Long Non-Coding RNAs and the Innate Immune Response
Abstract
:1. Introduction
2. Characteristics of lncRNAs
3. LncRNAs and mRNAs Share Similar Biogenesis Pathways
4. LncRNAs Are Expressed at Lower Levels Compared to mRNAs
5. LncRNAs Expression Is Cell and Tissue Specific
6. LncRNAs Show Poor Evolutionary Conservation
7. Subcellular Localisation of lncRNAs
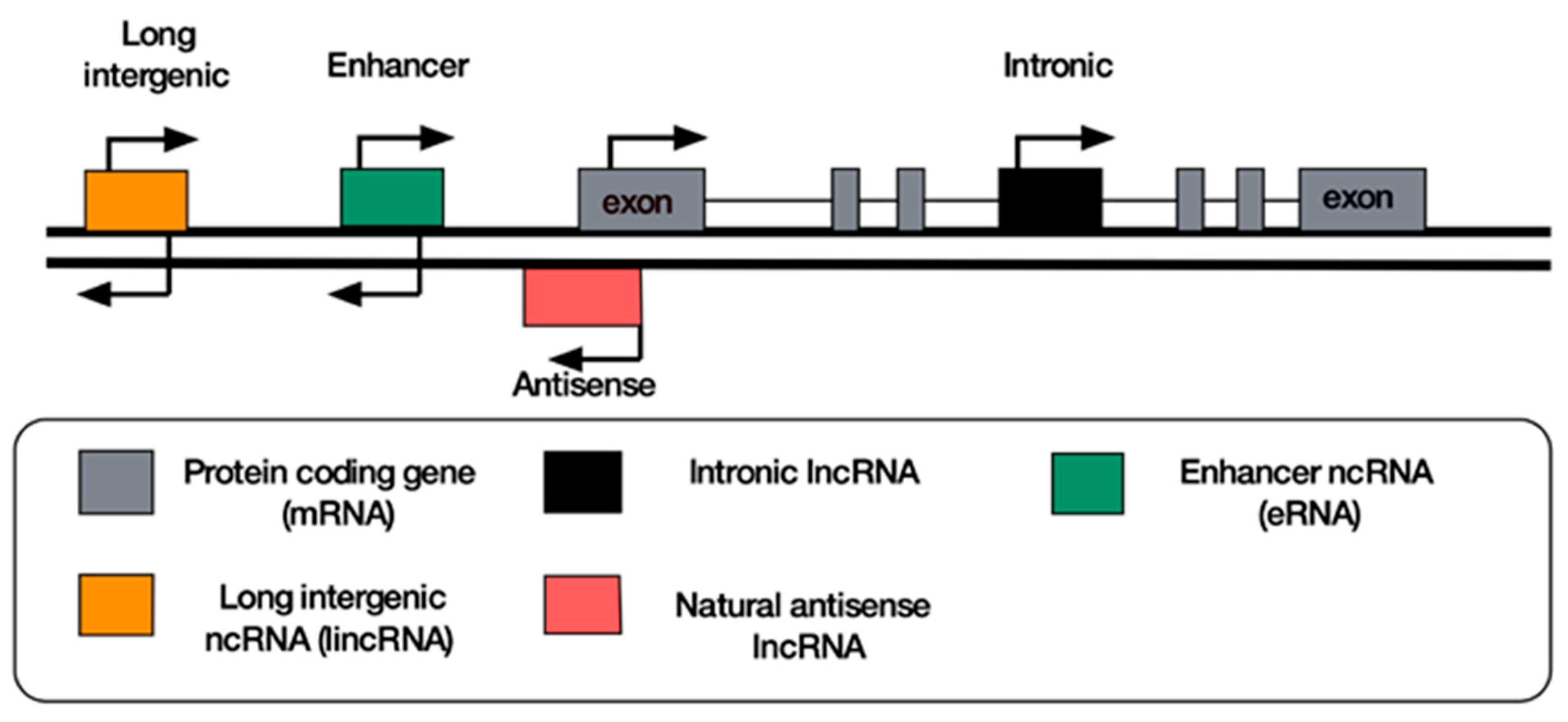
8. Classification of lncRNAs
9. Antisense lncRNAs
10. Long Intergenic Non-Coding RNAs
11. Enhancer RNAs
12. Intronic RNAs
13. circRNAs
14. Pseudogenes
15. LncRNAs and the Regulation of Biological Function
16. LncRNAs and the Innate Immune Response
17. LncRNAs in Hematopoietic Development, Differentiation and Apoptosis
18. LncRNAs in Monocytes and Macrophage Activation
19. Antisense lncRNAs in Monocytes and Macrophage Activation
20. LincRNAs in Monocytes and Macrophage Activation
21. LncRNAs in Dendritic Cells
22. LncRNAs in Fibroblasts
23. LncRNAs in Epithelial Cells
24. LncRNAs and Viral Infections
25. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef]
- Okazaki, Y.; Furuno, M.; Kasukawa, T.; Adachi, J.; Bono, H.; Kondo, S.; Nikaido, I.; Osato, N.; Saito, R.; Suzuki, H.; et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 2002, 420, 563–573. [Google Scholar] [PubMed]
- Gencode. GENCODE. Statistics about the Current GENCODE Release (version 29). 2019. Available online: https://www.gencodegenes.org/human/stats.html (accessed on 12 February 2019).
