Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far
Abstract
1. Approaches to Define RNA-Chromatin Interactions
- RIP-seq: RNA immunoprecipitation (RIP) exploits antibodies to pull down RNA bound to a given protein and the immunoprecipitated RNA subjected to high throughput sequencing (RIP-seq), thereby, enabling global identification of RNAs bound to protein of interest. Technical variants of this methods include native RIP-seq [72] and formaldehyde cross linked fRIP-seq [71].
- RIPiT-Seq: RNA: Protein immunoprecipitation in tandem (RIPiT) is suitable for RBPs with poor inherent ultraviolet (UV) crosslink ability. This method yields highly specific RNA binding footprints of any cellular RNPs and the resulted RNA footprints can then be combined with high-throughput sequencing (RIPiT-Seq) thereby providing a means to map the RNA binding sites of such RBPs [73]. This method has been used to identify and validate RNA binding pocket within WDR5 chromatin modifier [74].
- CLIP: Improves the specificity of RIP by UV crosslinking of RNA/protein complexes before extraction. This allows the removal of weakly bound RNA through stringent washing. The remaining RNA can then undergo reverse transcription and PCR amplification (or next generation sequencing). The main drawback of this method is the loss of a significant proportion of transcripts which are stalled at the cross-linking site resulting in truncated cDNAs. UV crosslinking can also introduce some bias as its ability to bind RNA to protein varies depending on the base/proximity of the reactive amino acids mediating the interaction. HITS-CLIP when CLIP is combined with high throughput next generation sequencing [75].
- iCLIP: Individual-nucleotide-resolution CLIP (iCLIP) was developed to enable the recovery of truncated cDNAs lost in conventional CLIP. The iCLIP protocol employs UV irradiation as a cross-linking source that preserves in vivo RNA-protein interactions through promoting covalent bonds at the sites of protein-RNA interactions. Following mild RNAse treatment, to obtain RNA fragments in an optimal size range, RNA-protein complexes are immunoprecipitated. The immunoprecipitated RNA is dephosphorylated to enable an adapter ligation to the 3′ end of the RNA and radioactive labelling at the 5′ end. This method includes SDS-PAGE separation and transfer to nitrocellulose membrane to capture radiolabelled, immunoprecipitated, crosslinked RNA-protein complexes. The captured RNA is then reverse transcribed into cDNA. Following cDNA circularization, restriction enzyme digestion to linearize the cDNA prior to PCR amplification and library preparation for high-throughput sequencing. Truncated cDNA represents the majority in the cDNA library and the position of the preceding nucleotide, after mapping to the genome, corresponds to the cross-linking site (Huppertz et al., 2014).
- eCLIP: Enhanced CLIP improves library preparation and circular ligation steps of iCLIP allowing greater power in filtering and mapping truncated sequences. eCLIP replaces the 5′ adaptor ligation with a 3′ cDNA ligation [76], whereas further improved eCLIP protocol Monitored eCLIP (meCLIP) uses both a 5′ ligation and a 3′ cDNA ligation [77].
- irCLIP: This is similar to iCLIP apart from the fact that it makes use of a biotinylated, fluorescent 3′ DNA adaptor [78].
- BrdU-CLIP: BrdU-CLIP built on the same principle as that of CLIP and iCLIP but employs a nucleotide analogue BrdUTP in reverse transcription to capture truncated and non-truncated cDNA products using BrdU antibody [79].
- GoldCLIP: Improved with a shortened iCLIP protocol that removes the SDS-PAGE separation and membrane transfer steps. RNPs are tagged with Halo-tag and overexpressed in cell line of interest. Halo-tagged protein-RNA complex affinity purified using Halo-ligand. Following denaturing washes, the purified RNAs subjected to high-throughput sequencing [80].
- PAR-CLIP: Photo-Activatable Ribonucleoside enhanced Cross-Linking and Immunoprecipitation (PAR-CLIP) is a modified CLIP method, where the introduction of photo-activated nucleosides in the media are taken up by cells and subsequently used for protein–RNA crosslinking thereby enabling the following advantages. First, PAR-CLIP shows in general 100- to 1,000-fold higher RNA recovery, in comparison to the conventional cross-linking at 254 nm. Secondly, UV radiation-induced T-to-C mutations characteristic of the cross-linked sites that have incorporated photo-activated nucleoside analogues. Based on this, PAR-CLIP exploits mutation analysis to improve the identification of precise RBP binding positions or footprint [81]. Studies using PAR-CLIP have identified that both EZH2 and JARID2 can directly interact with RNA in cells [82,83]. Their interaction with RNA is mutually exclusive and antagonistic to their ability to interact and bind to chromatin.
- fCLIP: Formaldehyde cross-linking, immunoprecipitation and sequencing (fCLIP) uses formaldehyde as a cross-linking reagent for CLIP to characterize the RNA binding protein binding regions on double stranded RNAs. dsRNAs are inefficiently crosslinked by UV, thus making it difficult to study the interactions between dsRNA binding proteins and their substrates. It has been used to characterize mapping of in vivo DROSHA cleavage sites at single nucleotide resolution [84].
- mRNA Interactome Capture (MIC):mRNA Interactome Capture (MIC) is an oligo dT-based capture of global mRNA protein-interactome from cells cross linked with complementary crosslinking chemistries: with UV (at 254 nm) or photoactivatable-ribonucleoside (4SU, 4 thiouridine)-enhanced crosslinking (PAR-CL) at 265 nm. This method characterized global mRNA proteome comprising novel RNA binding proteins, including metabolic enzymes. These two complementary chemistries allow a comparative analysis of the enriched RBPs. This investigation highlights the presence of intrinsically disordered structures in the large portion of the human proteome [85].
- RBDmap:RNA Binding Domain map (RBDmap) is an improved protocol of RIC, which finemaps the protein domains that interacts with mRNAs. UV irradiated cells were given stringent denaturing washes to purify the resulting covalently linked RBP–RNA complexes with oligo(dT) magnetic beads. As a defining modification to RIC, post elution the RBPs were subjected to partial proteolysis to retain only those protein regions that are bound to the RNA and are separated by a second oligo(dT) selection from the non-interacting peptides that are released into the supernatant. Mass-spectrometric analysis of the eluted and released peptides to calculate peptide intensity ratios between these fractions will determine the RNA-binding regions [86].
- OOPS: Orthogonal Organic Phase Separation (OOPS) is a method based on UV cross linking of cells at 254 nm followed by Acidic Guanidinium Thiocyanate-Phenol-Chloroform (AGPC) phase separation, where RNA and proteins fractionated into the upper aqueous phase and the lower organic phase, respectively. Whereas RNA-protein adducts, generated by UV crosslinking, separated into the aqueous-organic interface. This interface accumulated RNA-protein adducts at the interface represent reliable RNA binding proteins on global scale which are in specific interaction with RNA [87].
2. RNA Centric Methods to Study Global RNA-Chromatin Interactions
2.1. Chromatin Oligoaffinity Precipitation (ChOP)
2.2. RNA Antisense Purification (RAP)
2.3. Chromatin Isolation by RNA Purification (ChiRP)
2.4. Capture Hybridization Analysis of RNA Targets (CHART)
- CHART uses a two-step formaldehyde cross-linking approach to fix nuclei.
- RNase H sensitivity assay is used to identify regions in the target RNA that are accessible for hybridization with antisense oligonucleotides. A small number of short oligonucleotides that have been predetermined to interact with the RNA target are then used in CHART to enrich for RNA–chromatin complexes.
