Function and Evolution of Nematode RNAi Pathways
Abstract
:1. Introduction
2. General Aspects of the sRNA Pathways of C. elegans
3. The 21U-RNA Pathway
4. The 26G-RNA Pathway
4.1. ALG-3/4 Branch 26G-RNAs
4.2. ERGO-1 Branch 26G-RNAs
5. 22G-RNA Pathways: A Nexus of Gene Regulation
6. CSR-1 Pathway and Periodic An/Tn Clusters Inhibit PRG-1-Mediated Silencing
7. Parental Contribution of sRNAs
8. Cross- and Self-Regulation of RNAi-Like Pathways
9. Overview of RNAi-Like Pathways in Other Nematodes
10. Parallels between Nematode RNAi-Like Pathways and Metazoan piRNAs
10.1. Germline Expression
10.2. Function
10.3. sRNA Features
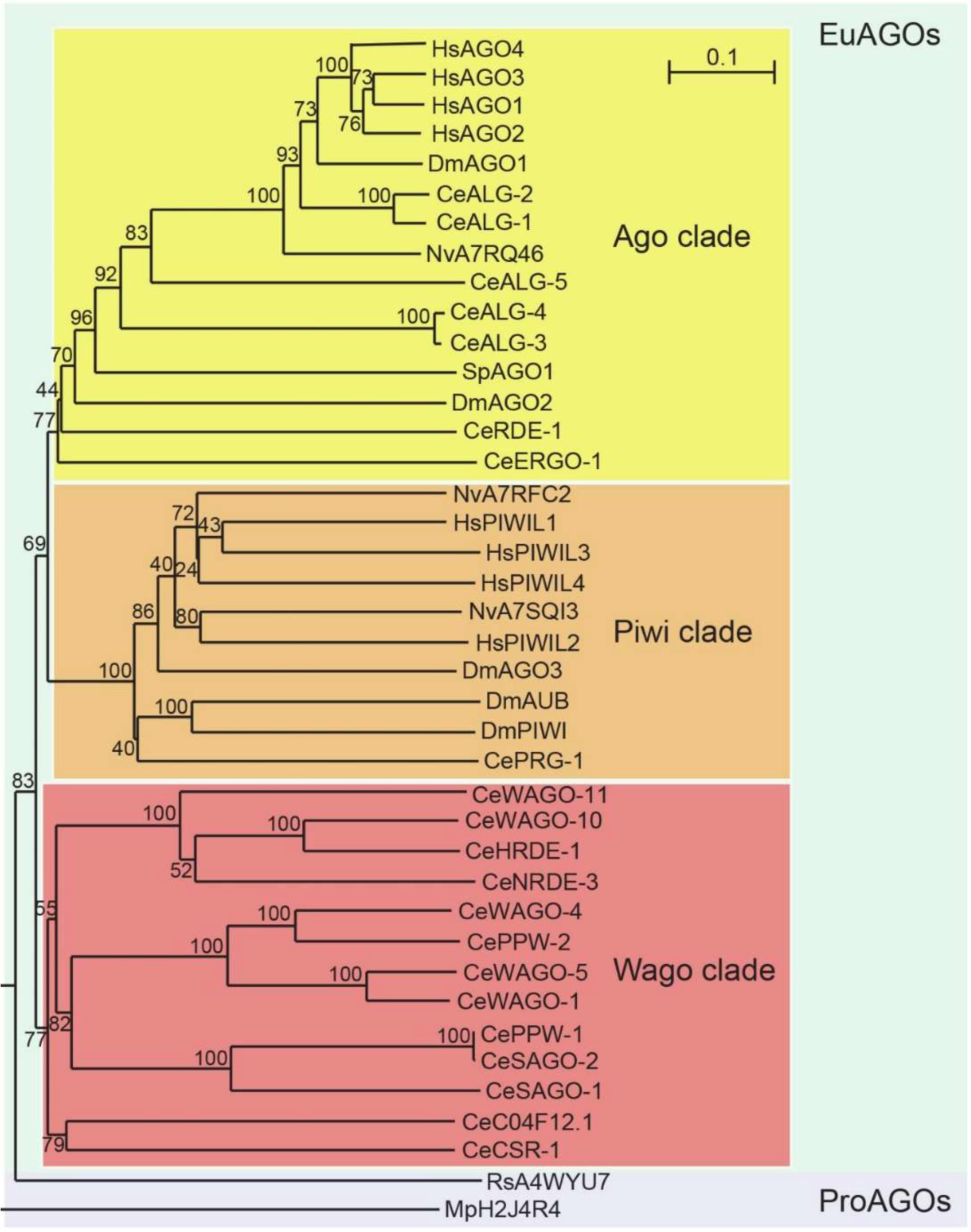
10.4. Sequence Homology of AGO Proteins
10.5. Evolutionarily Conserved Cofactors
11. Concluding Remarks
- Long piRNA precursor transcripts are typically transcribed by RNA PolII from large genomic regions known as piRNA clusters [8,18]. These clusters harbor degenerate TE copies, relics of old TE invasions. piRNA clusters are considered TE traps, in that once a TE jumps in a cluster at random, complementary piRNAs are produced and this will lead to the silencing of other homolog TE copies;
- After export of piRNA precursors to the cytoplasm, piRNA production starts by cleavage of piRNA precursors by Piwi AGOs and specialized endo- and exonucleases [18,127]. piRNA maturation is completed when Hen1 enzymes 2′-O-methylate the 3′ end of the piRNA. This modification is thought to provide stability to sRNAs;
- The so-called “ping-pong” amplification cycle involves typically two relaying Piwi AGOs [8,18]. The catalytic activity of one Piwi generates a piRNA that is accepted by another Piwi and this event is repeated in a loop. This feedforward loop allows for robust amplification of the piRNA pool and faithful silencing. The “ping-pong” amplification cycle seems to be an evolutionarily conserved mechanism in Piwi/piRNA pathways [127];
- Piwi-RISCs silence their targets both by post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) mechanisms [8,18,128]. PTGS is mainly dependent on target cleavage by Piwi AGOs, whereas TGS involves at least one Piwi AGO that is shuttled to the nucleus to target nascent RNAs. Nuclear Piwi AGOs are not sufficient for TGS. Interactions with other factors, such as histone methyltransferases, are required to establish repressive chromatin at target loci;
- Piwi/piRNA pathways function as an adaptive immune system against genetic parasites. Several features of adaptive immune systems are shared by Piwi/piRNA pathways, like the ability to recognize the threat, initiate a response, amplify the response and keep a memory of the response for further encounters. Memory of past encounters is embedded in piRNA clusters and is thus transmitted to the next generation. In addition, Piwi-RISCs may be directly inherited by the progeny in order to jump-start piRNA biogenesis in the next generation.