- Pertea, M.; Salzberg, S.L. Between a chicken and a grape: Estimating the number of human genes. Genome Biol. 2010, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Englert, M.; Felis, M.; Junker, V.; Beier, H. Novel upstream and intragenic control elements for the RNA polymerase III-dependent transcription of human 7SL RNA genes. Biochimie 2004, 86, 867–874. [Google Scholar] [CrossRef]
- Blythe, A.J.; Fox, A.H.; Bond, C.S. The ins and outs of lncRNA structure: How, why and what comes next? Biochim. Biophys. Acta 2016, 1859, 46–58. [Google Scholar] [CrossRef]
- Necsulea, A.; Necsulea, A.; Kaessmann, H.; Kaessmann, H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014, 15, 734–748. [Google Scholar] [CrossRef]
- Esteller, M.; Esteller, M.; Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yan, C.C.; Zhang, X.; You, Z.-H. Long non-coding RNAs and complex diseases: From experimental results to computational models. Brief. Bioinform. 2017, 18, 558–576. [Google Scholar] [CrossRef] [PubMed]
- Hon, C.-C.; Ramilowski, J.A.; Harshbarger, J.; Bertin, N.; Rackham, O.J.L.; Gough, J.; Denisenko, E.; Schmeier, S.; Poulsen, T.M.; Severin, J.; et al. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017, 543, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Natoli, G.; Andrau, J.-C. Noncoding transcription at enhancers: General principles and functional models. Annu. Rev. Genet. 2012, 46, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, L.; Chen, L.-L. Life without A tail: New formats of long noncoding RNAs. Int. J. Biochem. Cell Biol. 2014, 54, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Orera, J.; Messeguer, X.; Subirana, J.A.; Alba, M.M. Long non-coding RNAs as a source of new peptides. Elife 2014, 3, e03523. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Gloss, B.S.; Dinger, M.E. The specificity of long noncoding RNA expression. Biochim. Biophys. Acta 2016, 1859, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the Human genome is constrained: Variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014, 10, e1004525. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C.; et al. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef]
- Ulitsky, I.; Shkumatava, A.; Jan, C.H.; Sive, H.; Bartel, D.P. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011, 147, 1537–1550. [Google Scholar] [CrossRef]
- Johnsson, P.; Lipovich, L.; Grandér, D.; Morris, K.V. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta 2014, 1840, 1063–1071. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Hendrich, B.D.; Rupert, J.L.; Lafrenière, R.G.; Xing, Y.; Lawrence, J.; Willard, H.F. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 1992, 71, 527–542. [Google Scholar] [CrossRef]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the Cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Van Heesch, S.; van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.B.; de Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef]
- Ingolia, N.T.; Ingolia, N.T.; Lareau, L.F.; Lareau, L.F.; Weissman, J.S.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Kung, J.T.Y.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669. [Google Scholar] [CrossRef] [PubMed]
- Magistri, M.; Faghihi, M.A.; Laurent, G.S.; Wahlestedt, C. Regulation of chromatin structure by long noncoding RNAs: Focus on natural antisense transcripts. Trends Genet. 2012, 28, 389–396. [Google Scholar] [CrossRef]
- Faghihi, M.A.; Wahlestedt, C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009, 10, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Villegas, V.E.; Zaphiropoulos, P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015, 16, 3251–3266. [Google Scholar] [CrossRef]
