Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium
Abstract
:1. Introduction
2. Results
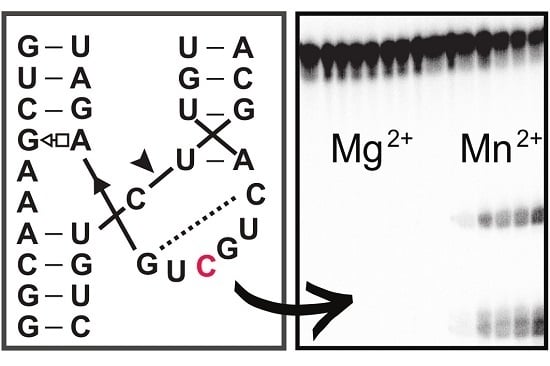
2.1. Varying Metal Ion Preference of a HHRz Variant
2.2. Effect of A6C Mutation on Another HHRz
3. Discussion
4. Materials and Methods
4.1. PCR Product of Wild-Type and Mutant Ribozymes
4.2. RNA Transcription
4.3. Kinetics of Cleavage
4.4. Measure of the Cleavage and kobs
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Altman, S. Nobel lecture. Enzymatic cleavage of RNA by RNA. Biosci. Rep. 1990, 10, 317–337. [Google Scholar] [CrossRef] [PubMed]
- Herschlag, D.; Cech, T.R. DNA cleavage catalysed by the ribozyme from Tetrahymena. Nature 1990, 344, 405–409. [Google Scholar] [CrossRef]
- Kim, J.J.; Kilani, A.F.; Zhan, X.; Altman, S.; Liu, F. The protein cofactor allows the sequence of an RNase P ribozyme to diversify by maintaining the catalytically active structure of the enzyme. RNA 1997, 3, 613–623. [Google Scholar]
- Cech, T.R. Structural biology. The ribosome is a ribozyme. Science 2000, 289, 878–879. [Google Scholar] [CrossRef]
- Michel, F.; Ferat, J.L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995, 64, 435–461. [Google Scholar] [CrossRef]
- Staley, J.P.; Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 1998, 92, 315–326. [Google Scholar] [CrossRef] [Green Version]
- Buzayan, J.M.; Gerlach, W.L.; Bruening, G. Satellite tobacco ringspot virus RNA: A subset of the RNA sequence is sufficient for autolytic processing. Proc. Natl. Acad. Sci. USA 1986, 83, 8859–8862. [Google Scholar] [CrossRef] [Green Version]
- Prody, G.A.; Bakos, J.T.; Buzayan, J.M.; Schneider, I.R.; Bruening, G. Autolytic processing of dimeric plant virus satellite RNA. Science 1986, 231, 1577–1580. [Google Scholar] [CrossRef]
- Saville, B.J.; Collins, R.A. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 1990, 61, 685–696. [Google Scholar] [CrossRef]
- Buzayan, J.M.; Gerlach, W.L.; Bruening, G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 1986, 323, 349–353. [Google Scholar] [CrossRef]
- Sharmeen, L.; Kuo, M.Y.; Dinter-Gottlieb, G.; Taylor, J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J. Virol. 1988, 62, 2674–2679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, C.H.; Riccitelli, N.J.; Ruminski, D.J.; Luptak, A. Widespread occurrence of self-cleaving ribozymes. Science 2009, 326, 953. [Google Scholar] [CrossRef] [Green Version]
- Roth, A.; Weinberg, Z.; Chen, A.G.; Kim, P.B.; Ames, T.D.; Breaker, R.R. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2014, 10, 56–60. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, Z.; Kim, P.B.; Chen, T.H.; Li, S.; Harris, K.A.; Lunse, C.E.; Breaker, R.R. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015, 11, 606–610. [Google Scholar] [CrossRef] [Green Version]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef]
- De la Pena, M.; Garcia-Robles, I.; Cervera, A. The Hammerhead Ribozyme: A Long History for a Short RNA. Molecules 2017, 22, 78. [Google Scholar] [CrossRef]
- De la Pena, M.; Garcia-Robles, I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA 2010, 16, 1943–1950. [Google Scholar] [CrossRef] [Green Version]
- Perreault, J.; Weinberg, Z.; Roth, A.; Popescu, O.; Chartrand, P.; Ferbeyre, G.; Breaker, R.R. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011, 7, e1002031. [Google Scholar] [CrossRef] [Green Version]
