p were present in classifiers for type 1 and type 2 diabetes | |
| Jiangs 2017 [89] | Human | miR130b-3p upregulated | PGC-1 downregulation | RT-qPCR | Placental trophoblastic cell line (Be Wo cells) | Impaired mitochondrial function and oxidative stress which affects foetal development | Inhibition of miR-130b-3p reverted effects found. |
| Ramya, 2017 [90] | Mice | miRNA-30 family miR-30b upregulated | Sirtuin gene downregulated | RNA microarrays/RT-qPCR | Neural stem cells | Decreased Sirt 1 protein: altered neuron/glia ratio | Diabetic induced NTDs via miRNAs |
| Shi 2017 [91] | Mice | Exosomal miRNA | RNA-Seq analysis | Blood | Maternal exosomal miRNAs in diabetes contribute to cardiac development deficiency leading to CHDs | Maternal exosomal miRNAs in diabetes could cross the maternal-foetal barrier | |
| Shyama sundar 2013 [92] | Mice | miR-200a, miR-200b, miR-466a-3p, miR-466d-3p Downregulated | Dcx and Pafah1b1 upregulated | RT-qPCR | Neural stem cells | Knock down of miRs increases gliogenesis and neurogenesis which if impaired may form the basis of NTDs | Hyperglycaemia alters epigenetic-reversible mechanisms in NSCs. |
| Wang 2017 [93] | Mice | miR192-2 upregulated | PGC-1 gene upregulated | RT-qPCR | Neuroepithelial cells | Less NTDs by diminishing autophagy | These regulate the teratogenicity of hyperglycaemia |
| Zhao 2017 [94] | Mice | miR-505-5p, miR-770-5p and miR-1a-1-5p differentially expressed | Association with diabetic embryopathy was sought | NGS | Embryos (9.5 days) | Putative target genes under-represented in a database of genes associated with cardiovascular and neural malformations | No differences in miRNA expression at 8.5 days |
| Zhao 2018 [95] | Mice | miR-27a upregulated | Nuclear factor erythroid 2-related factor 2 downregulated | RT-qPCR | Neural stem cells | Increased oxidative stress that suppresses Nuclear factor erythroid 2-related factor 2 and its responsive antioxidant enzymes resulting in diabetic embryopathy | Protein reduction also followed a (glucose) dose and time dependent-manner |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibarra, A.; Vega-Guedes, B.; Brito-Casillas, Y.; Wägner, A.M. Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application. Non-Coding RNA 2018, 4, 32. https://doi.org/10.3390/ncrna4040032
Ibarra A, Vega-Guedes B, Brito-Casillas Y, Wägner AM. Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application. Non-Coding RNA. 2018; 4(4):32. https://doi.org/10.3390/ncrna4040032
Chicago/Turabian StyleIbarra, Adriana, Begoña Vega-Guedes, Yeray Brito-Casillas, and Ana M. Wägner. 2018. "Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application" Non-Coding RNA 4, no. 4: 32. https://doi.org/10.3390/ncrna4040032
APA StyleIbarra, A., Vega-Guedes, B., Brito-Casillas, Y., & Wägner, A. M. (2018). Diabetes in Pregnancy and MicroRNAs: Promises and Limitations in Their Clinical Application. Non-Coding RNA, 4(4), 32. https://doi.org/10.3390/ncrna4040032