- Several lines of evidence suggest that Piwi/piRNA pathways and TEs are engaged in an evolutionary arms race, consistent with the “Red Queen” hypothesis [13,14,15,16]. Initially developed for host-parasite interactions, this theory may be applicable in the nucleic acid world to Piwi/piRNA pathways and TEs as genetic parasites. In this light, genetic changes beneficial to TEs are counteracted by genetic changes in piRNA pathway factors that eliminate or attenuate the TE advantage. In fact, the Drosophila genus seems to be rich in examples supporting an arms race between hosts and TEs [14,15,16]. Also, many factors involved in Piwi/piRNA pathways in diverse organisms are not evolutionarily conserved, suggesting that these pathways are evolving fast, potentially in response to TEs [13].
Funding
Acknowledgments
Conflicts of Interest
References
- Ågren, J.A.; Clark, A.G. Selfish genetic elements. PLoS Genet. 2018, 14, e1007700. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Wolf, Y.I. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu. Rev. Microbiol. 2017, 71, 233–261. [Google Scholar] [CrossRef]
- Schlee, M.; Hartmann, G. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 2016, 16, 566–580. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wang, Y.; Macfarlan, T.S. The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution. Trends Genet. 2017, 33, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef] [Green Version]
- Ketting, R.F. The Many Faces of RNAi. Dev. Cell 2011, 20, 148–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2018. [Google Scholar] [CrossRef]
- Swarts, D.C.; Makarova, K.; Wang, Y.; Nakanishi, K.; Ketting, R.F.; Koonin, E.V.; Patel, D.J.; van der Oost, J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014, 21, 743–753. [Google Scholar] [CrossRef] [Green Version]
- Madhani, H.D. The Frustrated Gene: Origins of Eukaryotic Gene Expression. Cell 2013, 155, 744–749. [Google Scholar] [CrossRef] [Green Version]
- Pearson, P.N. Red Queen Hypothesis. Encycl. Life Sci. 2001. [Google Scholar] [CrossRef]
- Obbard, D.J.; Gordon, K.H.J.; Buck, A.H.; Jiggins, F.M. The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009, 364, 99–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, W.H.; Hadfield, J.D.; Obbard, D.J. RNA-Interference Pathways Display High Rates of Adaptive Protein Evolution in Multiple Invertebrates. Genetics 2018, 208, 1585–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parhad, S.S.; Tu, S.; Weng, Z.; Theurkauf, W.E. Adaptive Evolution Leads to Cross-Species Incompatibility in the piRNA Transposon Silencing Machinery. Dev. Cell 2017, 43, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.; Wong, A.; Poh, Y.-P.; Theurkauf, W.E.; Jensen, J.D. Recurrent and recent selective sweeps in the piRNA pathway. Evolution 2013, 67, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Vermaak, D.; Henikoff, S.; Malik, H.S. Positive Selection Drives the Evolution of rhino, a Member of the Heterochromatin Protein 1 Family in Drosophila. PLoS Genet. 2005, 1, e9. [Google Scholar] [CrossRef]
- Olina, A.V.; Kulbachinskiy, A.V.; Aravin, A.A.; Esyunina, D.M. Argonaute Proteins and Mechanisms of RNA Interference in Eukaryotes and Prokaryotes. Biochem. Mosc. 2018, 83, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Fejes Tóth, K.; Aravin, A.A. piRNA Biogenesis in Drosophila melanogaster. Trends Genet. 2017, 33, 882–894. [Google Scholar] [CrossRef]
- Luteijn, M.J.; Ketting, R.F. PIWI-interacting RNAs: From generation to transgenerational epigenetics. Nat. Rev. Genet. 2013, 14, 523–534. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, dev161786. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perrière, G.; Gouy, M. WWW-query: An on-line retrieval system for biological sequence banks. Biochimie 1996, 78, 364–369. [Google Scholar] [CrossRef]
- Braukmann, F.; Jordan, D.; Miska, E. Artificial and natural RNA interactions between bacteria and C. elegans. RNA Biol. 2017, 14, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Marré, J.; Traver, E.C.; Jose, A.M. Extracellular RNA is transported from one generation to the next in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016, 113, 12496–12501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambros, V.; Lee, R.C.; Lavanway, A.; Williams, P.T.; Jewell, D. MicroRNAs and Other Tiny Endogenous RNAs in C. elegans. Curr. Biol. 2003, 13, 807–818. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.; Player, C.; Axtell, M.J.; Lee, W.; Nusbaum, C.; Ge, H.; Bartel, D.P. Large-Scale Sequencing Reveals 21U-RNAs and Additional MicroRNAs and Endogenous siRNAs in C. elegans. Cell 2006, 127, 1193–1207. [Google Scholar] [CrossRef]