- Seehafer, C.; Kalweit, A.; Steger, G.; Gräf, S.; Hammann, C. From alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA 2011, 17, 21–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammann, C.; Luptak, A.; Perreault, J.; de la Pena, M. The ubiquitous hammerhead ribozyme. RNA 2012, 18, 871–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahm, S.C.; Derrick, W.B.; Uhlenbeck, O.C. Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry 1993, 32, 13040–13045. [Google Scholar] [CrossRef] [PubMed]
- Hertel, K.J.; Herschlag, D.; Uhlenbeck, O.C. A kinetic and thermodynamic framework for the hammerhead ribozyme reaction. Biochemistry 1994, 33, 3374–3385. [Google Scholar] [CrossRef]
- Scott, W.G.; Horan, L.H.; Martick, M. The hammerhead ribozyme: Structure, catalysis, and gene regulation. Prog. Mol. Biol. Transl. Sci. 2013, 120, 1–23. [Google Scholar]
- Murray, J.B.; Seyhan, A.A.; Walter, N.G.; Burke, J.M.; Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998, 5, 587–595. [Google Scholar] [CrossRef] [Green Version]
- Nakano, S.-I.; Yamashita, H.; Tanabe, K.; Sugimoto, N. Bulky cations greatly increase the turnover of a native hammerhead ribozyme. RSC Adv. 2019, 9, 35820–35824. [Google Scholar] [CrossRef] [Green Version]
- Roychowdhury-Saha, M.; Burke, D.H. Extraordinary rates of transition metal ion-mediated ribozyme catalysis. RNA 2006, 12, 1846–1852. [Google Scholar] [CrossRef] [Green Version]
- Boots, J.L.; Canny, M.D.; Azimi, E.; Pardi, A. Metal ion specificities for folding and cleavage activity in the Schistosoma hammerhead ribozyme. RNA 2008, 14, 2212–2222. [Google Scholar] [CrossRef] [Green Version]
- Kisseleva, N.; Khvorova, A.; Westhof, E.; Schiemann, O. Binding of manganese(II) to a tertiary stabilized hammerhead ribozyme as studied by electron paramagnetic resonance spectroscopy. RNA 2005, 11, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Martick, M.; Lee, T.S.; York, D.M.; Scott, W.G. Solvent structure and hammerhead ribozyme catalysis. Chem. Biol. 2008, 15, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, J.A.; Uhlenbeck, O.C. Hammerhead redux: Does the new structure fit the old biochemical data? RNA 2008, 14, 605–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir, A.; Golden, B.L. Two Active Site Divalent Ions in the Crystal Structure of the Hammerhead Ribozyme Bound to a Transition State Analogue. Biochemistry 2016, 55, 633–636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiemann, O.; Fritscher, J.; Kisseleva, N.; Sigurdsson, S.T.; Prisner, T.F. Structural investigation of a high-affinity MnII binding site in the hammerhead ribozyme by EPR spectroscopy and DFT calculations. Effects of neomycin B on metal-ion binding. ChemBioChem 2003, 4, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Draper, D.E. A guide to ions and RNA structure. RNA 2004, 10, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riccitelli, N.J.; Delwart, E.; Luptak, A. Identification of minimal HDV-like ribozymes with unique divalent metal ion dependence in the human microbiome. Biochemistry 2014, 53, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Papp-Wallace, K.M.; Maguire, M.E. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 2006, 60, 187–209. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, C.E.; Weinberg, Z.; Hammann, C. Novel ribozymes: Discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019, 47, 9480–9494. [Google Scholar] [CrossRef]
- O’Rear, J.L.; Wang, S.; Feig, A.L.; Beigelman, L.; Uhlenbeck, O.C.; Herschlag, D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA 2001, 7, 537–545. [Google Scholar] [CrossRef] [Green Version]