- Kim, T.-K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar] [CrossRef]
- Chen, H.; Du, G.; Song, X.; Li, L. Non-coding Transcripts from Enhancers: New Insights into Enhancer Activity and Gene Expression Regulation. Genom. Proteom. Bioinform. 2017, 15, 201–207. [Google Scholar] [CrossRef]
- Liu, F. Enhancer-derived RNA: A Primer. Genom. Proteom. Bioinform. 2017, 15, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Ayupe, A.C.; Tahira, A.C.; Camargo, L.; Beckedorff, F.C.; Verjovski-Almeida, S.; Reis, E.M. Global analysis of biogenesis, stability and sub-cellular localization of lncRNAs mapping to intragenic regions of the human genome. RNA Biol. 2015, 12, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Boivin, V.; Deschamps-Francoeur, G.; Scott, M.S. Protein coding genes as hosts for noncoding RNA expression. Semin. Cell Dev. Biol. 2018, 75, 3–12. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Balakirev, E.S.; Ayala, F.J. Pseudogenes: Are They ‘Junk’ or Functional DNA? Annu. Rev. Genet. 2003, 37, 123–151. [Google Scholar] [CrossRef] [PubMed]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R.F. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 17, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Yang, W.; Wang, X.-J. Pseudogenes: Pseudo or real functional elements? J. Genet. Genom. 2013, 40, 171–177. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Cerase, A.; Pintacuda, G.; Tattermusch, A.; Avner, P. Xist localization and function: New insights from multiple levels. Genome Biol. 2015, 16, 166. [Google Scholar] [CrossRef] [PubMed]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, Z.-M.; Chang, Y.-N.; Hu, Z.-M.; Qi, H.-X.; Hong, W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene 2014, 547, 1–9. [Google Scholar] [CrossRef]
- Hadjicharalambous, M.R.; Roux, B.T.; Csomor, E.; Feghali-Bostwick, C.A.; Murray, L.A.; Clarke, D.L.; Lindsay, M.A. Long intergenic non-coding RNAs regulate human lung fibroblast function: Implications for idiopathic pulmonary fibrosis. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Pearson, M.J.; Philp, A.M.; Heward, J.A.; Roux, B.T.; Walsh, D.A.; Davis, E.T.; Lindsay, M.A.; Jones, S.W. Long Intergenic Noncoding RNAs Mediate the Human Chondrocyte Inflammatory Response and Are Differentially Expressed in Osteoarthritis Cartilage. Arthritis Rheumatol. 2016, 68, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.T.; Heward, J.A.; Donnelly, L.E.; Jones, S.W.; Lindsay, M.A. Catalog of Differentially Expressed Long Non-Coding RNA following Activation of Human and Mouse Innate Immune Response. Front. Immunol. 2017, 8, 1038. [Google Scholar] [CrossRef] [PubMed]
- Hadjicharalambous, M.R.; Roux, B.T.; Feghali-Bostwick, C.A.; Murray, L.A.; Clarke, D.L.; Lindsay, M.A. Long Non-coding RNAs Are Central Regulators of the IL-1β-Induced Inflammatory Response in Normal and Idiopathic Pulmonary Lung Fibroblasts. Front. Immunol. 2018, 9, 2906. [Google Scholar] [CrossRef]
- Krawczyk, M.; Emerson, B.M. p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. Elife 2014, 3, e01776. [Google Scholar] [CrossRef]
- Tong, X.; Gu, P.-C.; Xu, S.-Z.; Lin, X.-J. Long non-coding RNA-DANCR in human circulating monocytes: A potential biomarker associated with postmenopausal osteoporosis. Biosci. Biotechnol. Biochem. 2015, 79, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Zhang, S.; Yang, Z.; Lin, H.; Zhu, J.; Liu, L.; Wang, W.; Liu, S.; Liu, W.; Ma, Y.; et al. Self-Recognition of an Inducible Host lncRNA by RIG-I Feedback Restricts Innate Immune Response. Cell 2018, 173, 906–919.e13. [Google Scholar] [CrossRef] [PubMed]