- Cecere, G.; Zheng, G.X.Y.; Mansisidor, A.R.; Klymko, K.E.; Grishok, A. Promoters Recognized by Forkhead Proteins Exist for Individual 21U-RNAs. Mol. Cell 2012, 47, 734–745. [Google Scholar] [CrossRef]
- Conine, C.C.; Batista, P.J.; Gu, W.; Claycomb, J.M.; Chaves, D.A.; Shirayama, M.; Mello, C.C. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2010, 107, 3588–3593. [Google Scholar] [CrossRef] [Green Version]
- Gent, J.I.; Schvarzstein, M.; Villeneuve, A.M.; Gu, S.G.; Jantsch, V.; Fire, A.Z.; Baudrimont, A. A Caenorhabditis elegans RNA-Directed RNA Polymerase in Sperm Development and Endogenous RNA Interference. Genetics 2009, 183, 1297–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gent, J.I.; Lamm, A.T.; Pavelec, D.M.; Maniar, J.M.; Parameswaran, P.; Tao, L.; Kennedy, S.; Fire, A.Z. Distinct Phases of siRNA Synthesis in an Endogenous RNAi Pathway in C. elegans Soma. Mol. Cell 2010, 37, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Manoharan, A.P.; Harkins, T.T.; Bouffard, P.; Fitzpatrick, C.; Chu, D.S.; Thierry-Mieg, D.; Thierry-Mieg, J.; Kim, J.K. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 18674–18679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pak, J.; Fire, A. Distinct Populations of Primary and Secondary Effectors During RNAi in C. elegans. Science 2007, 315, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Sijen, T.; Steiner, F.A.; Thijssen, K.L.; Plasterk, R.H.A. Secondary siRNAs Result from Unprimed RNA Synthesis and Form a Distinct Class. Science 2007, 315, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Vasale, J.J.; Gu, W.; Thivierge, C.; Batista, P.J.; Claycomb, J.M.; Youngman, E.M.; Duchaine, T.F.; Mello, C.C.; Conte, D. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 3582–3587. [Google Scholar] [CrossRef] [Green Version]
- Aoki, K.; Moriguchi, H.; Yoshioka, T.; Okawa, K.; Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 2007, 26, 5007–5019. [Google Scholar] [CrossRef]
- Bagijn, M.P.; Goldstein, L.D.; Sapetschnig, A.; Weick, E.-M.; Bouasker, S.; Lehrbach, N.J.; Simard, M.J.; Miska, E.A. Function, Targets, and Evolution of Caenorhabditis elegans piRNAs. Science 2012, 337, 574–578. [Google Scholar] [CrossRef] [Green Version]
- Das, P.P.; Bagijn, M.P.; Goldstein, L.D.; Woolford, J.R.; Lehrbach, N.J.; Sapetschnig, A.; Buhecha, H.R.; Gilchrist, M.J.; Howe, K.L.; Stark, R.; et al. Piwi and piRNAs Act Upstream of an Endogenous siRNA Pathway to Suppress Tc3 Transposon Mobility in the Caenorhabditis elegans Germline. Mol. Cell 2008, 31, 79–90. [Google Scholar] [CrossRef]
- Lee, H.-C.; Gu, W.; Shirayama, M.; Youngman, E.; Conte, D., Jr.; Mello, C.C. C. elegans piRNAs Mediate the Genome-wide Surveillance of Germline Transcripts. Cell 2012, 150, 78–87. [Google Scholar] [CrossRef]
- Duchaine, T.F.; Wohlschlegel, J.A.; Kennedy, S.; Bei, Y.; Conte, D., Jr.; Pang, K.; Brownell, D.R.; Harding, S.; Mitani, S.; Ruvkun, G.; et al. Functional Proteomics Reveals the Biochemical Niche of C. elegans DCR-1 in Multiple Small-RNA-Mediated Pathways. Cell 2006, 124, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Shirayama, M.; Conte, D., Jr.; Vasale, J.; Batista, P.J.; Claycomb, J.M.; Moresco, J.J.; Youngman, E.M.; Keys, J.; Stoltz, M.J.; et al. Distinct Argonaute-Mediated 22G-RNA Pathways Direct Genome Surveillance in the C. elegans Germline. Mol. Cell 2009, 36, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.M.; Montgomery, T.A.; Breen, P.C.; Ruvkun, G. MUT-16 promotes formation of perinuclear Mutator foci required for RNA silencing in the C. elegans germline. Genes Dev. 2012, 26, 1433–1444. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.M.; Montgomery, B.E.; Breen, P.C.; Roovers, E.F.; Rim, Y.-S.; Ohsumi, T.K.; Newman, M.A.; van Wolfswinkel, J.C.; Ketting, R.F.; Ruvkun, G.; et al. MUT-14 and SMUT-1 DEAD Box RNA Helicases Have Overlapping Roles in Germline RNAi and Endogenous siRNA Formation. Curr. Biol. 2014, 24, 839–844. [Google Scholar] [CrossRef] [Green Version]