- Antisense oligonucleotide bound RNA–chromatin complexes are eluted using RNase H. This reduces nonspecific false positive binding events generated by direct binding of antisense oligonucleotide probes to DNA [92].
3. Non-RNA Centric Methods to Study Global RNA-Chromatin Interactions
3.1. Chromatin RNA Isolation by Sucrose Gradient Fractionation
3.2. Chromatin RNA Immunoprecipitation (ChRIP)
3.3. Profiling Interacting RNAs on Chromatin by Deep Sequencing (PIRCh-seq)
3.4. GRID-seq
3.5. MARGI-seq
3.6. ChAR-seq
4. Mechanisms by which lncRNA Targeted to Chromatin
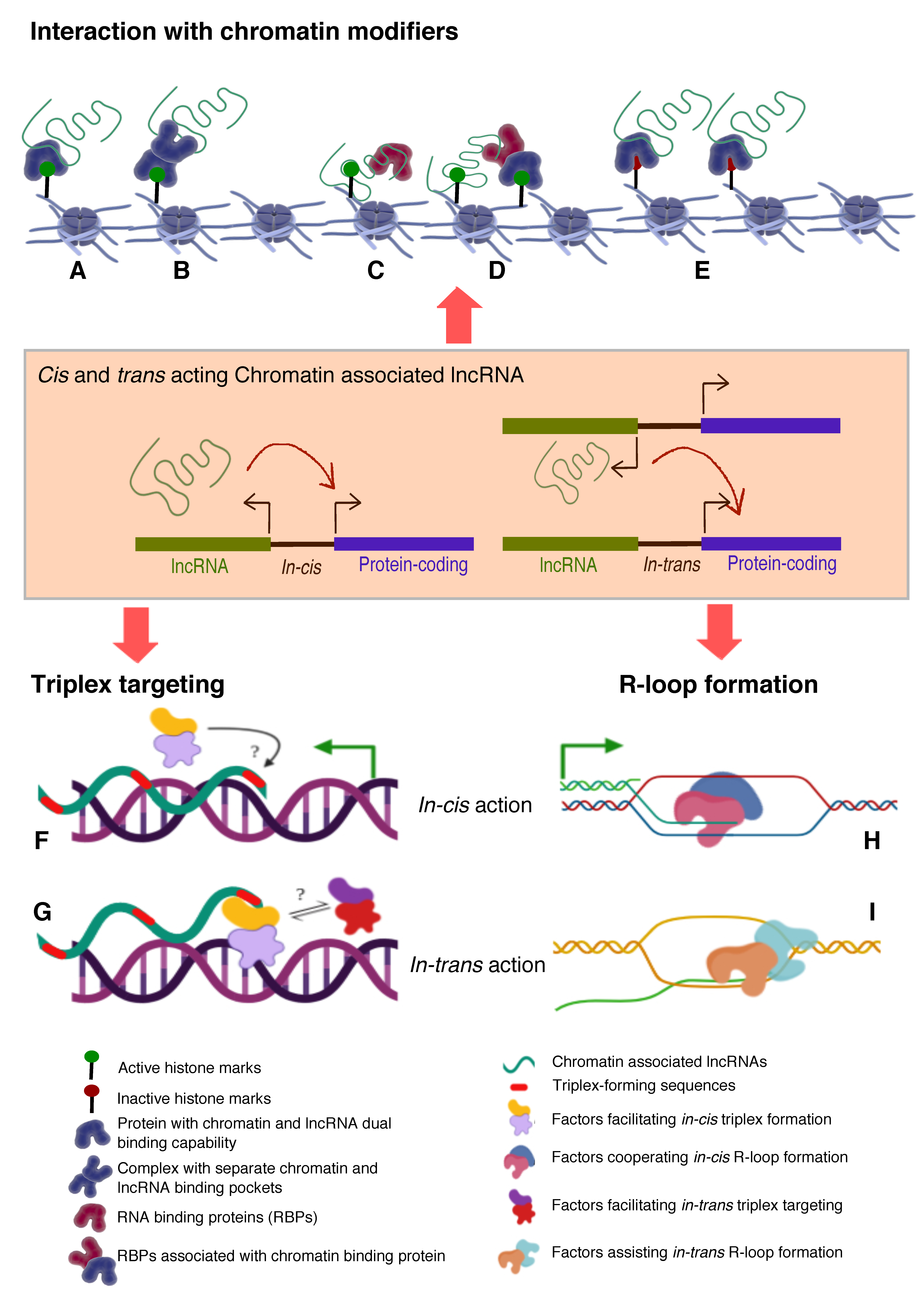
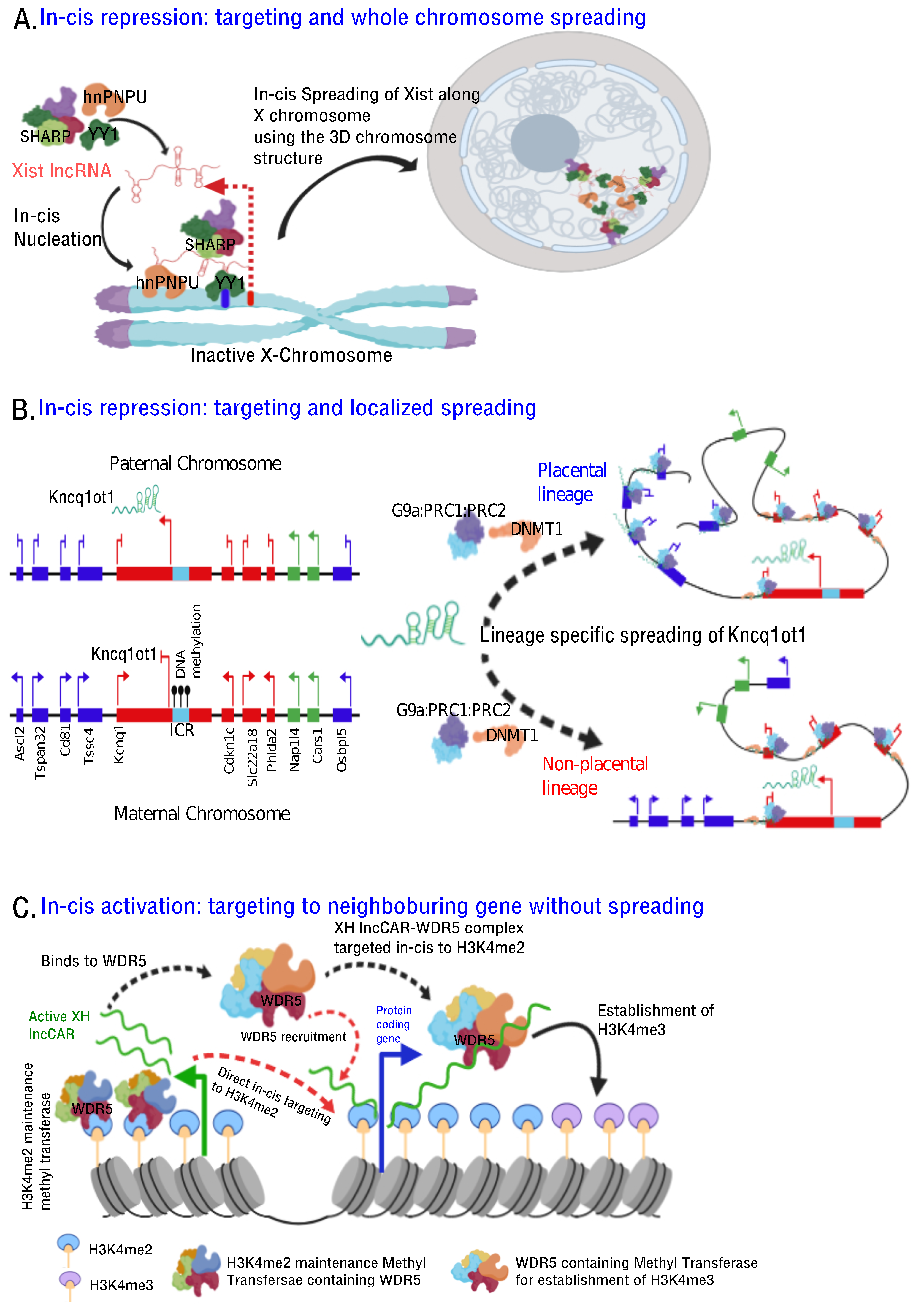
- Histone modifications, chromatin and DNA modifiers in the chromatin enrichment of lncRNAs: lncRNAs, which act as a scaffold and/or guide, are targeted to chromatin by proteins having dual RNA- and DNA binding capabilities like hnRNPK [71], PGC1α [105], PRC2 [65,66], YY1 [68,69], CTCF [70], DNMTs [106]. Alternatively, lncRNAs get targeted to chromatin by interacting with RNA binding proteins (RBPs) that facilitate interaction with additional DNA binding proteins, like hnRNPU [89] (Figure 2). It is important to emphasize here that both cis- and trans-acting lncRNAs can be targeted in this way, contrary to the prevailing view that cis acting chromatin bound lncRNAs are mostly coupled to transcription [107,108,109,110] In contrast to the actual definition of cis action being “on the same chromosome”, but over time it has been erroneously conceptualized as “action restricted to site of synthesis/transcription”. The best studies of cis regulation of chromatin bound lncRNAs comes from classical genomic imprinting loci where imprinted lncRNAs are monoallelically transcribed and are targeted to silence multiple genes on the same chromosome as exemplified from studies of mouse and human Kcnq1ot1 lncRNAs [29,30,32], Airn [42,111,112], Xist [42,112,113] etc. Chromatin targeting of H3K4me2 and WDR5 bound lncCARs (Active XH lncCARs) have been shown to be essential in maintaining active transcription of neighboring protein coding genes [28]. Chromatin targeting of active XH lncCARs occurs in part via WDR5 which has the potential to interact with both RNA and H3K4me2, an active histone chromatin mark. Thus, divergent transcription units enriched with H3K4me2 could recruit active XH lncCARs via WDR5. Similarly, recruitment of inactive CARs to their target genes could in part occur via EZH2, a PRC2 component with potential for the interactions with RNA and histone H3K27me3 [33].
- RNA:DNA triplex: Formation of triple helix nucleic acid structures involves Hoogsteen base-pairing interactions between RNA and the major groove of double-stranded DNA [114,115]. This RNA–DNA interaction has a stringent requirement for both polypurine sequence in DNA and a length restriction. Triplexes can form both in vitro and in vivo contexts and factors like GC content, extent of sequence complementarity, histone H3 tails, triplex target site (TTS) proximity to nucleosome entry site and open chromatin structure influence the stability of triplexes [116]. Multiple lncRNAs (having triplex forming sequence called Triplex Forming Oligonucleotides or TFOs) appear to use this mechanism to directly target specific complimentary sequences across the genome (Triplex Forming Regions or TFRs) to exert their regulatory functions (Figure 2, Table 3). Best examples of DNA:RNA triplex formation by lncRNA