- Kotzin, J.J.; Spencer, S.P.; McCright, S.J.; Kumar, D.B.U.; Collet, M.A.; Mowel, W.K.; Elliott, E.N.; Uyar, A.; Makiya, M.A.; Dunagin, M.C.; et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 2016, 537, 239–243. [Google Scholar] [CrossRef]
- Covarrubias, S.; Robinson, E.K.; Shapleigh, B.; Vollmers, A.; Katzman, S.; Hanley, N.; Fong, N.; McManus, M.T.; Carpenter, S. CRISPR/Cas-based screening of long non-coding RNAs (lncRNAs) in macrophages with an NF-κB reporter. J. Biol. Chem. 2017, 292, 20911–20920. [Google Scholar] [CrossRef]
- Cao, H.; Wahlestedt, C.; Kapranov, P. Strategies to Annotate and Characterize Long Noncoding RNAs: Advantages and Pitfalls. Trends Genet. 2018, 34, 704–721. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, G.; D’Osualdo, A.; Schubert, D.A.; Weber, A.; Bruscia, E.M.; Hartl, D. Cellular Innate Immunity: An Old Game with New Players. J. Innate Immun. 2017, 9, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Kondo, M. Lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. Immunol. Rev. 2010, 238, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, A.; He, X.C.; Thorvaldsen, J.L.; Sugimura, R.; Perry, J.M.; Tao, F.; Zhao, M.; Christenson, M.K.; Sanchez, R.; Yu, J.Y.; et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013, 500, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Jeong, M.; Sun, D.; Park, H.J.; Rodriguez, B.A.T.; Xia, Z.; Yang, L.; Zhang, X.; Sheng, K.; Darlington, G.J.; et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell 2015, 16, 426–438. [Google Scholar] [CrossRef]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Xie, N.; Tan, Z.; Banerjee, S.; Thannickal, V.J.; Abraham, E.; Liu, G. The human long noncoding RNA lnc-IL7R regulates the inflammatory response. Eur. J. Immunol. 2014, 44, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, B.; Servaas, N.H.; Rossato, M.; Tamassia, N.; Cassatella, M.A.; Cossu, M.; Beretta, L.; van der Kroef, M.; Radstake, T.R.D.J.; Bazzoni, F. The Long Non-coding RNA NRIR Drives IFN-Response in Monocytes: Implication for Systemic Sclerosis. Front Immunol. 2019, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Dave, R.K.; Dinger, M.E.; Andrew, M.; Askarian-Amiri, M.; Hume, D.A.; Kellie, S. Regulated expression of PTPRJ/CD148 and an antisense long noncoding RNA in macrophages by proinflammatory stimuli. PLoS ONE 2013, 8, e68306. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Yuan, L.; Tan, X.; Huang, D.; Wang, X.; Zheng, Z.; Mao, X.; Li, X.; Yang, L.; Huang, K.; et al. The LPS-inducible lncRNA Mirt2 is a negative regulator of inflammation. Nat. Commun. 2017, 8, 2049. [Google Scholar] [CrossRef] [PubMed]
- Castellanos-Rubio, A.; Fernandez-Jimenez, N.; Kratchmarov, R.; Luo, X.; Bhagat, G.; Green, P.H.R.; Schneider, R.; Kiledjian, M.; Bilbao, J.R.; Ghosh, S. A long noncoding RNA associated with susceptibility to celiac disease. Science 2016, 352, 91–95. [Google Scholar] [CrossRef]
- Chan, J.; Atianand, M.; Jiang, Z.; Carpenter, S.; Aiello, D.; Elling, R.; Fitzgerald, K.A.; Caffrey, D.R. Cutting Edge: A Natural Antisense Transcript, AS-IL1α, Controls Inducible Transcription of the Proinflammatory Cytokine IL-1α. J. Immunol. 2015, 195, 1359–1363. [Google Scholar] [CrossRef]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A Long Noncoding RNA Mediates Both Activation and Repression of Immune Response Genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.-Y.; Wang, Y.; Ma, S.; Chen, X.; Chen, J.; Su, C.-J.; Shibata, A.; Strauss-Soukup, J.K.; Drescher, K.M.; et al. LincRNA-Cox2 Promotes Late Inflammatory Gene Transcription in Macrophages through Modulating SWI/SNF-Mediated Chromatin Remodeling. J. Immunol. 2016, 196, 2799–2808. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, Z.; Liu, H.; Li, W.; Guo, X.; Zhang, Z.; Liu, Y.; Jia, L.; Li, Y.; Ren, Y.; et al. lincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ. 2019, 26, 130–145. [Google Scholar] [CrossRef]