- Yigit, E.; Batista, P.J.; Bei, Y.; Pang, K.M.; Chen, C.-C.G.; Tolia, N.H.; Joshua-Tor, L.; Mitani, S.; Simard, M.J.; Mello, C.C. Analysis of the C. elegans Argonaute Family Reveals that Distinct Argonautes Act Sequentially during RNAi. Cell 2006, 127, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Hammell, C.M.; Ambros, V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA 2006, 12, 589–597. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V.; Ruvkun, G. Recent Molecular Genetic Explorations of Caenorhabditis elegans MicroRNAs. Genetics 2018, 209, 651–673. [Google Scholar]
- Batista, P.J.; Ruby, J.G.; Claycomb, J.M.; Chiang, R.; Fahlgren, N.; Kasschau, K.D.; Chaves, D.A.; Gu, W.; Vasale, J.J.; Duan, S.; et al. PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Mol. Cell 2008, 31, 67–78. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Reinke, V. A C. elegans Piwi, PRG-1, Regulates 21U-RNAs during Spermatogenesis. Curr. Biol. 2008, 18, 861–867. [Google Scholar] [CrossRef]
- Simon, M.; Sarkies, P.; Ikegami, K.; Doebley, A.-L.; Goldstein, L.D.; Mitchell, J.; Sakaguchi, A.; Miska, E.A.; Ahmed, S. Reduced Insulin/IGF-1 Signaling Restores Germ Cell Immortality to Caenorhabditis elegans Piwi Mutants. Cell Rep. 2014, 7, 762–773. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Lee, H.-C.; Chaves, D.; Youngman, E.M.; Pazour, G.J.; Conte, D.; Mello, C.C. CapSeq and CIP-TAP Identify Pol II Start Sites and Reveal Capped Small RNAs as C. elegans piRNA Precursors. Cell 2012, 151, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Weick, E.-M.; Sarkies, P.; Silva, N.; Chen, R.A.; Moss, S.M.M.; Cording, A.C.; Ahringer, J.; Martinez-Perez, E.; Miska, E.A. PRDE-1 is a nuclear factor essential for the biogenesis of Ruby motif-dependent piRNAs in C. elegans. Genes Dev. 2014, 28, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Billi, A.C.; Freeberg, M.A.; Day, A.M.; Chun, S.Y.; Khivansara, V.; Kim, J.K. A Conserved Upstream Motif Orchestrates Autonomous, Germline-Enriched Expression of Caenorhabditis elegans piRNAs. PLoS Genet. 2013, 9, e1003392. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.; Kosalka, J.; Berkyurek, A.C.; Stempor, P.; Feng, X.; Mao, H.; Zeng, C.; Li, W.-J.; Yan, Y.-H.; Dong, M.-Q.; et al. The USTC co-opts an ancient machinery to drive piRNA transcription in C. elegans. Genes Dev. 2018. [Google Scholar] [CrossRef]
- Goh, W.-S.S.; Seah, J.W.E.; Harrison, E.J.; Chen, C.; Hammell, C.M.; Hannon, G.J. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev. 2014, 28, 797–807. [Google Scholar] [CrossRef]
- Kasper, D.M.; Wang, G.; Gardner, K.E.; Johnstone, T.G.; Reinke, V. The C. elegans SNAPc Component SNPC-4 Coats piRNA Domains and Is Globally Required for piRNA Abundance. Dev. Cell 2014, 31, 145–158. [Google Scholar] [CrossRef]
- Beltran, T.; Barruso, C.; Birkle, T.; Stevens, L.; Schwartz, H.T.; Sternberg, P.; Fradin, H.; Gunsalus, K.; Piano, F.; Martinez-Perez, E.; et al. Evolutionary analysis implicates RNA polymerase II pausing and chromatin structure in nematode piRNA biogenesis. bioRxiv 2018, 281360. [Google Scholar] [CrossRef]
- De Albuquerque, B.F.M.; Luteijn, M.J.; Rodrigues, R.J.C.; van Bergeijk, P.; Waaijers, S.; Kaaij, L.J.T.; Klein, H.; Boxem, M.; Ketting, R.F. PID-1 is a novel factor that operates during 21U-RNA biogenesis in Caenorhabditis elegans. Genes Dev. 2014, 28, 683–688. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.J.C.; de Jesus Domingues, A.M.; Hellmann, S.; Dietz, S.; de Albuquerque, B.; Renz, C.; Ulrich, H.D.; Butter, F.; Ketting, R. PETISCO is a novel protein complex required for 21U RNA biogenesis and embryonic viability. bioRxiv 2018, 463711. [Google Scholar] [CrossRef]
- Zeng, C.; Weng, C.; Wang, X.; Yan, Y.-H.; Li, W.-J.; Xu, D.; Hong, M.; Liao, S.; Feng, X.; Dong, M.-Q.; et al. Differential phase partition of a PICS complex is required for piRNA processing and chromosome segregation in C. elegans. bioRxiv 2018, 463919. [Google Scholar] [CrossRef]
- Tang, W.; Tu, S.; Lee, H.-C.; Weng, Z.; Mello, C.C. The RNase PARN-1 Trims piRNA 3′ Ends to Promote Transcriptome Surveillance in C. elegans. Cell 2016, 164, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Billi, A.C.; Alessi, A.F.; Khivansara, V.; Han, T.; Freeberg, M.; Mitani, S.; Kim, J.K. The Caenorhabditis elegans HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs. PLoS Genet. 2012, 8, e1002617. [Google Scholar] [CrossRef] [PubMed]