with specific DNA sequences include pRNA, which represses in cis the transcription of rRNA genes by targeting DNMT3b to their promoters [56], Fendrr which regulates developmental genes by recruiting the PRC2 complex [50], PARTICLE which regulates the expression of MAT2A in response to low-dose radiation [117], MEG3 which guides PRC2 to the regulatory regions of TGF-β pathway genes [33] and PAPAS which guides the CHD4/NuRD complex to the rDNA promoter [118]. Recently, a global approach mapped RNA: DNA triplexes genome-wide using protein free-nucleic acids, isolated from chromatin. This approach re-validated known triplex forming lncRNAs and also identified several novel candidate lncRNAs that may execute their actions via triplex formation [119]. Besides the latter experimental approach, a computational method called Triplex Domain Finder (TDF) has been developed to detect triplex forming regions in lncRNAs, and triplex target regions across the human genome. This method successfully validated DNA-binding domains of known triplex forming lncRNAs such as Fendrr, HOTAIR and MEG3 [101]. Two important aspects need to be considered about specificity of triplex formation mediated targeting of lncRNAs to chromatin. Firstly, there is generic sequence feature (polypurine stretch or TFOs) in lncRNAs that dictates its ability to form triplex at the genomic regions with TFRs. This still lacks one to one specificity. The question in that case remains whether any lncRNA with triplex forming capability can be targeted to all the “triplex targetable” i.e TFRs at genomic locations? and secondly, which factors initiate, promote and maintain triplex formation at target locations and that in principle is there a possibility of any difference between cis and trans targeting of triplexes lncRNAs (Figure 2)?
- R-loop formation: R-loops are three stranded RNA/DNA structures, which form co-transcriptionally at guanine-rich clusters (G-clusters) in the template strand during gene transcription [145,146]. It has been shown that RNAs containing four or more consecutive guanine residues near the 5’ end facilitates R-loop formation. R-loops, in the mammalian genome, predominantly seen at promoters and enhancers associated with GC-skewed sequences [147,148] and their formation and dynamics have been linked to transcriptional activities under physiological conditions [149,150] (Figure 2, Table 3). Recent evidence suggests that R-loop formation by lncRNAs seem to affect gene expression in cis through diverse mechanisms. For example, transcription of VIM-AS1 promotes the formation of R-loop structure that was found to promote transcriptional activation of its neighboring VIM gene and destabilization of R-loop structure affected VIM expression [140]. In another context, lncRNA GATA3-AS1 was found to be required for the formation of permissive chromatin marks H3K27 acetylation and H3K4 di/tri-methylation, at the GATA3-AS1-GATA3 locus. Mechanistically, GATA3-AS1 interacts with MLL1 methyltransferase and tethers to this gene locus via formation of DNA-RNA hybrid (R-loop) [151]. R-loop formation is a part of co-transcriptional process that targets nascent transcripts to chromatin in cis. Theoretically if any RNA with a GC-skewed sequences have the potential to form R-loop, then the pertinent question is how and in combination with which specific in-cis or trans- factors define the cis- and/or trans- mechanism of actions? (Figure 2).