- Elling, R.; Robinson, E.K.; Shapleigh, B.; Liapis, S.C.; Covarrubias, S.; Katzman, S.; Groff, A.F.; Jiang, Z.; Agarwal, S.; Motwani, M.; et al. Genetic Models Reveal cis and trans Immune-Regulatory Activities for lincRNA-Cox2. Cell Rep. 2018, 25, 1511–1524.e6. [Google Scholar] [CrossRef]
- Zhao, G.; Su, Z.; Song, D.; Mao, Y.; Mao, X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-κB. FEBS Lett. 2016, 590, 2884–2895. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Banerjee, S.; Guo, S.; Xie, N.; Ge, J.; Jiang, D.; Zörnig, M.; Thannickal, V.J.; Liu, G. Long noncoding RNA Malat1 regulates differential activation of macrophages and response to lung injury. JCI Insight 2019, 4, 176. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z.; Chao, T.-C.; Chao, T.C.; Chang, K.Y.; Chang, K.-Y.; Lin, N.; Lin, N.; Patil, V.S.; Patil, V.S.; et al. The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Iott, N.E.I.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014, 5, 3979. [Google Scholar]
- Atianand, M.K.; Hu, W.; Satpathy, A.T.; Shen, Y.; Ricci, E.P.; Alvarez-Dominguez, J.R.; Bhatta, A.; Schattgen, S.A.; McGowan, J.D.; Blin, J.; et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell 2016, 165, 1672–1685. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ming, Z.; Gong, A.-Y.; Wang, Y.; Chen, X.; Hu, G.; Zhou, R.; Shibata, A.; Swanson, P.C.; Chen, X.-M. A long noncoding RNA, lincRNA-Tnfaip3, acts as a coregulator of NF-κB to modulate inflammatory gene transcription in mouse macrophages. FASEB J. 2017, 31, 1215–1225. [Google Scholar] [CrossRef]
- Yang, C.-A.; Li, J.-P.; Yen, J.-C.; Lai, I.-L.; Ho, Y.-C.; Chen, Y.-C.; Lan, J.-L.; Chang, J.-G. lncRNA NTT/PBOV1 Axis Promotes Monocyte Differentiation and Is Elevated in Rheumatoid Arthritis. Int. J. Mol. Sci. 2018, 19, 2806. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, X.; Xie, M.; Liu, M.; Ye, M.; Li, M.; Chen, X.-M.; Li, X.; Zhou, R. The NF-κB-Responsive Long Noncoding RNA FIRRE Regulates Posttranscriptional Regulation of Inflammatory Gene Expression through Interacting with hnRNPU. J. Immunol. 2017, 199, 3571–3582. [Google Scholar] [CrossRef]
- Hu, W.; Yuan, B.; Flygare, J.; Lodish, H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 2011, 25, 2573–2578. [Google Scholar] [CrossRef]
- Buenrostro, J.D.; Giresi, P.G.; Zaba, L.C.; Chang, H.Y.; Greenleaf, W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 2013, 10, 1213–1218. [Google Scholar] [CrossRef]
- Clark, G.J.; Angel, N.; Kato, M.; López, J.A.; MacDonald, K.; Vuckovic, S.; Hart, D.N. The role of dendritic cells in the innate immune system. Microbes Infect. 2000, 2, 257–272. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Zheng, Y.; Jin, X.; Liu, M.; Li, S.; Zhao, Q.; Liu, X.; Wang, Y.; Shi, M.; et al. The Long Noncoding RNA MALAT1 Induces Tolerogenic Dendritic Cells and Regulatory T Cells via miR155/Dendritic Cell-Specific Intercellular Adhesion Molecule-3 Grabbing Nonintegrin/IL10 Axis. Front. Immunol. 2018, 9, 1847. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, Y.; Han, Y.; Lin, L.; Wu, C.; Xu, S.; Jiang, Z.; Xu, J.; Liu, Q.; Cao, X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 2014, 344, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Hernández, L.A.; Gómez-Olivares, J.L.; Buentello-Volante, B.; Lucio, V.M.B. Fibroblasts: The Unknown Sentinels Eliciting Immune Responses Against Microorganisms. Eur. J. Microbiol. Immunol. 2017, 7, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Rapicavoli, N.A.; Qu, K.; Zhang, J.; Mikhail, M.; Laberge, R.-M.; Chang, H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife 2013, 2, e00762. [Google Scholar] [CrossRef] [PubMed]