- Kamminga, L.M.; van Wolfswinkel, J.C.; Luteijn, M.J.; Kaaij, L.J.T.; Bagijn, M.P.; Sapetschnig, A.; Miska, E.A.; Berezikov, E.; Ketting, R.F. Differential Impact of the HEN1 Homolog HENN-1 on 21U and 26G RNAs in the Germline of Caenorhabditis elegans. PLoS Genet. 2012, 8, e1002702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montgomery, T.A.; Rim, Y.-S.; Zhang, C.; Dowen, R.H.; Phillips, C.M.; Fischer, S.E.J.; Ruvkun, G. PIWI Associated siRNAs and piRNAs Specifically Require the Caenorhabditis elegans HEN1 Ortholog henn-1. PLoS Genet. 2012, 8, e1002616. [Google Scholar] [CrossRef] [Green Version]
- De Albuquerque, B.F.M.; Placentino, M.; Ketting, R.F. Maternal piRNAs Are Essential for Germline Development following De Novo Establishment of Endo-siRNAs in Caenorhabditis elegans. Dev. Cell 2015, 34, 448–456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, C.M.; Brown, K.C.; Montgomery, B.E.; Ruvkun, G.; Montgomery, T.A. piRNAs and piRNA-Dependent siRNAs Protect Conserved and Essential C. elegans Genes from Misrouting into the RNAi Pathway. Dev. Cell 2015, 34, 457–465. [Google Scholar] [CrossRef]
- Shen, E.-Z.; Chen, H.; Ozturk, A.R.; Tu, S.; Shirayama, M.; Tang, W.; Ding, Y.-H.; Dai, S.-Y.; Weng, Z.; Mello, C.C. Identification of piRNA Binding Sites Reveals the Argonaute Regulatory Landscape of the C. elegans Germline. Cell 2018, 172, 937–951. [Google Scholar] [CrossRef]
- Zhang, D.; Tu, S.; Stubna, M.; Wu, W.-S.; Huang, W.-C.; Weng, Z.; Lee, H.-C. The piRNA targeting rules and the resistance to piRNA silencing in endogenous genes. Science 2018, 359, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Almeida, M.V.; Dietz, S.; Redl, S.; Karaulanov, E.; Hildebrandt, A.; Renz, C.; Ulrich, H.D.; König, J.; Butter, F.; Ketting, R.F. GTSF-1 is required for formation of a functional RNA-dependent RNA Polymerase complex in Caenorhabditis elegans. EMBO J. 2018, 37, e99325. [Google Scholar] [CrossRef]
- Thivierge, C.; Makil, N.; Flamand, M.; Vasale, J.J.; Mello, C.C.; Wohlschlegel, J.D.C., Jr.; Duchaine, T.F. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat. Struct. Mol. Biol. 2012, 19, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Blumenfeld, A.L.; Jose, A.M. Reproducible features of small RNAs in C. elegans reveal NU RNAs and provide insights into 22G RNAs and 26G RNAs. RNA 2016, 22, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Welker, N.C.; Maity, T.S.; Ye, X.; Aruscavage, P.J.; Krauchuk, A.A.; Liu, Q.; Bass, B.L. Dicer’s Helicase Domain Discriminates dsRNA Termini to Promote an Altered Reaction Mode. Mol. Cell 2011, 41, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Pavelec, D.M.; Lachowiec, J.; Duchaine, T.F.; Smith, H.E.; Kennedy, S. Requirement for the ERI/DICER Complex in Endogenous RNA Interference and Sperm Development in Caenorhabditis elegans. Genetics 2009, 183, 1283–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmer, F.; Tijsterman, M.; Parrish, S.; Koushika, S.P.; Nonet, M.L.; Fire, A.; Ahringer, J.; Plasterk, R.H.A. Loss of the Putative RNA-Directed RNA Polymerase RRF-3 Makes C. elegans Hypersensitive to RNAi. Curr. Biol. 2002, 12, 1317–1319. [Google Scholar] [CrossRef]
- Welker, N.C.; Pavelec, D.M.; Nix, D.A.; Duchaine, T.F.; Kennedy, S.; Bass, B.L. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 2010, 16, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Conine, C.C.; Moresco, J.J.; Gu, W.; Shirayama, M.; Conte, D.; Yates, J.R.; Mello, C.C. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 2013, 155, 1532–1544. [Google Scholar] [CrossRef] [PubMed]
- Wallach, E.E.; Kandeel, F.R.; Swerdloff, R.S. Role of temperature in regulation of spermatogenesis and the use of heating as a method for contraception. Fertil. Steril. 1988, 49, 1–23. [Google Scholar] [CrossRef]
- Almeida, M.V.; de Jesus Domingues, A.M.; Ketting, R.F. Maternal and zygotic gene regulatory effects of endogenous RNAi pathways. bioRxiv 2018, 453266. [Google Scholar] [CrossRef]
- Zhuang, J.J.; Hunter, C.P. Tissue-specificity of Caenorhabditis elegans Enhanced RNAi Mutants. Genetics 2011, 188, 235–237. [Google Scholar] [CrossRef]