5. lncRNA-Dependent Mechanisms in Chromatin Organization
6. Conclusions and Future Outlook
Funding
Conflicts of Interest
References
- Gilbert, W. Origin of Life—The RNA World. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Beadle, G.W.; Tatum, E.L. Genetic Control of Biochemical Reactions in Neurospora. Proc. Natl. Acad. Sci. USA 1941, 27, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar] [PubMed]
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Pheasant, M.; Mattick, J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 2007, 29, 288–299. [Google Scholar] [CrossRef]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar]
- Mattick, J.S. The central role of RNA in human development and cognition. FEBS Lett. 2011, 585, 1600–1616. [Google Scholar] [CrossRef]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef]
- Li, W.; Notani, D.; Ma, Q.; Tanasa, B.; Nunez, E.; Chen, A.Y.; Merkurjev, D.; Zhang, J.; Ohgi, K.; Song, X.; et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013, 498, 516–520. [Google Scholar] [CrossRef]
- NE, I.I.; Heward, J.A.; Roux, B.; Tsitsiou, E.; Fenwick, P.S.; Lenzi, L.; Goodhead, I.; Hertz-Fowler, C.; Heger, A.; Hall, N.; et al. Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 2014, 5, 3979. [Google Scholar]
- Ding, M.; Liu, Y.; Liao, X.; Zhan, H.; Liu, Y.; Huang, W. Enhancer RNAs (eRNAs): New Insights into Gene Transcription and Disease Treatment. J. Cancer 2018, 9, 2334–2340. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.A.; Ares, M., Jr. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2006, 103, 3262–3267. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kim, D.; Morris, K.V. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc. Natl. Acad. Sci. USA 2007, 104, 12422–12427. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Kapranov, P.; Ozsolak, F.; Kim, S.W.; Foissac, S.; Lipson, D.; Hart, C.; Roels, S.; Borel, C.; Antonarakis, S.E.; Monaghan, A.P.; et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature 2010, 466, 642–646. [Google Scholar] [CrossRef]
- Taft, R.J.; Glazov, E.A.; Cloonan, N.; Simons, C.; Stephen, S.; Faulkner, G.J.; Lassmann, T.; Forrest, A.R.; Grimmond, S.M.; Schroder, K.; et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009, 41, 572–578. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef]
- Montes, M.; Nielsen, M.M.; Maglieri, G.; Jacobsen, A.; Hojfeldt, J.; Agrawal-Singh, S.; Hansen, K.; Helin, K.; van de Werken, H.J.G.; Pedersen, J.S.; et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat. Commun. 2015, 6, 6967. [Google Scholar] [CrossRef]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Anandakumar, S.; Vijayakumar, S.; Arumugam, N.; Gromiha, M.M. Mammalian Mitochondrial ncRNA Database. Bioinformation 2015, 11, 512–513. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bianchessi, V.; Badi, I.; Bertolotti, M.; Nigro, P.; D’Alessandra, Y.; Capogrossi, M.C.; Zanobini, M.; Pompilio, G.; Raucci, A.; Lauri, A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J. Mol. Cell Cardiol. 2015, 81, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Kramer, M.C.; Liang, D.; Tatomer, D.C.; Gold, B.; March, Z.M.; Cherry, S.; Wilusz, J.E. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015, 29, 2168–2182. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Subhash, S.; Mishra, K.; Akhade, V.S.; Kanduri, M.; Mondal, T.; Kanduri, C. H3K4me2 and WDR5 enriched chromatin interacting long non-coding RNAs maintain transcriptionally competent chromatin at divergent transcriptional units. Nucleic Acids Res. 2018, 46, 9384–9400. [Google Scholar] [CrossRef]
- Chiesa, N.; De Crescenzo, A.; Mishra, K.; Perone, L.; Carella, M.; Palumbo, O.; Mussa, A.; Sparago, A.; Cerrato, F.; Russo, S.; et al. The KCNQ1OT1 imprinting control region and non-coding RNA: New properties derived from the study of Beckwith-Wiedemann syndrome and Silver-Russell syndrome cases. Hum. Mol. Genet. 2012, 21, 10–25. [Google Scholar] [CrossRef]
- Mohammad, F.; Pandey, R.R.; Nagano, T.; Chakalova, L.; Mondal, T.; Fraser, P.; Kanduri, C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell Biol. 2008, 28, 3713–3728. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Vaid, R.; Enroth, S.; Uday, S.; Reinius, B.; Mitra, S.; Mohammed, A.; James, A.R.; Hoberg, E.; et al. MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015, 6, 7743. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Karreth, F.A.; Tay, Y.; Perna, D.; Ala, U.; Tan, S.M.; Rust, A.G.; DeNicola, G.; Webster, K.A.; Weiss, D.; Perez-Mancera, P.A.; et al. In vivo identification of tumor- suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell 2011, 147, 382–395. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Wu, H.; Ni, P.; Gu, Z.; Qiao, Y.; Chen, N.; Sun, F.; Fan, Q. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010, 38, 5366–5383. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Kanduri, C. Long noncoding RNAs: Lessons from genomic imprinting. Biochim. Biophys. Acta 2016, 1859, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, F.; Mondal, T.; Guseva, N.; Pandey, G.K.; Kanduri, C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 2010, 137, 2493–2499. [Google Scholar] [CrossRef] [PubMed]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Grote, P.; Wittler, L.; Hendrix, D.; Koch, F.; Wahrisch, S.; Beisaw, A.; Macura, K.; Blass, G.; Kellis, M.; Werber, M.; et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 2013, 24, 206–214. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Gius, D.; Onyango, P.; Muldoon-Jacobs, K.; Karp, J.; Feinberg, A.P.; Cui, H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008, 451, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Marin-Bejar, O.; Marchese, F.P.; Athie, A.; Sanchez, Y.; Gonzalez, J.; Segura, V.; Huang, L.; Moreno, I.; Navarro, A.; Monzo, M.; et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013, 14, R104. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, X.J.; Ji, Y.X.; Zhang, P.; Deng, K.Q.; Gong, J.; Ren, S.; Wang, X.; Chen, I.; Wang, H.; et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016, 22, 1131–1139. [Google Scholar] [CrossRef]
- Mayer, C.; Schmitz, K.M.; Li, J.; Grummt, I.; Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010, 24, 2264–2269. [Google Scholar] [CrossRef]
- Mondal, T.; Rasmussen, M.; Pandey, G.K.; Isaksson, A.; Kanduri, C. Characterization of the RNA content of chromatin. Genome Res. 2010, 20, 899–907. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Wang, C.Y.; Jegu, T.; Chu, H.P.; Oh, H.J.; Lee, J.T. SMCHD1 Merges Chromosome Compartments and Assists Formation of Super-Structures on the Inactive X. Cell 2018, 174, 406–421. [Google Scholar] [CrossRef]
- McGrath, J.; Solter, D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 1984, 37, 179–183. [Google Scholar] [CrossRef]