- Zgheib, C.; Hodges, M.M.; Hu, J.; Liechty, K.W.; Xu, J. Long non-coding RNA Lethe regulates hyperglycemia-induced reactive oxygen species production in macrophages. PLoS ONE 2017, 12, e0177453. [Google Scholar] [CrossRef] [PubMed]
- Schleimer, R.P.; Kato, A.; Kern, R.; Kuperman, D.; Avila, P.C. Epithelium: At the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 2007, 120, 1279–1284. [Google Scholar] [CrossRef]
- Tong, Q.; Gong, A.-Y.; Zhang, X.-T.; Lin, C.; Ma, S.; Chen, J.; Hu, G.; Chen, X.-M. LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of Mi-2/NuRD-mediated epigenetic histone modifications. FASEB J. 2016, 30, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Morchikh, M.; Cribier, A.; Raffel, R.; Amraoui, S.; Cau, J.; Severac, D.; Dubois, E.; Schwartz, O.; Bennasser, Y.; Benkirane, M. HEXIM1 and NEAT1 Long Non-coding RNA Form a Multi-subunit Complex that Regulates DNA-Mediated Innate Immune Response. Mol. Cell 2017, 67, 387–399.e5. [Google Scholar] [CrossRef]
- Imamura, K.; Takaya, A.; Ishida, Y.I.; Fukuoka, Y.; Taya, T.; Nakaki, R.; Kakeda, M.; Imamachi, N.; Sato, A.; Yamada, T.; et al. Diminished nuclear RNA decay upon Salmonella infection upregulates antibacterial noncoding RNAs. EMBO J. 2018, 37, e97723. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Chen, S.; Tian, R.; Huang, X.; Deng, R.; Xue, B.; Qin, Y.; Xu, Y.; Wang, J.; Guo, M.; et al. Long Noncoding RNA ITPRIP-1 Positively Regulates the Innate Immune Response through Promotion of Oligomerization and Activation of MDA5. J. Virol. 2018, 92, 146. [Google Scholar] [CrossRef] [PubMed]



| LncRNA Name | Stimuli | Cell Type | Function | Mechanism | Refs |
|---|---|---|---|---|---|
| H19 | N/A | Mouse hematopoietic stem cells | Maintains HSC quiescence | Regulates the Igf2-Igfr1 pathway via the translocation of FOX3 to the cytoplasm | [69] |
| LncHSC-1/2 | N/A | Mouse hematopoietic stem cells | LncHSC-1 regulates myeloid differentiation and LncHSC-2 cell self-renewal and differentiation | LncHSC-2 recruits the hematopoietic TF E2A to its binding sites | [70] |
| PACER | PMA and LPS | Human monocytes (U937) | Promotes COX2 expression | Binds to the repressive p50 NF-κΒ subunit of COX2 promoter to enable p300 HAT recruitment in order to increase histone acetylation and initiate the assembly of RNAP-II complexes | [61] |
| Morrbid | N/A | Human and mouse monocytes, neutrophils, eosinophils | Controls the lifespan of monocytes, neutrophils and eosinophils | Regulates Bcl2l11 (Bim) transcription by promoting PCR2 enrichment at its promoter and deposition of H3K27me3 | [64] |
| THRIL | PMA Pam3CSK4 | Human monocytes (THP-1) | Regulates the expression of the innate-associated mediators TNF-α, CCL1, IL-8, CSF1 and CXC10 | Forms a functional lncRNA-hnRNPL complex in order to regulate TNF-α transcription by binding to its promoter | [84] |
| LincRNA-COX2 | Pam3CSK4 | Mouse bone marrow derived macrophages | Regulates the expression of several immune genes | Interacts with the nuclear proteins hnRNP-A/B and hnRNP-A2/B1 | [6,78,79,80,81,99] |
| LPS | Regulates the NLRP3 inflammasome sensor and ASC adaptor as well as autophagy | Binds to p65 NF-κΒ subunit to promote its transcription. It was also found to regulate TRIF-mediated autophagy via caspase-1 activation | |||