- Fischer, S.E.J.; Montgomery, T.A.; Zhang, C.; Fahlgren, N.; Breen, P.C.; Hwang, A.; Sullivan, C.M.; Carrington, J.C.; Ruvkun, G. The ERI-6/7 helicase acts at the first stage of an siRNA amplification pathway that targets recent gene duplications. PLoS Genet. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Newman, M.A.; Ji, F.; Fischer, S.E.J.; Anselmo, A.; Sadreyev, R.I.; Ruvkun, G. The surveillance of pre-mRNA splicing is an early step in C. elegans RNAi of endogenous genes. Genes Dev. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xu, F.; Mao, H.; Ji, J.; Yin, M.; Feng, X.; Guang, S. Nuclear RNAi Contributes to the Silencing of Off-Target Genes and Repetitive Sequences in Caenorhabditis elegans. Genetics 2014, 197, 121–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumesic, P.A.; Natarajan, P.; Chen, C.; Drinnenberg, I.A.; Schiller, B.J.; Thompson, J.; Moresco, J.J.; Yates, J.R.; Bartel, D.P.; Madhani, H.D. Stalled Spliceosomes Are a Signal for RNAi-Mediated Genome Defense. Cell 2013, 152, 957–968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.E.J.; Butler, M.D.; Pan, Q.; Ruvkun, G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 2008, 455, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Homolka, D.; Chen, K.-M.; Sachidanandam, R.; Fauvarque, M.-O.; Pillai, R.S. Recruitment of Armitage and Yb to a transcript triggers its phased processing into primary piRNAs in Drosophila ovaries. PLoS Genet. 2017, 13, e1006956. [Google Scholar] [CrossRef] [PubMed]
- Vourekas, A.; Zheng, K.; Fu, Q.; Maragkakis, M.; Alexiou, P.; Ma, J.; Pillai, R.S.; Mourelatos, Z.; Wang, P.J. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015, 29, 617–629. [Google Scholar] [CrossRef]
- Ashe, A.; Sapetschnig, A.; Weick, E.-M.; Mitchell, J.; Bagijn, M.P.; Cording, A.C.; Doebley, A.-L.; Goldstein, L.D.; Lehrbach, N.J.; Le Pen, J.; et al. piRNAs Can Trigger a Multigenerational Epigenetic Memory in the Germline of C. elegans. Cell 2012, 150, 88–99. [Google Scholar] [CrossRef]
- Luteijn, M.J.; van Bergeijk, P.; Kaaij, L.J.T.; Almeida, M.V.; Roovers, E.F.; Berezikov, E.; Ketting, R.F. Extremely stable Piwi-induced gene silencing in Caenorhabditis elegans. EMBO J. 2012, 31, 3422–3430. [Google Scholar] [CrossRef] [Green Version]
- Shirayama, M.; Seth, M.; Lee, H.-C.; Gu, W.; Ishidate, T.; Conte, D., Jr.; Mello, C.C. piRNAs Initiate an Epigenetic Memory of Nonself RNA in the C. elegans Germline. Cell 2012, 150, 65–77. [Google Scholar] [CrossRef]
- Buckley, B.A.; Burkhart, K.B.; Gu, S.G.; Spracklin, G.; Kershner, A.; Fritz, H.; Kimble, J.; Fire, A.; Kennedy, S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012, 489, 447–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guang, S.; Bochner, A.F.; Pavelec, D.M.; Burkhart, K.B.; Harding, S.; Lachowiec, J.; Kennedy, S. An Argonaute Transports siRNAs from the Cytoplasm to the Nucleus. Science 2008, 321, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Mao, H.; Zhu, C.; Zong, D.; Weng, C.; Yang, X.; Huang, H.; Liu, D.; Feng, X.; Guang, S. The Nrde Pathway Mediates Small-RNA-Directed Histone H3 Lysine 27 Trimethylation in Caenorhabditis elegans. Curr. Biol. 2015, 25, 2398–2403. [Google Scholar] [CrossRef] [Green Version]
- Guang, S.; Bochner, A.F.; Burkhart, K.B.; Burton, N.; Pavelec, D.M.; Kennedy, S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 2010, 465, 1097–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhart, K.B.; Guang, S.; Buckley, B.A.; Wong, L.; Bochner, A.F.; Kennedy, S. A Pre-mRNA–Associating Factor Links Endogenous siRNAs to Chromatin Regulation. PLoS Genet. 2011, 7, e1002249. [Google Scholar] [CrossRef]
- Sapetschnig, A.; Sarkies, P.; Lehrbach, N.J.; Miska, E.A. Tertiary siRNAs Mediate Paramutation in C. elegans. PLoS Genet. 2015, 11, e1005078. [Google Scholar] [CrossRef] [PubMed]
- Claycomb, J.M.; Batista, P.J.; Pang, K.M.; Gu, W.; Vasale, J.J.; van Wolfswinkel, J.C.; Chaves, D.A.; Shirayama, M.; Mitani, S.; Ketting, R.F.; et al. The Argonaute CSR-1 and Its 22G-RNA Cofactors Are Required for Holocentric Chromosome Segregation. Cell 2009, 139, 123–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seth, M.; Shirayama, M.; Gu, W.; Ishidate, T.; Conte, D., Jr.; Mello, C.C. The C. elegans CSR-1 Argonaute Pathway Counteracts Epigenetic Silencing to Promote Germline Gene Expression. Dev. Cell 2013, 27, 656–663. [Google Scholar] [CrossRef]