- Barlow, D.P.; Stoger, R.; Herrmann, B.G.; Saito, K.; Schweifer, N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 1991, 349, 84–87. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Zemel, S.; Tilghman, S.M. Parental imprinting of the mouse H19 gene. Nature 1991, 351, 153–155. [Google Scholar] [CrossRef] [PubMed]
- Borsani, G.; Tonlorenzi, R.; Simmler, M.C.; Dandolo, L.; Arnaud, D.; Capra, V.; Grompe, M.; Pizzuti, A.; Muzny, D.; Lawrence, C.; et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature 1991, 351, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Nagano, T.; Fraser, P. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm. Genome 2009, 20, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Rojas, C.; Hernandez, A.J.; Sarma, K.; Lee, J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 2014, 55, 171–185. [Google Scholar] [CrossRef]
- Beltran, M.; Yates, C.M.; Skalska, L.; Dawson, M.; Reis, F.P.; Viiri, K.; Fisher, C.L.; Sibley, C.R.; Foster, B.M.; Bartke, T.; et al. The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res. 2016, 26, 896–907. [Google Scholar] [CrossRef]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef]
- Belak, Z.R.; Ovsenek, N. Assembly of the Yin Yang 1 transcription factor into messenger ribonucleoprotein particles requires direct RNA binding activity. J. Biol. Chem. 2007, 282, 37913–37920. [Google Scholar] [CrossRef]
- Belak, Z.R.; Ficzycz, A.; Ovsenek, N. Biochemical characterization of Yin Yang 1-RNA complexes. Biochem. Cell Biol. 2008, 86, 31–36. [Google Scholar] [CrossRef]
- Sun, S.; Del Rosario, B.C.; Szanto, A.; Ogawa, Y.; Jeon, Y.; Lee, J.T. Jpx RNA activates Xist by evicting CTCF. Cell 2013, 153, 1537–1551. [Google Scholar] [CrossRef]
- Hendrickson, D.G.; Kelley, D.R.; Tenen, D.; Bernstein, B.; Rinn, J.L. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016, 17, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Ricci, E.P.; Moore, M.J. RIPiT-Seq: A high-throughput approach for footprinting RNA:protein complexes. Methods 2014, 65, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.W.; Flynn, R.A.; Chen, Y.; Qu, K.; Wan, B.; Wang, K.C.; Lei, M.; Chang, H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife 2014, 3, e02046. [Google Scholar] [CrossRef] [PubMed]
- Licatalosi, D.D.; Mele, A.; Fak, J.J.; Ule, J.; Kayikci, M.; Chi, S.W.; Clark, T.A.; Schweitzer, A.C.; Blume, J.E.; Wang, X.; et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 2008, 456, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, E.L.; Pratt, G.A.; Shishkin, A.A.; Gelboin-Burkhart, C.; Fang, M.Y.; Sundararaman, B.; Blue, S.M.; Nguyen, T.B.; Surka, C.; Elkins, K.; et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 2016, 13, 508–514. [Google Scholar] [CrossRef] [PubMed]
- Hocq, R.; Paternina, J.; Alasseur, Q.; Genovesio, A.; Le Hir, H. Monitored eCLIP: High accuracy mapping of RNA-protein interactions. Nucleic Acids Res. 2018, 46, 11553–11565. [Google Scholar] [CrossRef]
- Zarnegar, B.J.; Flynn, R.A.; Shen, Y.; Do, B.T.; Chang, H.Y.; Khavari, P.A. irCLIP platform for efficient characterization of protein-RNA interactions. Nat. Methods 2016, 13, 489–492. [Google Scholar] [CrossRef]
- Weyn-Vanhentenryck, S.M.; Mele, A.; Yan, Q.; Sun, S.; Farny, N.; Zhang, Z.; Xue, C.; Herre, M.; Silver, P.A.; Zhang, M.Q.; et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014, 6, 1139–1152. [Google Scholar] [CrossRef]
- Gu, J.; Wang, M.; Yang, Y.; Qiu, D.; Zhang, Y.; Ma, J.; Zhou, Y.; Hannon, G.J.; Yu, Y. GoldCLIP: Gel-omitted Ligation-dependent CLIP. Genom. Proteom. Bioinform. 2018, 16, 136–143. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M., Jr.; Jungkamp, A.C.; Munschauer, M.; et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Son, J.; Shen, S.S.; Reinberg, D.; Bonasio, R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct Mol. Biol. 2013, 20, 1258–1264. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S.; Bonasio, R.; Saldana-Meyer, R.; Yoshida, T.; Son, J.; Nishino, K.; Umezawa, A.; Reinberg, D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 2014, 53, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, V.N. fCLIP-seq for transcriptomic footprinting of dsRNA-binding proteins: Lessons from DROSHA. Methods 2019, 152, 3–11. [Google Scholar] [CrossRef]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar] [CrossRef]
- Castello, A.; Frese, C.K.; Fischer, B.; Jarvelin, A.I.; Horos, R.; Alleaume, A.M.; Foehr, S.; Curk, T.; Krijgsveld, J.; Hentze, M.W. Identification of RNA-binding domains of RNA-binding proteins in cultured cells on a system-wide scale with RBDmap. Nat. Protoc. 2017, 12, 2447–2464. [Google Scholar] [CrossRef]
- Queiroz, R.M.L.; Smith, T.; Villanueva, E.; Marti-Solano, M.; Monti, M.; Pizzinga, M.; Mirea, D.M.; Ramakrishna, M.; Harvey, R.F.; Dezi, V.; et al. Comprehensive identification of RNA-protein interactions in any organism using orthogonal organic phase separation (OOPS). Nat. Biotechnol. 2019, 37, 169–178. [Google Scholar] [CrossRef]
- Mariner, P.D.; Walters, R.D.; Espinoza, C.A.; Drullinger, L.F.; Wagner, S.D.; Kugel, J.F.; Goodrich, J.A. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol. Cell 2008, 29, 499–509. [Google Scholar] [CrossRef]
- Hacisuleyman, E.; Goff, L.A.; Trapnell, C.; Williams, A.; Henao-Mejia, J.; Sun, L.; McClanahan, P.; Hendrickson, D.G.; Sauvageau, M.; Kelley, D.R.; et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014, 21, 198–206. [Google Scholar] [CrossRef]
- Sun, L.; Goff, L.A.; Trapnell, C.; Alexander, R.; Lo, K.A.; Hacisuleyman, E.; Sauvageau, M.; Tazon-Vega, B.; Kelley, D.R.; Hendrickson, D.G.; et al. Long noncoding RNAs regulate adipogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 3387–3392. [Google Scholar] [CrossRef]
- Simon, M.D.; Wang, C.I.; Kharchenko, P.V.; West, J.A.; Chapman, B.A.; Alekseyenko, A.A.; Borowsky, M.L.; Kuroda, M.I.; Kingston, R.E. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 20497–20502. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.D.; Pinter, S.F.; Fang, R.; Sarma, K.; Rutenberg-Schoenberg, M.; Bowman, S.K.; Kesner, B.A.; Maier, V.K.; Kingston, R.E.; Lee, J.T. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 2013, 504, 465–469. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Subhash, S.; Kanduri, C. Chromatin RNA Immunoprecipitation (ChRIP). Methods Mol. Biol. 2018, 1689, 65–76. [Google Scholar] [PubMed]
- Fang, J.; Ma, Q.; Chu, C.; Huang, B.; Li, L.; Cai, P.; Batista, P.J.; Tolentino, K.E.M.; Xu, J.; Li, R.; et al. Functional classification of noncoding RNAs associated with distinct histone modifications by PIRCh-seq. bioRxiv 2019, 667881. [Google Scholar] [CrossRef]
- Li, X.; Zhou, B.; Chen, L.; Gou, L.T.; Li, H.; Fu, X.D. GRID-seq reveals the global RNA-chromatin interactome. Nat. Biotechnol. 2017, 35, 940–950. [Google Scholar] [CrossRef]
- Sridhar, B.; Rivas-Astroza, M.; Nguyen, T.C.; Chen, W.; Yan, Z.; Cao, X.; Hebert, L.; Zhong, S. Systematic Mapping of RNA-Chromatin Interactions In Vivo. Curr Biol. 2017, 27, 602–609. [Google Scholar] [CrossRef]