| Multiple | LincRNA-Cox2 deficient mice and macrophages | Regulates the expression of several immune genes | Functions as an eRNA to regulate the activity of the COX2 gene but also demonstrates in trans-regulation of immune-associated genes in vivo | ||
| LPS | Mouse macrophages (RAW 264.7 and primary peritoneal) | Regulates the expression of NF-κΒ-regulated inflammatory genes | Interacts with the SWI/SNF complex to regulate the assembly of NF-κΒ subunits and chromatin remodelling | ||
| TLR4 ligand | Mouse bone-marrow-derived dendritic cells | N/A | Demonstrates NF-κΒ-dependent expression | ||
| TNF-α | Murine intestinal epithelial cells (IEC4.1 cell line) | Regulates the expression of the Il12b gene | Demonstrates NF-κΒ-dependent expression and promotes the recruitment of the Mi-2/NuRD complex to the Il12b promoter | ||
| AS-IL1α | TLR ligands and Listeria monocytogenes | Mouse bone marrow derived macrophages | Regulates IL-1α transcription | Facilitates RNAP-II recruitment to the IL-1α locus and demonstrates NF-κΒ-dependent expression | [77] |
| IL1β-eRNA IL1β-RBT46 | LPS | Human monocytes (THP-1 and primary) | Regulate the expression of IL-1β and CXCL8 | Demonstrate NF-κΒ-dependent expression | [85] |
| Lnc-IL7R | LPS | Human monocytes (THP-1) | Regulates the expression of the inflammatory mediators IL-6, IL-8, E-selectin and VCAM-1 | Regulates deposition of H3K27me3 at the promoters of the E-selectin and VCAM-1 genes | [72] |
| Ptprj-as1 | LPS Pam3Cys | Mouse bone marrow derived macrophages and RAW 264.7 | N/A | N/A | [74] |
| IL7AS | LPS | Human monocytes (THP-1), mouse macrophages (RAW 264.7) | Regulates IL-6 expression and release | Demonstrates NF-κΒ-dependent expression | [59,60] |
| IL-1β | Human epithelial cells (A549 cell line) Human lung fibroblasts (primary) | ||||
| Linc-EPS | Multiple | Mouse bone marrow derived macrophages | Represses the inflammatory response by inhibiting IRGs expression | Interacts with hnRNPL via a CANACA motif in its 3′ region and regulates nucleosome positioning at IRG promoters | [86] |
| LincRNA-Tnfaip3 | LPS | Mouse macrophages (RAW 264.7 and primary mouse peritoneal macrophages) | Regulates the expression of several NF-κΒ mediated inflammatory genes | Directly interacts with Hmgb1 and NF-κΒ to form a functional complex to regulate Hmgb1-mediated histone modifications | [87] |
| Lnc-13 | LPS | Human monocytes (primary and U937, THP-1) and mouse bone marrow derived macrophages | Suppresses the expression of several immune-associated genes | Demonstrates NF-κΒ-dependent expression and interacts with Hdac1 on chromatin and hnRNPD to regulate gene expression | [76] |
| NRIR | LPS | Human monocytes (primary) | Regulates the expression of several interferon-stimulated genes and protein release of CXCL10 and CCL8 | Demonstrates type I IFN-dependent expression | [73] |
| NTT | N/A | Peripheral blood mononuclear cells (PBMCs) Human monocytes (THP-1) | Regulates cell cycle G1 arrest and differentiation as well as expression of IL-10 and CXCL10 | Interacts with the TF C/EBPβ and the promoter of its neighbouring gene PBOV1 via hnRNP-U | [88] |
| Mirt2 | LPS | Peritoneal macrophages (C57BL/6 mice) HEK293T and RAW264.7 cells | Regulates macrophage polarisation and aberrant inflammatory activity | Inhibits TRAF6 Lys63-mediated ubiquitination and the activation of the MAPK and NF-κΒ pathways | [75] |
| Lnc-Lsm3b | Viral RNA molecules | Mouse macrophages (peritoneal, RAW 264.7), L929 and HEK293T cell lines | Inactivates late RIG-1 innate activity and type I IFNs production | Acts as a decoy by saturating RIG-1 binding sites to inhibit inflammation and to prevent tissue host damage | [63] |