- Seth, M.; Shirayama, M.; Tang, W.; Shen, E.-Z.; Tu, S.; Lee, H.-C.; Weng, Z.; Mello, C.C. The Coding Regions of Germline mRNAs Confer Sensitivity to Argonaute Regulation in C. elegans. Cell Rep. 2018. [Google Scholar] [CrossRef] [PubMed]
- Wedeles, C.J.; Wu, M.Z.; Claycomb, J.M. Protection of Germline Gene Expression by the C. elegans Argonaute CSR-1. Dev. Cell 2013, 27, 664–671. [Google Scholar] [CrossRef] [PubMed]
- Frøkjær-Jensen, C.; Jain, N.; Hansen, L.; Davis, M.W.; Li, Y.; Zhao, D.; Rebora, K.; Millet, J.R.M.; Liu, X.; Kim, S.K.; et al. An Abundant Class of Non-coding DNA Can Prevent Stochastic Gene Silencing in the C. elegans Germline. Cell 2016, 166, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Minkina, O.; Hunter, C.P. Stable Heritable Germline Silencing Directs Somatic Silencing at an Endogenous Locus. Mol. Cell 2017, 65, 659–670. [Google Scholar] [CrossRef] [PubMed]
- Schott, D.; Yanai, I.; Hunter, C.P. Natural RNA interference directs a heritable response to the environment. Sci. Rep. 2014, 4, 7387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stoeckius, M.; Grün, D.; Rajewsky, N. Paternal RNA contributions in the Caenorhabditis elegans zygote. EMBO J. 2014, 33, 1740–1750. [Google Scholar] [CrossRef] [Green Version]
- Wan, G.; Fields, B.D.; Spracklin, G.; Shukla, A.; Phillips, C.M.; Kennedy, S. Spatiotemporal regulation of liquid-like condensates in epigenetic inheritance. Nature 2018, 557, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Feng, X.; Chen, X.; Weng, C.; Yan, Q.; Xu, T.; Hong, M.; Guang, S. A Cytoplasmic Argonaute Protein Promotes the Inheritance of RNAi. Cell Rep. 2018, 23, 2482–2494. [Google Scholar] [CrossRef] [PubMed]
- Stoeckius, M.; Maaskola, J.; Colombo, T.; Rahn, H.-P.; Friedländer, M.R.; Li, N.; Chen, W.; Piano, F.; Rajewsky, N. Large-scale sorting of C. elegans embryos reveals the dynamics of small RNA expression. Nat. Methods 2009, 6, 745–751. [Google Scholar] [CrossRef]
- Houri-Ze’evi, L.; Korem, Y.; Sheftel, H.; Faigenbloom, L.; Toker, I.A.; Dagan, Y.; Awad, L.; Degani, L.; Alon, U.; Rechavi, O. A Tunable Mechanism Determines the Duration of the Transgenerational Small RNA Inheritance in C. elegans. Cell 2016, 165, 88–99. [Google Scholar] [CrossRef]
- Pak, J.; Maniar, J.M.; Mello, C.C.; Fire, A. Protection from Feed-Forward Amplification in an Amplified RNAi Mechanism. Cell 2012, 151, 885–899. [Google Scholar] [CrossRef] [Green Version]
- Dalzell, J.J.; McVeigh, P.; Warnock, N.D.; Mitreva, M.; Bird, D.M.; Abad, P.; Fleming, C.C.; Day, T.A.; Mousley, A.; Marks, N.J.; et al. RNAi Effector Diversity in Nematodes. PLoS Negl. Trop. Dis. 2011, 5, e1176. [Google Scholar] [CrossRef]
- Holz, A.; Streit, A. Gain and Loss of Small RNA Classes—Characterization of Small RNAs in the Parasitic Nematode Family Strongyloididae. Genome Biol. Evol. 2017, 9, 2826–2843. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Selkirk, M.E.; Jones, J.T.; Blok, V.; Boothby, T.; Goldstein, B.; Hanelt, B.; Ardila-Garcia, A.; Fast, N.M.; Schiffer, P.M.; et al. Ancient and Novel Small RNA Pathways Compensate for the Loss of piRNAs in Multiple Independent Nematode Lineages. PLoS Biol. 2015, 13, e1002061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Montgomery, T.A.; Qi, Y.; Ruvkun, G. High-throughput sequencing reveals extraordinary fluidity of miRNA, piRNA, and siRNA pathways in nematodes. Genome Res. 2013, 23, 497–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Czech, B.; Crunk, A.; Wallace, A.; Mitreva, M.; Hannon, G.J.; Davis, R.E. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011. [Google Scholar] [CrossRef] [PubMed]
- De Wit, E.; Linsen, S.E.V.; Cuppen, E.; Berezikov, E. Repertoire and evolution of miRNA genes in four divergent nematode species. Genome Res. 2009. [Google Scholar] [CrossRef] [PubMed]
- Tu, S.; Wu, M.Z.; Wang, J.; Cutter, A.D.; Weng, Z.; Claycomb, J.M. Comparative functional characterization of the CSR-1 22G-RNA pathway in Caenorhabditis nematodes. Nucleic Acids Res. 2014. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Blaxter, M. Functional diversification of Argonautes in nematodes: An expanding universe. Biochem. Soc. Trans. 2013, 41, 881–886. [Google Scholar] [CrossRef]