- Bell, J.C.; Jukam, D.; Teran, N.A.; Risca, V.I.; Smith, O.K.; Johnson, W.L.; Skotheim, J.M.; Greenleaf, W.J.; Straight, A.F. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 2018, 7. [Google Scholar]
- Werner, M.S.; Sullivan, M.A.; Shah, R.N.; Nadadur, R.D.; Grzybowski, A.T.; Galat, V.; Moskowitz, I.P.; Ruthenburg, A.J. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 2017, 24, 596–603. [Google Scholar] [CrossRef]
- Mahat, D.B.; Kwak, H.; Booth, G.T.; Jonkers, I.H.; Danko, C.G.; Patel, R.K.; Waters, C.T.; Munson, K.; Core, L.J.; Lis, J.T. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat. Protoc. 2016, 11, 1455–1476. [Google Scholar] [CrossRef]
- Kuo, C.C.; Hanzelmann, S.; Senturk Cetin, N.; Frank, S.; Zajzon, B.; Derks, J.P.; Akhade, V.S.; Ahuja, G.; Kanduri, C.; Grummt, I.; et al. Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res. 2019, 47, e32. [Google Scholar] [CrossRef] [PubMed]
- Hezroni, H.; Koppstein, D.; Schwartz, M.G.; Avrutin, A.; Bartel, D.P.; Ulitsky, I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015, 11, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.M.; Kim, S.O.; Inoue, K.; Smola, M.J.; Lee, D.M.; Schertzer, M.D.; Wooten, J.S.; Baker, A.R.; Sprague, D.; Collins, D.W.; et al. Functional classification of long non-coding RNAs by k-mer content. Nat. Genet. 2018, 50, 1474–1482. [Google Scholar] [CrossRef]
- Monsalve, M.; Wu, Z.; Adelmant, G.; Puigserver, P.; Fan, M.; Spiegelman, B.M. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 2000, 6, 307–316. [Google Scholar] [CrossRef]
- Di Ruscio, A.; Ebralidze, A.K.; Benoukraf, T.; Amabile, G.; Goff, L.A.; Terragni, J.; Figueroa, M.E.; De Figueiredo Pontes, L.L.; Alberich-Jorda, M.; Zhang, P.; et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 2013, 503, 371–376. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Ebisuya, M.; Yamamoto, T.; Nakajima, M.; Nishida, E. Ripples from neighbouring transcription. Nat. Cell Biol. 2008, 10, 1106–1113. [Google Scholar] [CrossRef]
- Martens, J.A.; Laprade, L.; Winston, F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 2004, 429, 571–574. [Google Scholar] [CrossRef]
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference—A crash course. Trends Genet. 2005, 21, 339–345. [Google Scholar] [CrossRef]
- Rougeulle, C.; Heard, E. Antisense RNA in imprinting: Spreading silence through Air. Trends Genet. 2002, 18, 434–437. [Google Scholar] [CrossRef]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Raefski, A.S.; O’Neill, M.J. Identification of a cluster of X-linked imprinted genes in mice. Nat. Genet. 2005, 37, 620–624. [Google Scholar] [CrossRef] [PubMed]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar] [CrossRef]
- Li, Y.; Syed, J.; Sugiyama, H. RNA-DNA Triplex Formation by Long Noncoding RNAs. Cell Chem. Biol. 2016, 23, 1325–1333. [Google Scholar] [CrossRef]
- Maldonado, R.; Schwartz, U.; Silberhorn, E.; Langst, G. Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells. Mol. Cell 2019, 73, 1243–1254 e1246. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- Zhao, Z.; Senturk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef]
- Senturk Cetin, N.; Kuo, C.C.; Ribarska, T.; Li, R.; Costa, I.G.; Grummt, I. Isolation and genome-wide characterization of cellular DNA:RNA triplex structures. Nucleic Acids Res. 2019, 47, 2306–2321. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, K.; Zhao, Y.; Zhang, S.; Wang, X.; Li, Y.; Lu, L.; Chen, X.; Chen, F.; Bao, X.; et al. Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 2015, 6, 10026. [Google Scholar] [CrossRef]
- Ng, S.Y.; Johnson, R.; Stanton, L.W. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012, 31, 522–533. [Google Scholar] [CrossRef]
- Ng, S.Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Pintacuda, G.; Wei, G.; Roustan, C.; Kirmizitas, B.A.; Solcan, N.; Cerase, A.; Castello, A.; Mohammed, S.; Moindrot, B.; Nesterova, T.B.; et al. hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 2017, 68, 955–969 e910. [Google Scholar] [CrossRef]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef]
- Sasaki, Y.T.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 106, 2525–2530. [Google Scholar] [CrossRef]
- Lin, R.; Maeda, S.; Liu, C.; Karin, M.; Edgington, T.S. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007, 26, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Song, D.Y.; Zong, X.; Shevtsov, S.P.; Hearn, S.; Fu, X.D.; Dundr, M.; Prasanth, K.V. SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol. Biol. Cell 2012, 23, 3694–3706. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 Regulates Expression of the Proto-oncogene SPHK1 via Triplex-Mediated Changes in Chromatin Structure. Mol. Cell 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Blank-Giwojna, A.; Postepska-Igielska, A.; Grummt, I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019, 26, 2904–2915 e2904. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef]
- Liu, F.; Marquardt, S.; Lister, C.; Swiezewski, S.; Dean, C. Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 2010, 327, 94–97. [Google Scholar] [CrossRef]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013, 340, 619–621. [Google Scholar] [CrossRef]
- Boque-Sastre, R.; Soler, M.; Oliveira-Mateos, C.; Portela, A.; Moutinho, C.; Sayols, S.; Villanueva, A.; Esteller, M.; Guil, S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc. Natl. Acad. Sci. USA 2015, 112, 5785–5790. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5’ to 3’ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef]
- Iglesias, N.; Redon, S.; Pfeiffer, V.; Dees, M.; Lingner, J.; Luke, B. Subtelomeric repetitive elements determine TERRA regulation by Rap1/Rif and Rap1/Sir complexes in yeast. EMBO Rep. 2011, 12, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Graf, M.; Bonetti, D.; Lockhart, A.; Serhal, K.; Kellner, V.; Maicher, A.; Jolivet, P.; Teixeira, M.T.; Luke, B. Telomere Length Determines TERRA and R-Loop Regulation through the Cell Cycle. Cell 2017, 170, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Lieber, M.R. G clustering is important for the initiation of transcription-induced R-loops in vitro, whereas high G density without clustering is sufficient thereafter. Mol. Cell Biol. 2009, 29, 3124–3133. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Zhang, Z.; Lu, Z.; Hsieh, C.L.; Lieber, M.R. Competition between the RNA transcript and the nontemplate DNA strand during R-loop formation in vitro: A nick can serve as a strong R-loop initiation site. Mol. Cell Biol. 2010, 30, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, J.Y.; Zhang, X.; Gu, Y.; Xiao, R.; Shao, C.; Tang, P.; Qian, H.; Luo, D.; Li, H.; et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol. Cell 2017, 68, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Ginno, P.A.; Lott, P.L.; Christensen, H.C.; Korf, I.; Chedin, F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol. Cell 2012, 45, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Stork, C.T.; Bocek, M.; Crossley, M.P.; Sollier, J.; Sanz, L.A.; Chedin, F.; Swigut, T.; Cimprich, K.A. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. Elife 2016, 5. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J.Y.; Huang, Y.J.; Gu, Y.; Qiu, J.; Qian, H.; Shao, C.; Zhang, X.; Hu, J.; Li, H.; et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Mol. Cell 2018, 69, 412–425. [Google Scholar] [CrossRef]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F., 3rd; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.K.; Blanco, M.; Jackson, C.; Aznauryan, E.; Ollikainen, N.; Surka, C.; Chow, A.; Cerase, A.; McDonel, P.; Guttman, M. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 2016, 354, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zeitz, M.J.; Wang, H.; Niu, B.; Ge, S.; Li, W.; Cui, J.; Wang, G.; Qian, G.; Higgins, M.J.; et al. Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 2014, 204, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Bose, D.A.; Donahue, G.; Reinberg, D.; Shiekhattar, R.; Bonasio, R.; Berger, S.L. RNA Binding to CBP Stimulates Histone Acetylation and Transcription. Cell 2017, 168, 135–149 e122. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Yi, X.; Li, X.; Hu, C.; Wang, J.; Bai, L.; Czajkowsky, D.M.; Shao, Z. The non-coding RNA composition of the mitotic chromosome by 5’-tag sequencing. Nucleic Acids Res. 2016, 44, 4934–4946. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 2013, 19, 429–442. [Google Scholar] [CrossRef]
- Davidovich, C.; Cech, T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA 2015, 21, 2007–2022. [Google Scholar] [CrossRef]
- Ringrose, L. Noncoding RNAs in Polycomb and Trithorax Regulation: A Quantitative Perspective. Annu. Rev. Genet. 2017, 51, 385–411. [Google Scholar] [CrossRef]
- Portoso, M.; Ragazzini, R.; Brencic, Z.; Moiani, A.; Michaud, A.; Vassilev, I.; Wassef, M.; Servant, N.; Sargueil, B.; Margueron, R. PRC2 is dispensable for HOTAIR-mediated transcriptional repression. EMBO J. 2017, 36, 981–994. [Google Scholar] [CrossRef]
- Blanco, M.R.; Guttman, M. Re-evaluating the foundations of lncRNA-Polycomb function. EMBO J. 2017, 36, 964–966. [Google Scholar] [CrossRef]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484 e421. [Google Scholar] [CrossRef] [PubMed]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.E.; Manteiga, J.C.; Henninger, J.E.; Sabari, B.R.; Dall’Agnese, A.; Hannett, N.M.; Spille, J.H.; Afeyan, L.K.; Zamudio, A.V.; Shrinivas, K.; et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019, 572, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Shrinivas, K.; Sabari, B.R.; Coffey, E.L.; Klein, I.A.; Boija, A.; Zamudio, A.V.; Schuijers, J.; Hannett, N.M.; Sharp, P.A.; Young, R.A.; et al. Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 2019, 75, 549–561. [Google Scholar] [CrossRef]

| Long Noncoding RNA | Function | Site of Action | Technique/Approach | Interacting Proteins | Ref. |
|---|---|---|---|---|---|
| lncRNA interaction with chromatin modifying complexes | |||||
| Kcnq1ot1 | Lineage-specific transcriptional silencing at the imprinted Kcnq1 locus | In-cis, bi-directional | Detection: Allelic RT-PCR Interaction: ChOP, ChRIP, allelic-ChIP, RIP | G9a, PRC2, DNMT1 (lineage specific interaction) | [30,32,43] |
| Airn | Lineage specific transcriptional silencing of imprinted genes Igf2r and Slc22a2/3. | In-cis, bi-directional | Detection: Allelic RT-PCR, RNA in-situ Interaction: RNA TRAP, RIP, allelic ChIP | Slc22a2/3 silencing: G9a-Airn complex Igf2r silencing: Transcriptional interference | [44,45] |
| HOTAIR | Transcriptional silencing of HOXD locus | In-trans | Detection: ChIP-chip of chromatin state maps (H3K4me3 and H3K27me3) in differentiating skin fibroblast cells Interaction: ChIRP-seq, Native RIP, IP | PRC2 (EZH2) LSD1 recruiting them to target gene promoters (simultaneous interaction) | [46,47] |
| HOTTIP | Homeotic gene activation at HOXA locus | In-cis | Detection: ChIP-chip of chromatin state maps in differentiating fibroblast cells, 5C Interaction: Native RIP, IP | WDR5 | [48] |
| Braveheart | Activation of cardiovascular progenitor | In-trans | Detection: RNA-seq from mouse ESCs and differentiated tissues representing all three germ layers Interaction: In-vitro biotin-RNA pull-down, RIP | PRC2(SUZ12) | [49] |
| Fendrr | Differentiation of tissues derived from lateral mesoderm | In-trans | Detection: RNA-seq, ChIP-seq Interaction: RNA co-IP | EZH2, SUZ12 (PRC2), WDR5 (simultaneous interaction) | [50] |
| ANRIL | Controlling cellular senescence by transcriptional silencing | In-cis antisense | Detection: qPCR, RNA-FISH Interaction: CLIP | CBX7 (PRC1) | [51,52] |
| Linc-Pint | Epigenetic regulation via p53 response | In-trans | Detection: Custom tiling microarrays Interaction: RIP-seq | PRC2 | [53] |
| Chaer | Epigenetic regulator of cardiac hypertrophy | In-trans | Detection: RNA-seq from pressure overload-induced mouse failing heart Interaction: RIP, Tagged RNA pull-down | EZH2 (PRC2) (66-mer motif of Chaer interacts in mTORC1 dependent manner) | [54] |
| pRNA | Regulation of CpG methylation at the rRNA genes | In-cis | Detection: RNase A treatment followed by immunofluorescence and ChIP for NoRC in NIH3T3 cells Interaction: Indirect evidence | DNMT3b recruited to DNA: RNA triplex at target promoter | [55,56] |
| ChRIP-seq (Act-D Treatment) | PIRCh-seq | GRID-seq | MARGI-seq | ChAR-seq | ||
|---|---|---|---|---|---|---|
| Active XH lncCARs | Inactive lncCARs | |||||
| Cell lines used | BT-549 | BT-549 | H9, HFF, mESC, MEF and mNPC | MBA-MB-231, mESC, S2 | hESC, HEK | CME-W1-cl8+ |
| Organism | Human | Human | Human and Mouse | Human, Mouse and Drosophila | Human | Drosophila |
| Number of cells/chromatin required | 50–60 µg chromatin per IP | 50–60 µg chromatin per IP | 10–20 µg chromatin per IP | 5–10 µg chromatin per library | 10,000–20,000 µg chromatin per library | 100–400 million Drosophila cells per library |
| Crosslinking | 1% Formaldehyde | UV and 1% Formaldehyde | 1% Glutaraldehyde | Formaldehyde and DSG | 1% Formaldehyde | 1% Formaldehyde |
| Probes or oligos | Antibody based | Antibody based | Antibody based | Customized biotinylated bivalent linker | Ligation based: customized linker DNA. | Biotinylated oligonucleotide bridge (linker DNA) |
| Technical limitations | Chromatin fragment size | Chromatin fragment size | Chromatin fragment size | Frequency of AluI restriction sites in the genome | Specificity of linker ligation to RNA and the proximity of bound RNA to free DNA ends (fragment size) | Specificity of bridge ligation to RNA and the proximity of bound RNAs to free DNA ends (fragment size) |
| Number of chromatin bound ncRNAs | 209 | 276 | 258 | 72 (7.36%) | Not provided | Less ncRNA and abundant mRNAs |
| Overrepresented class of ncRNAs | 191 lncRNAs out of 209 ncRNAs | lncRNAs and novel transcripts (“cuffs”) | 247 lncRNAs out of 258 ncRNAs | 32 lncRNAs MALAT1, NEAT1 and U2 snRNA, roX2, snoRNAs | Not provided | 18% snoRNA 19% snRNA |