| MALAT1 | PMA, LPS | Human monocytes (THP-1), mouse macrophages (RAW 264.7) | Regulates the expression of inflammatory genes such as IL-6 and TNF-α | Interacts with NF-κΒ p50/p65 subunits to inhibit NF-κΒ DNA binding activity | [82,83,93] |
| PMA, LPS, IL-4 | Mouse macrophages (BMDM), human monocytes (PBMCs, THP-1) | Regulates LPS-mediated M1 macrophage activation and IL-4-mediated M2 differentiation and pro-fibrotic phenotype | Demonstrates Clec16a-dependent expression and regulation of mitochondrial pyruvate carriers | ||
| LPS | Mouse bone-marrow-derived dendritic cells | Induces increased tolerogenic activity of DCs | Enhances DC-SIGN expression, IL-10 production and acts as an miR-155 sponge | ||
| FIRRE | LPS | Human macrophages (U937) Human intestinal epithelial cells (SW480) Mouse macrophages (RAW264.7) | Regulates expression of several inflammatory genes | Demonstrates NF-κΒ-dependent expression and interacts with hnRNPU to regulate mRNA stability by targeting their AREs | [89] |
| LincRNA-AK170409 LincRNA-Cox2 | TLR ligands | Immortalized murine bone marrow–derived macrophages (iBMDM) | Both regulate NF-κΒ-dependent signalling | Both lncRNAs demonstrate NF-κΒ-dependent activity - LincRNA-COX2 promotes IκΒα degradation | [65] |
| Lnc-DC | N/A | Human conventional dendritic cells | Regulates DC differentiation | Binds directly to STAT3 to prevent dephosphorylation of Y705 by SHP1 | [94] |
| Lethe | TNF-α, IL-1β, dexamethasone | MEF lines (mouse embryonic fibroblasts) | Regulates the expression of several NF-κΒ mediated inflammatory genes | Interacts with the RelA (p65) subunit of NF-κΒ to inhibit DNA binding and gene activation | [96,97] |
| Glucose | Mouse macrophages (RAW 264.7 and bone marrow derived macrophages) | Regulates ROS production and NOX2 gene expression | Interacts with the NF-κΒ p65 subunit to control its translocation to the nucleus | ||
| MIR3142HG | IL-1β | Human lung fibroblasts (primary) | Regulates CCL2 and IL-8 mRNA and protein release | Demonstrates NF-κΒ-dependent expression | [60] |
| NEAT1 | Influenza, HSV-1, poly I:C | Human epithelial cells (A549 cell line) and HeLa cells | Regulates expression of IL-8 | Interacts and relocates SFPQ from the IL8 promoter to the paraspeckles | [100,101] |
| Multiple | HUVEC cells (Human umbilical vein endothelial cells), HEK293 cells, HeLa and 293 T cells | Regulates the DNA-mediated innate immune response | Interacts with HEXIM1 to form the HDP-RNP complex which is required for the cGAS-STING-IRF3 pathway | ||
| LncITPRIP-1 | Viral infections | Huh7, Huh7.5, Huh7.5.1-MAVS, FL-neo, and HEK293T cells | Promotes the activation of the innate immune response | Binds to the C-terminus of MDA5 and promotes its oligomerisation to enhance IFN signalling and production | [103] |
| NEAT1v2 eRNA07573 | Salmonella infection | HeLa cells | Regulate expression of immune-associated genes and response to antibacterial defence | Inhibit levels of the exosome/NEXT components and demonstrate elevated transcript stability | [102] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadjicharalambous, M.R.; Lindsay, M.A. Long Non-Coding RNAs and the Innate Immune Response. Non-Coding RNA 2019, 5, 34. https://doi.org/10.3390/ncrna5020034
Hadjicharalambous MR, Lindsay MA. Long Non-Coding RNAs and the Innate Immune Response. Non-Coding RNA. 2019; 5(2):34. https://doi.org/10.3390/ncrna5020034
Chicago/Turabian StyleHadjicharalambous, Marina R., and Mark A. Lindsay. 2019. "Long Non-Coding RNAs and the Innate Immune Response" Non-Coding RNA 5, no. 2: 34. https://doi.org/10.3390/ncrna5020034
APA StyleHadjicharalambous, M. R., & Lindsay, M. A. (2019). Long Non-Coding RNAs and the Innate Immune Response. Non-Coding RNA, 5(2), 34. https://doi.org/10.3390/ncrna5020034