- Lau, M.W.; Trachman, R.J.; Ferre-D’Amare, A.R. A divalent cation-dependent variant of the glmS ribozyme with stringent Ca2+ selectivity co-opts a preexisting nonspecific metal ion-binding site. RNA 2017, 23, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Mir, A.; Chen, J.; Robinson, K.; Lendy, E.; Goodman, J.; Neau, D.; Golden, B.L. Two Divalent Metal Ions and Conformational Changes Play Roles in the Hammerhead Ribozyme Cleavage Reaction. Biochemistry 2015, 54, 6369–6381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachas, S.T.; Ferre-D’Amare, A.R. Convergent Use of Heptacoordination for Cation Selectivity by RNA and Protein Metalloregulators. Cell Chem. Biol. 2018, 25, 962–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, S.; Das, R. Primerize-2D: Automated primer design for RNA multidimensional chemical mapping. Bioinformatics 2017, 33, 1405–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Bcep176 | Mouse Gut HHRz | |||||
|---|---|---|---|---|---|---|
| WT(C6) | WT(A6) | Mutant(A6C) | ||||
| [mM] | Mg2+ | Mn2+ | Mg2+ | Mn2+ | Mg2+ | Mn2+ |
| 0.001 | ND | ND | ND | <10−6 | ND | ND |
| 0.003 | ND | ND | 1.2x10−5 | 0.0025 | ND | ND |
| 0.01 | ND | ND | 0.00070 | 0.031 | ND | 0.00093 |
| 0.03 | ND | 0.00012 | 0.019 | 0.096 | ND | 0.0070 |
| 0.1 | ND | 0.017 | 0.056 | 0.45 | <10−6 | 0.056 |
| 0.3 | 0.0018 | 0.057 | 0.30 | 0.39 | 3.6 × 10−6 | 0.18 |
| 1 | 0.0041 | 0.31 | ND | ND | 0.067 | 0.24 |
| 3 | 0.051 | 0.29 | ND | ND | 0.14 | 0.37 |
| 10 | 0.041 | ND | ND | ND | 0.33 | ND |
| Seq ID | Sequences |
|---|---|
| Bcep176 (C6)(Bcep176_Rev1 + Bcep176_Rev2) | ggAAUAGGUCGAAACGGCGGGAGGAAGACGUAGUAACGGCCCGCUGUCUGCACGUUAUGCGUGUACUGCUGAGAUCAGCGCCA |
| Bcep176 (C6A) (Bcep176_Rev2 + Bcep176_Rev4) | ggAAUAGGUCGAAACGGCGGGAGGAAGACGUAGUAACGGCCCGCUGUCUGCACGUUAUGCGUGUACUGaUGAGAUCAGCGCCA |
| Bcep176 (GUUU)(GAAA → GTTT) (Bcep176_Rev1 + Bcep176_Rev3) | ggAAUAGGUCguuuCGGCGGGAGGAAGACGUAGUAACGGCCCGCUGUCUGCACGUUAUGCGUGUACUGCUGAGAUCAGCGCCA |
| Bcep176_Rev1 | TGGCGCTGATCTCAGCAGTACACGCATAACGTGCAGACAGCGGGCCGTTACTACGTCTT |
| Bcep176_Rev2 | TAATACGACTCACTATAGGAATAGGTCGAAACGGCGGGAGGAAGACGTAGTAACGGCCC |
| Bcep176_Rev3 | TAATACGACTCACTATAGGAATAGGTCGTTTCGGCGGGAGGAAGACGTAGTAACGGCCC |
| Bcep176_Rev4 | TGGCGCTGATCTCAACAGTACACGCATAACGTGCAGACAGCGGGCCGTTACTACGTCTT |
| Rz mouse gut_HHRz (A6), (Rz mouse gut_fw + Rz mouse gut_rev) | ggUACCGAAUAAAUCCCCUGAUGAGCAACGGUGAGAGCCGGCGAAACUACCCAAACAAGGGUAGUCGGGAUAGUACCAUAA |
| Rz mouse gut_fw | TTCTAATACGACTCACTATAGGTACCGAATAAATCCCCTGaTGAGCAACGGTGAGAGCC |
| Rz mouse gut_rev | TTATGGTACTATCCCGACTACCCTTGTTTGGGTAGTTTCGCCGGCTCTCACCGTTGC |
| Rz mouse gut_HHRz (A6C), (Rz mouse gut_mutated_fw + Rz mouse gut_rev) | ggUACCGAAUAAAUCCCCUGcUGAGCAACGGUGAGAGCCGGCGAAACUACCCAAACAAGGGUAGUCGGGAUAGUACCAUAA |
| Rz_mouse gut_mutated_fw | TTCTAATACGACTCACTATAGGTACCGAATAAATCCCCTGcTGAGCAACGGTGAGAGCC |
| Complementary primer (to prevent cleavage during transcription) | GTAGTTTCGCCGGCTCTCACCGTTGCTCATCAGGGGATTTATTCGGTACC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naghdi, M.R.; Boutet, E.; Mucha, C.; Ouellet, J.; Perreault, J. Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium. Non-Coding RNA 2020, 6, 14. https://doi.org/10.3390/ncrna6010014
Naghdi MR, Boutet E, Mucha C, Ouellet J, Perreault J. Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium. Non-Coding RNA. 2020; 6(1):14. https://doi.org/10.3390/ncrna6010014
Chicago/Turabian StyleNaghdi, Mohammad Reza, Emilie Boutet, Clarisse Mucha, Jonathan Ouellet, and Jonathan Perreault. 2020. "Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium" Non-Coding RNA 6, no. 1: 14. https://doi.org/10.3390/ncrna6010014
APA StyleNaghdi, M. R., Boutet, E., Mucha, C., Ouellet, J., & Perreault, J. (2020). Single Mutation in Hammerhead Ribozyme Favors Cleavage Activity with Manganese over Magnesium. Non-Coding RNA, 6(1), 14. https://doi.org/10.3390/ncrna6010014