| Relation with steady state levels of nuclear expression | Chromatin enrichment of active lncCARs independent of steady state nuclear levels | Information not provided | lncRNAs overrepresented as compared to mRNAs or other ncRNAs that generally has higher expression. | Positively correlated | Positively correlated | Positively correlated |
| Nascent transcript enrichment | Actinomycin D treated cells were used for the assay. Functionally characterized active XH lncCARs were validated for transcription independent chromatin enrichment | Not mentioned | Less compared to GRID-seq [96], CAR [57] and CPE [99] (Chromatin pellet extract) data | Nascent transcripts are enriched | In HEK cell pxRNA peaks detected in 69.1% of all the transcription start sites. DiRNA peaks detected in 61% of all the transcription start sites | Yes. Positive correlation with (Permissive nuclear Run-On sequencing) PRO-seq data [100] |
| Mechanism of action | Active XH lncCARs regulate transcription in cis (FOXD3-AS1, HOXC13-AS, GATA6-AS1 and HOXC-AS2) | One of the inactive CARs Meg3 regulates TGF-ß pathway genes in trans via triplex formation | Validated chromatin targeting of lnc-Nr2f1 | No | No | No |
| References | [28] | [33] | [95] | [96] | [97] | [98] |
| Long Noncod RNA | Function | Site of Action | Technique/Approach | Interacting Protein | Ref. |
|---|---|---|---|---|---|
| lncRNAs in interaction with RBPs with dual DNA and RNA-binding specificities | |||||
| linc-YY1 | YY1-mediated regulation of myogenesis | In-trans affects the eviction of YY1/PRC2 from the YY1 target genes | Detection: Poly A+ RNA seq from proliferating and differentiating C2C12 cells Interaction: Native IP using biotinylated in vitro-synthesized RNA. RIP suggested interaction with EZH2, SUZ12 and YY1 | 386–851 bp region of the linc-YY1 interacts most efficiently with YY1. | [120] |
| RMST | REST dependent regulation of pluripotency and neuronal differentiation via SOX2 pathway | In-trans | Detection: Custom designed micro-array. Total RNA obtained from hESCs differentiated into neural progenitors and neurons Interaction: Biotinylated antisense oligo-based RNA pull down, f-RIP | hnRNPA2/B1 and SOX2 REST dependent Neuronal differentiation. Dictates the binding of SOX2 at the target gene promoters, implicated in neurogenesis. | [121,122] |
| LUNAR1 | Notch regulated enhancing of IGF1 signaling | In-cis (looping as eRNA locus) | Detection: RNA-seq generated from multiple human T-ALL cell lines and primary leukemia samples Interaction: Hi-C, ChIRP, ChIP | Interacts with IGF1R intronic enhancer to recruit Mediator and RNAP2 | [123] |
| linc-p21 | Transcriptional repressor in p53-dependent response | In-trans | Detection: RNA-seq from genetically modified cell lines for knockdown and restorable p53 levels Interaction: Biotinylated antisense oligo-based RNA pull down, f-RIP, RIP | hnRNP-K: 780 nt region at the 5′ end of lincRNA-p21interacts with hnRNPK1 | [124] |
| SAMMSON | Regulation of mitochondrial homeostasis and metabolism | In-trans | Detection: GWAS of chromosome 3p melanoma specific focal amplification (Copy Number gain) from clinical SNP array data (TCGA) Interaction: RAP–MS, ChIRP, RAP-western blotting | p32 | [125] |
| lncRNAs in higher-order structures | |||||
| Xist | X chromosome inactivation | In-cis | Detection: Allelic RT-PCR, RNA-FISH Interaction: RIP, RIP-seq, RAP, RAP–MS, ChIRP-MS | EZH2 (PRC2) SHARP, SAF-A and LBR. hnRNPK binds to a 600 nt region of Xist RNA and recruits Polycomb-initiating complex PCGF3/5-PRC1 Xist lncRNA binds 81 proteins. Protein Spen interacts via RepA region | [31,67] [58] [126] [127] |
| Firre | Role in adipogenesis; by mediating inter-chromosomal interactions | In-cis but colocalize with spatially proximal trans genomic locations | Detection: PolyA+ RNA-seq of primary brown and white adipocytes, preadipocytes, and cultured adipocytes Interaction: RAP, ChIRP, CHART, ChOP | Interacts with hnRNPU through a 156-bp repeat RNA domain | [89,90] |
| NEAT1 | Nucleation and maintenance of paraspeckles | In-trans | Detection: qRT-PCR of HeLa cell nuclei fractionated by sucrose step-gradient centrifugation, Co-localization with paraspeckle protein PSF(SFPQ), PSP1, and p54. Custom microarray from (C2C12 cells) myoblast differentiation stages Interaction: UV-RIP, qRT-PCR of co-IP | Paraspeckle proteins | [128,129,130] |
| MALAT1 | Splicing, CeRNA | In-trans | Detection: Subtractive hybridization in cancer cell lines Interaction: Co-RNA-FISH, qRT-PCR of co-IP | SRSF1 to regulate splicing of mRNAs | [131,132,133,134] |
| LncRNAs forming R-loops and triple helixes | |||||
| MEG3 | Tumor suppressor lncRNA, Transcriptional repression of TGF-β pathway genes via triplex formation | In-trans | Detection: ChRIP-seq using antibodies against H3K27me3 and EZH2 from BT549 cell line Interaction: ChRIP, RIP, ChOP-seq, EMSA | EZH2(PRC2) | [33,57,94,101] |
| Khps1 | Transcriptional activation of SPHK1 via triplex mediated chromatin changes | In-cis | Detection: RNA-seq (not clear of exactly what) Interaction: RIP, Triplex forming EMSA | p300/CBP | [135,136] |
| COOLAIR | Regulates seed dormancy and flowering time through the regulation of FLC expression and flowering | In-cis | Detection: qRT-PCR, RNA-seq Interaction: R-loop foot printing, ChIRP | AtNDX binds to COOLAIR promoter to stabilize R-loops | [137,138]; [139] |
| VIM-AS1 | Promote transcriptional activation of the VIM1 gene | In-cis | Detection: Colon cancer hyper methylation associated positive correlation with divergent VIM1 gene Interaction: R-loop foot printing, EMSA, RNase H1 assay, DRIP with S9.6 | Interacts with single stranded DNA(R-loop) to enhance NF-κB binding at the VIM1 promoter | [140] |
| TERRA | Maintenance of short telomeric structure by regulating the rate of replicative senescence | Detection: qRT-PCR after releasing G1-arrested cells into the cell cycle Interaction: DRIP with S9.6, EMSA | Interacts with telomeric DNA forming R-loops that promotes homology directed repair at very short telomeres by excluding Rif2 mediated RNase H2 recruitment | [141,142,143]; [144] | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, K.; Kanduri, C. Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Non-Coding RNA 2019, 5, 54. https://doi.org/10.3390/ncrna5040054
Mishra K, Kanduri C. Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Non-Coding RNA. 2019; 5(4):54. https://doi.org/10.3390/ncrna5040054
Chicago/Turabian StyleMishra, Kankadeb, and Chandrasekhar Kanduri. 2019. "Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far" Non-Coding RNA 5, no. 4: 54. https://doi.org/10.3390/ncrna5040054
APA StyleMishra, K., & Kanduri, C. (2019). Understanding Long Noncoding RNA and Chromatin Interactions: What We Know So Far. Non-Coding RNA, 5(4), 54. https://doi.org/10.3390/ncrna5040054