- Gao, F.; Liu, X.; Wu, X.-P.; Wang, X.-L.; Gong, D.; Lu, H.; Xia, Y.; Song, Y.; Wang, J.; Du, J.; et al. Differential DNA methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis. Genome Biol. 2012, 13, R100. [Google Scholar] [CrossRef] [Green Version]
- Ishidate, T.; Ozturk, A.R.; Durning, D.J.; Sharma, R.; Shen, E.; Chen, H.; Seth, M.; Shirayama, M.; Mello, C.C. ZNFX-1 Functions within Perinuclear Nuage to Balance Epigenetic Signals. Mol. Cell 2018, 70, 639–649. [Google Scholar] [CrossRef]
- Jehn, J.; Gebert, D.; Pipilescu, F.; Stern, S.; Kiefer, J.S.T.; Hewel, C.; Rosenkranz, D. PIWI genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Commun. Biol. 2018, 1, 137. [Google Scholar] [CrossRef]
- Lewis, S.H.; Quarles, K.A.; Yang, Y.; Tanguy, M.; Frézal, L.; Smith, S.A.; Sharma, P.P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2017, 2, 174–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akkouche, A.; Mugat, B.; Barckmann, B.; Varela-Chavez, C.; Li, B.; Raffel, R.; Pélisson, A.; Chambeyron, S. Piwi Is Required during Drosophila Embryogenesis to License Dual-Strand piRNA Clusters for Transposon Repression in Adult Ovaries. Mol. Cell 2017, 66, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Dönertas, D.; Sienski, G.; Brennecke, J. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 2013, 27, 1693–1705. [Google Scholar] [CrossRef] [Green Version]
- Ohtani, H.; Iwasaki, Y.W.; Shibuya, A.; Siomi, H.; Siomi, M.C.; Saito, K. DmGTSF1 is necessary for Piwi-piRISC-mediated transcriptional transposon silencing in the Drosophila ovary. Genes Dev. 2013, 27, 1656–1661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemoto, N.; Yoshimura, T.; Miyazaki, S.; Tashiro, F.; Miyazaki, J. Gtsf1l and Gtsf2 Are Specifically Expressed in Gonocytes and Spermatids but Are Not Essential for Spermatogenesis. PLoS ONE 2016, 11, e0150390. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Watanabe, T.; Kuramochi-Miyagawa, S.; Takemoto, N.; Shiromoto, Y.; Kudo, A.; Kanai-Azuma, M.; Tashiro, F.; Miyazaki, S.; Katanaya, A.; et al. Mouse GTSF1 is an essential factor for secondary piRNA biogenesis. EMBO Rep. 2018, 19, e42054. [Google Scholar] [CrossRef] [PubMed]
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790. [Google Scholar] [CrossRef]
- Sato, K.; Siomi, M.C. Two distinct transcriptional controls triggered by nuclear Piwi-piRISCs in the Drosophila piRNA pathway. Curr. Opin. Struct. Biol. 2018, 53, 69–76. [Google Scholar] [CrossRef]
- Blaxter, M. Nematodes: The Worm and Its Relatives. PLoS Biol. 2011, 9, e1001050. [Google Scholar] [CrossRef]



| Clade I-II | Clade III | Clade IV | Clade V | ||
|---|---|---|---|---|---|
| DCR-1 | |||||
| RdRPs | RRF-3 | ||||
| EGO-1/RRF-1 | |||||
| Argonautes | ALG-1/2 | ||||
| CSR-1 | |||||
| HRDE-1 | |||||
| NRDE-3 | |||||
| ERGO-1 | |||||
| ALG-3 | |||||
| PRG-1 | |||||
| HENN-1 | |||||
| sRNA classes | 21U-RNA | ||||
| 22G-RNA | * | ||||
| 26G-RNA | |||||
| miRNA |
| Metazoan piRNAs | 21U-RNAs | 26G-RNAs | |||
|---|---|---|---|---|---|
| Expression | Predominantly germline and embryos | Germline and embryos | Germline and embryos | ||
| Length (in nucleotides) | 23–35 | 21 | ~26 | ||
| 5′ Bias | U | U | G | ||
| Phenotype | Mutants are sterile | Viable; transgenerational germline mortality | Some mutants cause sterility at higher temperatures | ||
| Cofactors | Piwi clade Argonautes | ||||
| Hen1 enzymes | |||||
| Gtsf1 proteins | |||||
| Armitage/MOV1-L1/ ERI-6/7 | |||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, M.V.; Andrade-Navarro, M.A.; Ketting, R.F. Function and Evolution of Nematode RNAi Pathways. Non-Coding RNA 2019, 5, 8. https://doi.org/10.3390/ncrna5010008
Almeida MV, Andrade-Navarro MA, Ketting RF. Function and Evolution of Nematode RNAi Pathways. Non-Coding RNA. 2019; 5(1):8. https://doi.org/10.3390/ncrna5010008
Chicago/Turabian StyleAlmeida, Miguel Vasconcelos, Miguel A. Andrade-Navarro, and René F. Ketting. 2019. "Function and Evolution of Nematode RNAi Pathways" Non-Coding RNA 5, no. 1: 8. https://doi.org/10.3390/ncrna5010008
APA StyleAlmeida, M. V., Andrade-Navarro, M. A., & Ketting, R. F. (2019). Function and Evolution of Nematode RNAi Pathways. Non-Coding RNA, 5(1), 8. https://doi.org/10.3390/ncrna5010008






